Abstract
Magnetosomes comprise a magnetic nanocrystal surrounded by a lipid bilayer membrane. These unique prokaryotic organelles align inside magnetotactic bacterial cells and serve as an intracellular compass allowing the bacteria to navigate along the geomagnetic field in aquatic environments. Cryoelectron tomography of Magnetospirillum strains has revealed that the magnetosome chain is surrounded by a network of filaments that may be composed of MamK given that the filaments are absent in the mamK mutant cells. The process of the MamK filament assembly is unknown. Here we prove the authenticity of the MamK filaments and show that MamK exhibits linear distribution inside Magnetospirillum sp. cells even in the area without magnetosomes. The mamK gene alone is sufficient to direct the synthesis of straight filaments in Escherichia coli, and one extremity of the MamK filaments is located at the cellular pole. By using dual fluorescent labeling of MamK, we found that MamK nucleates at multiple sites and assembles into mosaic filaments. Time-lapse experiments reveal that the assembly of the MamK filaments is a highly dynamic and kinetically asymmetrical process. MamK bundles might initiate the formation of a new filament or associate to one preexistent filament. Our results demonstrate the mechanism of biogenesis of prokaryotic cytoskeletal filaments that are structurally and functionally distinct from the known MreB and ParM filaments. In addition to positioning magnetosomes, other hypothetical functions of the MamK filaments in magnetotaxis might include anchoring magnetosomes and being involved in magnetic reception.
Keywords: assembly, magnetosomes, prokaryote, magnetic reception
For some time, eukaryotic cells have been known to use cytoskeletal polymers and molecular motors to establish their asymmetrical shapes, to transport intracellular constituents, and to drive their motility (1). Molecular motors interact with actin filaments and microtubules to move cargo as well as to generate tension in the cytoskeleton (2). Until recently, the lack of a cytoskeleton and intracellular organelles has been one of the defining features of prokaryotes. However, bacteria do have homologs of the eukaryotic cytoskeleton (3). At present, two kinds of actin-like prokaryotic proteins, MreB and ParM, have been extensively studied. MreB is an ancestor of eukaryotic actin and is found in almost all rod-shaped bacteria (4–6). MreB forms helical actin-like filaments lying underneath the cytoplasmic membrane, which might be the master organizer of the spatial distribution of proteins involved in the establishment and maintenance of rod morphology (7–9). MreB appears to form two-stranded filaments similar to F-actin, except that the strands do not twist around each other. The double-stranded filaments further associate into pairs and larger bundles (for a review, see ref. 10). In vitro MreB filament assembles with nucleation and polymerization rates that are much faster than those of eukaryotic actin (11). The ParM protein binds specifically to the ParR–parC complex, and the ParM filament assembly depends on this complex (12). In vivo, ParM filaments form a bundle that extends the length of the bacterium, with plasmid DNA localized at each ends, and polymerization of ParM has been postulated to provide enough force to push plasmids to opposite poles of the cell (12, 13). Purified ParM polymerized in an ATP-dependent manner (13) into two-stranded helical filaments similar to conventional actin filaments (14). Electron microscopy of polymeric ParM revealed well separated, individual filaments (13, 14).
In addition to actin-like MreB protein, magnetotactic bacteria possess unique intracellular organelles, the magnetosomes (15). Magnetosomes are generated by membrane invagination and biomineralization to form a single domain magnetite or greigite nanocrystal inside of each vesicle (15–17). Magnetosomes assemble into a linear chain to create a magnetic dipole moment that allows the bacteria to navigate along the geomagnetic field (known as magnetotaxis). The mechanisms ensuring the correct orientation of individual magnetic particles in the chain and anchoring the dipole chain inside of the cells remain elusive. Recent electron cryotomography studies suggest that the alignment of the magnetosomes depends on a filamentous structure (16, 17). The mamK gene is located on the magnetosome island and encodes an actin homolog (18). Deletion of the mamK gene leads to disappearance of the filaments, which can be restored by introducing the functional MamK–GFP fusion protein (17). Therefore, MamK might be the essential component of the filaments. Here, by using immunogold staining of ultrathin frozen sections and fluorescence microscopy, we proved the authenticity of the MamK filaments in Magnetospirillum sp. and found that the mamK gene alone is sufficient to direct the assembly of straight mosaic filaments in Escherichia coli. The properties and the biosynthesis process of the MamK filaments are different from those of MreB and ParM and seem to be optimally adapted for the magnetotaxis.
Results and Discussion
Authenticity and Distribution of MamK Filaments in Magnetospirillum magneticum.
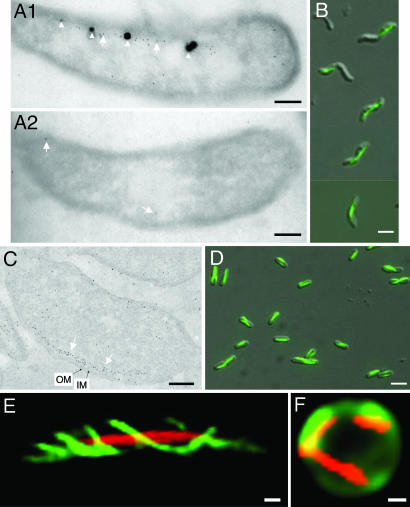
Recent cryoelectron tomography studies have shown that the depletion of the mamK gene abolishes the formation of the filaments (17); hence, MamK may be the main component of the magnetosome-associated filaments. By using anti-MamK antibody-based immunogold staining of ultrathin frozen sections of the M. magneticum AMB-1 cells, we found that the gold particles aligned with the magnetosomes (Fig. 1A1). The magnetosome island undergoes frequent rearrangements, and spontaneous mutants affected in magnetosome formation arise at a frequency of up to 10−2 under certain conditions (19). In our laboratory, we obtained a nonmagnetic AMB-1 spontaneous mutant that lost the mamK gene as revealed by PCR amplification analysis. Immunoblot analyses showed that this mutant did not contain MamK and confirmed the specificity of the polyclonal antibody used; a single polypeptide with a molecular size expected for MamK (≈35 kDa) was detected only in the wild-type strain, but it was absent from the mutant (Fig. 5, which is published as supporting information on the PNAS web site). Immunogold staining of the ultrathin frozen section of this mutant showed that gold particles were detected only sporadically in the samples (Fig. 1A2). These findings proved the authenticity of the MamK filaments observed under electron cryotomography.
Fig. 1.
The mamK gene is sufficient for directing assembly of MamK filaments in E. coli. (A1, A2, and C) Immunogold staining of ultrathin frozen sections using anti-MamK antibodies shows the position of MamK in the M. magneticum sp. AMB-1 wild-type strain (A1), spontaneous mam mutant (A2), and E. coli TG1/p6020 (C). The 7-nm gold particles are indicated by white arrows, magnetosomes are indicated by white arrowheads, and inner (IM) and outer membranes (OM) are indicated by black arrows. (B and D) Overlaid Nomarski and fluorescence images of the MamK–GFP expressed from p2020 in AMB-1 (B) or from p6020 in E. coli (D). (E and F) MamK–mCherry (red color) was coexpressed with MreB–YFP (green color in E) in the wild-type strain or coexpressed with periplasmic GFP (green color in F) in the mreB mutant cell. Folded GFP was exported into the periplasm via the Tat pathway as described in ref. 33. (Scale bars: B, D, and F, 3 μm; A1, A2, C, E and F, 0.3 μm.)
The MamK proteins are homologous to the eukaryotic actins and prokaryotic actin-like proteins but form a phylogenetic branch distinct from MreB-, ParM-, or eukaryotic actin-protein families (17). Because the MamK filament is present only in magnetotactic bacteria, it may have a peculiar function dedicated to the magnetotaxis. Interestingly, the gold particles aligned in areas without magnetosomes (Fig. 1A1), which implies that MamK filament localization is independent of the magnetosomes. This observation also would suggest that the MamK filaments might serve as tracks for moving and positioning the magnetosomes, similar to the eukaryotic cytoskeletal tracks used for intracellular cargo traffic (2). However, a motor protein required for traffic of organelles on the cytoskeletal tracks has not been revealed in magnetotactic bacteria, and the mechanism of magnetosome traffic remains enigmatic. Nevertheless, Scheffel et al. (16) have recently provided a clue for understanding this process. They found that, whereas the magnetosome vesicles are always arranged along the MamK-like filament, empty vesicles and immature magnetosomes are predominantly located at the ends of chains in wild-type cells, and mature magnetosomes are mostly found at midcell. In contrast, in the absence of the MamJ protein, empty vesicles and those containing immature crystals are scattered throughout the cytoplasm and are dissociated from the filaments. Scheffel et al. (16) have proposed that the MamJ protein connects magnetosome vesicles to the MamK filament and that it is involved in the MamK-dependent, dynamic localization of the magnetosomes at midcell. Our observation and the postulated MamK function are consistent with this hypothesis.
MamK Alone Is Sufficient for Directing Synthesis of Straight Filaments Structurally and Functionally Distinct from the MreB Filaments.
To elucidate the structure and the function of the MamK filaments, we assessed the capacity of the mamK gene for directing filament synthesis in the naturally nonmagnetotactic model bacterium E. coli. Recently, Komeili et al. (17) reported that MamK–GFP remains functional and that the fluorescence appears as a straight line along the inner curvature of the M. magneticum AMB-1 spiral cells. However, the mechanism of MamK filament assembly remains elusive. We constructed a plasmid p2020, which directs the synthesis of MamK-GFP fusion with an identical sequence to that synthesized from the plasmid pAK22 as described by Komeili et al. (see Materials and Methods). Expression of mamK–gfp fusion from the plasmid p2020 resulted in the appearance of linear fluorescence in the M. magneticum AMB-1 strain (Fig. 1B), which is consistent with the early report of the MamK–GFP expression from pAK22 in the AMB-1 strain (17). The same filamentous fluorescence pattern also was observed in E. coli strain TG1/p2020 (Fig. 5). To study the kinetics of the MamK–GFP polymerization, the expression of mamK–gfp must be tightly controlled. Therefore, we constructed MamK–GFP fusion in a derivative of the plasmid pBAD24. In the resulting plasmid, p6020, mamK–gfp expression is tightly controlled by an arabinose-inducible promoter. When expressed in E. coli, MamK–GFP fluorescence appeared as straight lines along the longitudinal axis in almost all of the cells when the expression was induced by arabinose (Fig. 1D). Some lines showed slight twisting and changes from one side of a cell to the opposite side of another attached cell, but they did not appear as a helical structure. Immunoblot analyses confirmed the specificity of the polyclonal antibody with E. coli; a single polypeptide with molecular size expected for MamK–GFP (≈60 kDa) was detected only in the strain expressing the fusion protein, but it was absent from the strain containing the empty plasmid (Fig. 5). Immunogold staining of ultrathin frozen sections of the E. coli cells showed that most gold particles were organized roughly in a chain along the cytoplasmic membrane (Fig. 1C), which is fully consistent with the MamK–GFP fluorescence distribution. As in Magnetospirillum cells (Fig. 1A1), no highly organized filamentous structures were evident under the conditions used, although both the inner and the outer membranes were obvious (Fig. 1C). These results show that the mamK gene alone is sufficient to direct the synthesis and assembly of MamK into a linear structure in a heterologous organism without assistance of other magnetosome-specific proteins.
In contrast to MamK filaments, the ParM polymerization requires the presence of the cognate ParR and parC in a host cell (13). parC is a stretch of centromeric DNA, and ParR is a repressor protein that binds to the parC locus (20). Polymerization of ParM filaments with plasmid DNA localized at each end seems to provide enough force to push plasmids to opposite poles of the cells (12, 13). Because polymerization of MamK filaments is independent of other magnetosome-specific proteins and of magnetosomes, the MamK filaments might use a different mechanism than ParM for the segregation of magnetosomes.
MamK is homologous to the rod-shaped determination actin MreB. We compared the structure and function of the two actin-like cytoskeletal proteins. As anticipated, MreB fused to yellow fluorescent protein (YFP) displayed helical filaments in wild-type cells (green color in Fig. 1E). The coexpressed MamK–mCherry appeared as straight filaments in the same cells. Therefore, MamK and MreB could coexist as distinct filaments. The MamK–mCherry filaments remained evident in the spherical mreB cells of which the cellular periphery was shown by the periplasmic GFP (Fig. 1F). These data indicate that the assembly of the MamK filaments is independent of MreB and that MamK could not restore the rod-shaped morphology of the mutant under the conditions used.
Dynamic Process of MamK Filament Assembly.
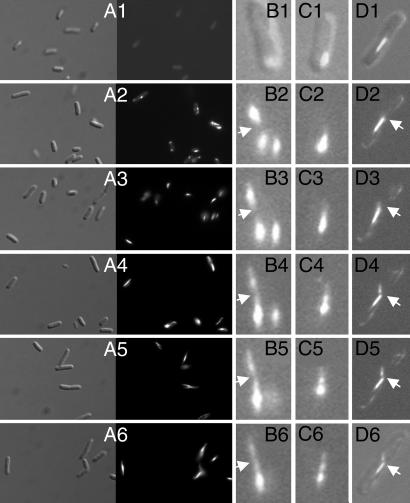
Assembly of actin filaments is a multiple-step process including nucleation, elongation, and remodeling. In eukaryotic cells, both actin filaments and microtubules are structurally and kinetically polarized so that one end of the polymer elongates faster than the other (21). We first analyzed the MamK assembly by collecting cells at different time points after the induction of the mamK expression. Twenty minutes after the induction, cells were uniformly fluorescent (Fig. 2A1), suggesting that the fluorescence signal spreads to the entire cytoplasm. Fluorescence foci started to be evident 10 min later in ≈10% of cells. Forty minutes after the induction, almost all cells contained one, two, or more fluorescence foci (Fig. 2A2). The fluorescence foci converted to almond (Fig. 2A3), which elongated into filaments with time (Fig. 2 A4–A6).
Fig. 2.
Kinetic of MamK–GFP synthesis and assembly. Cells in LB liquid cultures were collected 20 (A1), 40 (A2), 60 (A3), 80 (A4), 100 (A5), and 120 (A6) min after induction of the MamK–GFP synthesis from p6020 and inspected under fluorescence microscope as Nomarski (Left) or fluorescence (Right) images. Alternatively, after induction, cells were fixed in agarose on slides and inspected at time intervals of 15 min, with the first image taken 30 min after the induction. (B1, C1, D1, and D6) Overlaid Nomarski and fluorescence images. (B2–B6, C2–C6, and D2–D5) Fluorescence images.
Further details of MamK filament assembly were obtained by using time-lapse microscopy of growing cells, and images were taken every 15 min. A fluorescent focus appeared 30 min after the induction of MamK–GFP expression (Fig. 2 B1 and C1). As the cells were fixed in agarose, the kinetics of MamK–GFP expression and folding was different from the liquid cultures described in Fig. 2 A1–A6. Additional foci appeared 45 min after induction in the same cell or in another septating cell (Fig. 2B2). Short threads were evident 75 min after the induction (Fig. 2 B4 and C4) and elongated proportionally with the time to reach a filamentous appearance (Fig. 2 B4–B6 and C4–C6; see Movies 1 and 2, which are published as supporting information on the PNAS web site). The filament might exhibit different intensity in different regions (Fig. 2 B6, C5, and C6). Intriguingly, the short thread on the right side of the cell in Fig. 2B4 seemed to dissociate and reassociate to the left filament (Fig. 2B6). In a septating cell, the old filament seemed to end at the septum, with short smear fibers on another side of the septum (Fig. 2D3), which initiated the assembly of a new filament without alignment with the old one (Fig. 2 D3–D6; see Movie 3, which is published as supporting information on the PNAS web site). Clearly, the MamK polymerization is highly dynamic and kinetically asymmetrical, and the whole network is continuously rearranged. Similarly, in vitro assembly of ParM filaments in the presence of hydrolysable ATP showed that ATP–ParM filaments may abruptly switch from bidirectional elongation to rapid, endwise complete disassembly (22).
Dual Fluorescent Labeling Reveals a Mosaic Structure of the MamK Filaments.
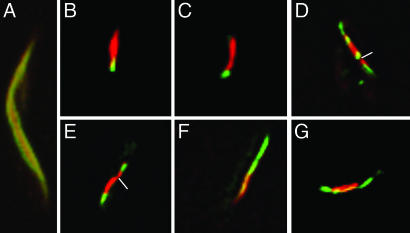
To address the questions of whether the MamK filament has a polarity and how it polymerizes, we used dual fluorescence to label MamK. When MamK was synthesized by simultaneous induction of red MamK–mCherry and green MamK–GFP expression, the MamK-filaments appeared yellow (Fig. 3A), indicating that the two fusion proteins were capable of assembling into the same filament. When MamK–mCherry was first synthesized for 1 h and then MamK–GFP was synthesized for an additional hour, ≈24% of cells had red filaments, 17% had green filaments, 53% had both colors, and 6% had yellow filaments. Among the dual-color filaments, ≈20% were with red color on one side and green on another side (Fig. 3 B and C). Therefore, later-synthesized green filaments seem to be added at one end of the preexistent red filaments. In contrast, ≈70% of the dual-color filaments were composed of green and red segments, and most were asymmetrical (Fig. 3 D and E). The two colors might merge together to form a continuous filament (Fig. 3 F and G). This mosaic structure is consistent with the observation that MamK nucleates at multiple sites and that short filamentous modules assemble to make the long filament.
Fig. 3.
Dual color labeling of the MamK filaments. MamK–mCherry and MamK–GFP were coexpressed in E. coli either simultaneously (A) or expressed first as red MamK–mCherry for 1 h and then green MamK–GFP for another hour (B–G).
Recently, electronic cryotomographs revealed that networks of short filamentous bundles of 200–250 nm in length run parallel to four or five individual magnetosomes along the magnetosome chain and associate to make a long filament (17). Our time-lapse experiments showed that MamK bundles may associate to one preexistent filament or initiate the formation of a new filament in a septating cell. Such an assembly mechanism leads to the mosaic feature of the MamK filaments. To understand the significance of such a feature requires further studies.
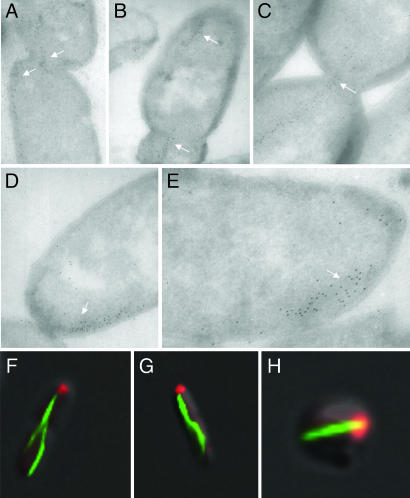
To gain more details about filaments in the septum region, ultrathin frozen immunostaining images were analyzed. Most MamK filaments seemed to be ended at the septa, with the extremity slightly across the septum (Fig. 4A–C). Short filamentous cables might nucleate at these points and make new filaments in the daughter cells. In addition, whereas one filament ended at the septum in one daughter cell, another filament was formed at the old pole in another daughter cell (Fig. 4B). Mosaic structure and the avoidance of deeply crossing over the septum imply an ingenious architecture of the MamK filaments that could play a role in positioning the magnetic dipole chain.
Fig. 4.
Polar localization of the MamK–GFP filament extremity. Localization of the MamK–GFP filaments in E. coli cells are revealed by immunogold staining (A–E, white arrows) or green fluorescence (F–H). Coexpressed IcsA–mCherry is shown by red fluorescence in the wild-type (F and G) and mreB mutant cell (H).
One Extremity of the MamK Filaments Is Located at the Cell Pole.
The annotation of the MamK sequence from the genome of Magnetospirillum magnetotacticum MS-1 does not include the corresponding first 25 residues of MamK from the M. magneticum AMB-1 strain (GenBank accession no. ZP_00054405.2; see Fig. 6, which is published as supporting information on the PNAS web site). This segment encompasses the first phosphate-binding motif, which is conserved among all actin-like proteins (Fig. 6) (23). We constructed the fusion MamK26–347–GFP from mamK of M. magneticum AMB-1, which lacks the first 25 residues corresponding to the segment missing from ZP_00054405.2 sequence. Unlike the full-length MamK–GFP, MamK26–347–GFP appeared only as fluorescent foci at the poles of the cells (data not shown). Therefore, the truncated MamK26–347 was unable to polymerize into filaments, suggesting that ATP-binding is essential for the MamK filament assembly. However, the truncated MamK26–347–GFP might possess sufficient information for being targeted to the polar area.
Notably, immunogold staining revealed that one extremity of MamK filaments located at the pole or septum (future pole; see Fig. 4 A–E). Bacterial cells are often polarized, exhibiting specialized structures at or near the poles of the cell (for reviews, see refs. 24 and 25). One structure is the organized arrays of membrane receptor, which govern chemosensory behavior in swimming bacteria via a phosphor-relay system (26). In Shigella sp., the polar protein IcsA mediates assembly of an actin tail inside infected mammalian cells (27). Polar localization of this protein seems to depend on the cytoskeletal MreB filament (28, 29). When coexpressed with IcsA507–620–mCherry (a cytoplasmic old pole marker), one extremity of the MamK–GFP filament was located close to IcsA in the rod-shaped wild-type strain (Fig. 4 F and G) as well as in the spherical mreB mutant cells (Fig. 4H). In the spherical cells, the localization of the MamK–GFP filament extremities is not restricted by geometric parameters. The colocalization of the MamK–GFP filament end with IcsA is therefore meaningful, but the biological implication is elusive.
Using Earth's magnetic field for orientation, navigation, and homing are critical traits expressed by organisms ranging from bacteria to higher vertebrates (30). How to convert a magnetic torque into a biochemical signal is the central mechanistic question of magnetoreception. A linear chain of single-domain magnetic crystals has been extracted from tissues in the frontal region of the sockeye salmon, Oncorhynchus nerka (31). It has been proposed that magnetosomes are anchored via a cytoskeletal filament to a mechanically activated transmembrane ion channel and torque from the magnetosomes could cause the transient opening of the channel, leading to membrane depolarization (30). Cytoskeletal filaments might underpin the magnetoreception in both vertebrates and magnetotactic bacteria. Recent cryoelectron microscopic results show that MamJ is required for the alignment of magnetosomes to the MamK filaments (16). The next questions that should be studied include understanding how magnetosomes are fixed on MamK filaments and whether the MamK filaments are involved in magnetoreception.
Materials and Methods
Bacterial Strains, Plasmids, and Growth.
The strains used in this study were E. coli MC1000 [araD139 (ara, leu)7697 lacX74 galU galK atrA] and its derivative YLS3 (mreB) (29); E. coli TG1 [Δ(lac-pro) supE thi hsdD5/F′ traD36 proA+B+ lacIq lacZΔM15]; and M. magneticum sp. AMB-1 (American Type Culture Collection 700264) (17).
Plasmid ptac-IcsA507–620–mCherry (pAWY-3) (28) encodes a segment of IcsA fused to the mCherry fluorescent protein under the control of the tac promoter. Plasmid plac-YFP–MreB (pLE7) (32) expresses hybrid protein consisting of MreB fused to the C-terminal of YFP under the control of the lac promoter. The pRR–GFP (33) encodes GFP fused to a twin-arginine signal peptide under the control of the arabinose promoter of pBAD24. The plasmid pAK20 encodes mamA–gfp and was described by Komeili et al. (34).
To study the kinetics of MamK–GFP polymerization, the expression of this fusion protein must be tightly controlled. The mamK–gfp fusion was constructed based on the plasmid pRR–GFP, and its expression was under the control of pBAD promoter. The M. magneticum AMB-1 mamK gene was amplified by PCR with the primers PMKREN-F (5′-tatgaattcatatgtggattgatctgttagcacgcgaacgg-3′) and PMKRBN-R (5′-agcggatccgctagcctgaccggaaacgtcaccaagctgacacc-3′) and the Expand High Fidelity PCR System according to the manufacturer's instructions (Roche, Basel, Switzerland). The amplified fragment was purified, double-digested by EcoRI and NheI, and cloned into the corresponding sites of the plasmid pRR–GFP (33), resulting in plasmid p6020. In addition, the amplified mamK was double-digested by EcoRI and BamHI and cloned into the corresponding sites of the plasmid pAWY-3 (28), resulting in plasmid ptac-MamK–mCherry. These two plasmids can be used for dual fluorescent labeling of MamK. To express the MamK–GFP fusion in M. magneticum strain AMB-1, the mamK gene was amplified by using primers 28MKEN-F (5′-tatgaattcatatgagtgaaggtgaaggccaggcc-3′) and MKSBN-R (5′-agcggatccgctagccgagccggagacgtctccaagctgacgcc-3′) and cloned at the EcoRI and BamHI sites in pAK20. The resulting plasmid, p2020, is almost the same as the plasmid pAK22 described by Komeili et al. (17). The only difference between the two plasmids is that two bases (AT) were inserted between the EcoRI site and the mamK start codon in p2020 compared with pAK22. However, the amino acid sequences of the MamK–GFP synthesized from these two plasmids are identical.
E. coli strains were routinely grown in LB medium (35). As required, 100 μg/ml ampicillin (Amp), 30 μg/ml chloramphenicol (Cm), 0.2% (wt/vol) glucose, 0.2% (wt/vol) arabinose, or 0.5 mM isopropyl β-d-thiogalactoside (IPTG) were added. Cultures of M. magneticum sp. AMB-1 were grown microaerobically in enriched Magnetospirillum growth medium (36).
Fluorescence, Electronic Microscopy, and Time-Lapse Imaging.
Overnight cultures were diluted 1:100 for the wild-type strain and 1:50 for the YLS-3 mutant in LB/Amp/glucose medium and incubated at 37°C with shaking for 3 h. Cells were centrifuged, washed once with LB/Amp medium, and resuspended in LB/Amp with 0.2% arabinose and/or 0.5 mM IPTG, and grown at 23°C for 1 or 2 h with shaking. Samples were examined directly under fluorescence microscope or after fixation in 0.25% agarose on slides. Images and z stack of 25–33 images were captured with a step distance of 0.15 μm with an Axiovert 200M (Zeiss, Göttingen, Germany) connected with an ORCA ER camera (Hamamatsu, Tokyo, Japan). Image restoration was obtained by deconvolution with Huygens Essential software (Scientific Volume Imaging, Hilversum, The Netherlands). Three-dimensional visualization was performed with Imaris software package (Bitplane, Zürich, Switzerland).
For time-lapse experiments, the TG1/p6020 strain was induced with 0.2% arabinose and grown at room temperature on 0.25% agarose pads, and Nomarski and fluorescence images of the same field were collected at 15-min intervals. For kinetic experiments, the TG1/p6020 strain was induced with 0.2% arabinose and grown at 23°C with shaking. Samples were collected at 20-min intervals, and cells were fixed in 2% paraformaldehyde in PBS (140 mM NaCl/3 mM KCl/8 mM Na2HPO4/1.5 mM KH2PO4) for 15 min at 23°C and washed twice in PBS. For dual fluorescent labeling of MamK protein, the TG1/p6020 (pBAD-MamK–GFP)/ptac-MamK–mCherry strain was induced in LB/Amp/Cm with 0.5 mM IPTG and grown at 23°C for 1 h with shaking. Cells were centrifuged, washed once with LB/Amp/Cm medium, resuspended in LB/Amp/Cm with 0.2% arabinose, and incubated for an additional hour at 23°C with shaking. For simultaneous dual fluorescent labeling, the strain was induced in LB/Amp/Cm with 0.5 mM IPTG and 0.2% arabinose and grown at 23°C with shaking. Immunogold staining of ultrathin sections was performed by using 7-nm gold-conjugated protein A as described by Anba et al. (37). Antiserum used is polyclonal rabbit anti-MamK at dilution at 1:1,500.
Supplementary Material
Acknowledgments
We thank Christopher Lefèvre for analyzing the mamK gene of the spontaneous mutant, M. Goldberg (Massachusetts General Hospital, Cambridge, MA) for the plasmid pAWY-3, and Y. L. Shih (University of Connecticut Health Center, Farmington, CT) for the mreB mutant strain and plasmid pLE7. This work was supported by Human Frontier Science Program Grant RGP0035/2004-C.
Abbreviations
- Amp
ampicillin
- Cm
chloramphenicol
- IPTG
isopropyl β-d-thiogalactoside
- YFP
yellow fluorescent protein.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Pollard TD. Nature. 2003;422:741–745. doi: 10.1038/nature01598. [DOI] [PubMed] [Google Scholar]
- 2.Schliwa M, Woehlke G. Nature. 2003;422:759–765. doi: 10.1038/nature01601. [DOI] [PubMed] [Google Scholar]
- 3.Gitai Z. Cell. 2005;120:577–586. doi: 10.1016/j.cell.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 4.van den Ent F, Amos LA, Lowe J. Nature. 2001;413:39–44. doi: 10.1038/35092500. [DOI] [PubMed] [Google Scholar]
- 5.Cabeen MT, Jacobs-Wagner C. Nat Rev Microbiol. 2005;3:601–610. doi: 10.1038/nrmicro1205. [DOI] [PubMed] [Google Scholar]
- 6.Moller-Jensen J, Lowe J. Curr Opin Cell Biol. 2005;17:75–81. doi: 10.1016/j.ceb.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 7.Jones LJ, Carballido-Lopez R, Errington J. Cell. 2001;104:913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- 8.Daniel RA, Errington J. Cell. 2003;113:767–776. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- 9.Divakaruni AV, Loo RR, Xie Y, Loo JA, Gober JW. Proc Natl Acad Sci USA. 2005;102:18602–18607. doi: 10.1073/pnas.0507937102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amos LA, van den Ent F, Lowe J. Curr Opin Cell Biol. 2004;16:24–31. doi: 10.1016/j.ceb.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Esue O, Cordero M, Wirtz D, Tseng Y. J Biol Chem. 2005;280:2628–2635. doi: 10.1074/jbc.M410298200. [DOI] [PubMed] [Google Scholar]
- 12.Moller-Jensen J, Borch J, Dam M, Jensen RB, Roepstorff P, Gerdes K. Mol Cell. 2003;12:1477–1487. doi: 10.1016/s1097-2765(03)00451-9. [DOI] [PubMed] [Google Scholar]
- 13.Moller-Jensen J, Jensen RB, Lowe J, Gerdes K. EMBO J. 2002;21:3119–3127. doi: 10.1093/emboj/cdf320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van den Ent F, Moller-Jensen J, Amos LA, Gerdes K, Lowe J. EMBO J. 2002;21:6935–6943. doi: 10.1093/emboj/cdf672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bazylinski DA, Frankel RB. Nat Rev Microbiol. 2004;2:217–230. doi: 10.1038/nrmicro842. [DOI] [PubMed] [Google Scholar]
- 16.Scheffel A, Gruska M, Faivre D, Linaroudis A, Plitzko JM, Schuler D. Nature. 2006;440:110–114. doi: 10.1038/nature04382. [DOI] [PubMed] [Google Scholar]
- 17.Komeili A, Li Z, Newman DK, Jensen GJ. Science. 2006;311:242–245. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- 18.Grünberg K, Wawer C, Tebo BM, Schuler D. Appl Environ Microbiol. 2001;67:4573–4582. doi: 10.1128/AEM.67.10.4573-4582.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ullrich S, Kube M, Schubbe S, Reinhardt R, Schuler D. J Bacteriol. 2005;187:7176–7184. doi: 10.1128/JB.187.21.7176-7184.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dam M, Gerdes K. J Mol Biol. 1994;236:1289–1298. doi: 10.1016/0022-2836(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 21.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD. Molecular Biology of the Cell. New York: Garland; 1994. [Google Scholar]
- 22.Garner EC, Campbell CS, Mullins RD. Science. 2004;306:1021–1025. doi: 10.1126/science.1101313. [DOI] [PubMed] [Google Scholar]
- 23.Bork P, Sander C, Valencia A. Proc Natl Acad Sci USA. 1992;89:7290–7294. doi: 10.1073/pnas.89.16.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shapiro L, McAdams HH, Losick R. Science. 2002;298:1942–1946. doi: 10.1126/science.1072163. [DOI] [PubMed] [Google Scholar]
- 25.Janakiraman A, Goldberg MB. Trends Microbiol. 2004;12:518–525. doi: 10.1016/j.tim.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 26.Maddock JR, Shapiro L. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 27.Goldberg MB, Barzu O, Parsot C, Sansonetti PJ. J Bacteriol. 1993;175:2189–2196. doi: 10.1128/jb.175.8.2189-2196.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nilsen T, Yan AW, Gale G, Goldberg MB. J Bacteriol. 2005;187:6187–6196. doi: 10.1128/JB.187.17.6187-6196.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shih YL, Kawagishi I, Rothfield L. Mol Microbiol. 2005;58:917–928. doi: 10.1111/j.1365-2958.2005.04841.x. [DOI] [PubMed] [Google Scholar]
- 30.Kirschvink JL, Walker MM, Diebel CE. Curr Opin Neurobiol. 2001;11:462–467. doi: 10.1016/s0959-4388(00)00235-x. [DOI] [PubMed] [Google Scholar]
- 31.Mann S, Sparks N, Walker M, Kirschvink J. J Exp Biol. 1988;140:35–49. doi: 10.1242/jeb.140.1.35. [DOI] [PubMed] [Google Scholar]
- 32.Shih YL, Le T, Rothfield L. Proc Natl Acad Sci USA. 2003;100:7865–7870. doi: 10.1073/pnas.1232225100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santini C-L, Bernadac A, Zhang M, Chanal A, Ize B, Blanco C, Wu L-F. J Biol Chem. 2001;276:8159–8164. doi: 10.1074/jbc.C000833200. [DOI] [PubMed] [Google Scholar]
- 34.Komeili A, Vali H, Beveridge TJ, Newman DK. Proc Natl Acad Sci USA. 2004;101:3839–3844. doi: 10.1073/pnas.0400391101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller JH. Experiments in Molecular Genetics. Woodbury, NY: Cold Spring Harbor Lab Press; 1972. [Google Scholar]
- 36.Yang C, Takeyama H, Tanaka T, Matsunaga T. Enzyme Microb Technol. 2001;29:13–19. doi: 10.1016/s0141-0229(01)00343-x. [DOI] [PubMed] [Google Scholar]
- 37.Anba J, Bernadac A, Pages J-M, Lazdunski C. Biol Cell. 1984;50:273–278. doi: 10.1111/j.1768-322x.1984.tb00274.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.