Abstract
Nonhomologous end-joining (NHEJ) repairs DNA double-strand breaks created by ionizing radiation or V(D)J recombination of the immunoglobulin genes. The breaks often leave mismatched or nonligatable ends, and NHEJ must repair the breaks with high efficiency and minimal nucleotide loss. Here, the NHEJ proteins Ku, DNA-dependent protein kinase catalytic subunit, XRCC4/Ligase IV, and Cernunnos/XRCC4-like factor joined mismatched and noncohesive DNA ends in the absence of processing factors. Depending on the mismatch, Cernunnos stimulated joining 8- to 150-fold. For substrates with a blunt end and a 3′ overhanging end, Ku, XRCC4/Ligase IV, and Cernunnos ligated the 3′ overhanging hydroxyl group to the 5′ phosphate of the blunt end, leaving the other strand unjoined. This activity provides a mechanism for retaining 3′ overhang sequences, as observed during V(D)J recombination in vivo. Thus, Cernunnos/XRCC4-like factor promotes a mismatched end (MEnd) DNA ligase activity to facilitate joining and to preserve DNA sequence. Furthermore, MEnd ligase activity may have applications in recombinant DNA technology.
Keywords: DNA repair, nonhomologous end-joining, V(D)J recombination
The nonhomologous end-joining (NHEJ) pathway is conserved in eukaryotes, from yeast to humans. Without requiring homologous DNA, NHEJ repairs DNA double-strand breaks produced by xenobiotic agents, such as topoisomerase II inhibitors and ionizing radiation, or by the cellular pathway for V(D)J recombination of the immunoglobulin genes (1). Even when the structure of the DNA ends prevents ligation, NHEJ processes the ends and repairs the breaks with high efficiency and minimal nucleotide loss. For ligatable ends, such as the blunt signal ends created by V(D)J recombination, NHEJ suppresses processing and repairs the breaks directly. Thus, NHEJ optimizes the preservation of DNA sequence, but the mechanism is not understood.
Core proteins for NHEJ include Ku, DNA-dependent protein kinase catalytic subunit (DNA-PKcs), and XRCC4/Ligase IV (XL). Ku consists of two subunits, Ku70 and Ku86, binds with high affinity to DNA ends (2, 3), and recruits DNA-PKcs to the ends (4). DNA-PKcs brings the ends together into a synaptic complex and then undergoes activation of its kinase domain to facilitate later steps in the joining reaction (5). XL performs the final ligation step (6).
When the DNA ends cannot be ligated directly, NHEJ uses nuclease and polymerase activities to process the ends. The exo/endonuclease Artemis interacts with DNA-PKcs. Activation of the DNA-PKcs kinase facilitates Artemis endonuclease activity, which opens the hairpin ends created during V(D)J recombination (7–9). DNA polymerases mu and lambda interact with Ku and XL in vitro (10) and add nucleotides to the immunoglobulin light chain and heavy chain junctions, respectively, during V(D)J recombination (11, 12).
Two groups recently identified a new NHEJ protein, designated Cernunnos or XRCC4-like factor (XLF). Buck et al. (13) studied cells from individuals with growth retardation, microcephaly, and immunodeficiency and found them to be sensitive to ionizing radiation and defective in V(D)J recombination, indicating a defect in NHEJ. They cloned “Cernunnos” by transfecting the mutant cells with a library of cDNA expression vectors and screening for restoration of ionizing radiation resistance and V(D)J recombination. Ahnesorg et al. (14) isolated XLF from a yeast two-hybrid screen for proteins that interacted with XRCC4. Transfection of XLF cDNA restored resistance to ionizing radiation and V(D)J recombination to cells from a patient with combined immunodeficiency. Both groups reported that cells from affected individuals had mutations in the Cernunnos/XLF gene. For simplicity, we will refer to the protein as Cernunnos for the remainder of this paper.
Cernunnos was weakly homologous to XRCC4, but the amino acid sequence failed to suggest a potential function. Using purified proteins, we discovered that Cernunnos, acting in concert with Ku, stimulated XL to ligate one of the two strands from mismatched DNA ends. Furthermore, Cernunnos biased the choice of the ligated strand to optimize preservation of DNA sequence.
Results
Cernunnos Promotes Joining of Mismatched Ends and Noncohesive Ends.
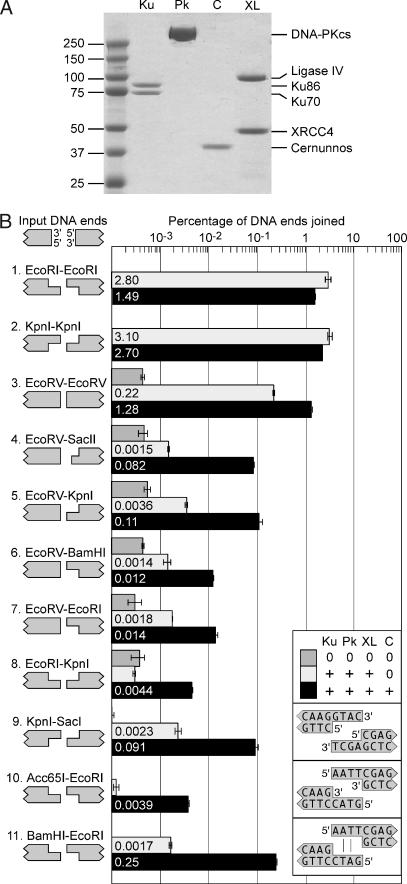
We purified Ku, DNA-PKcs, XL, and Cernunnos to apparent homogeneity (Fig. 1A) and tested their ability to join cohesive 5′ ends (EcoRI–EcoRI) or cohesive 3′ ends (KpnI–KpnI). To measure joining efficiency, we used quantitative PCR, which amplified junctions produced from one pair of ends. The PCR primers eliminated signals from competing junctions, such as those produced from intramolecular ligation into circular monomers or intermolecular ligation of other ends. Ku, DNA-PKcs, and XL joined EcoRI–EcoRI ends and KpnI–KpnI ends with efficiencies of 1.2–2.2% (Fig. 1B, rows 1 and 2). However, the addition of Cernunnos had no effect on joining efficiency.
Fig. 1.
Cernunnos stimulates the joining of mismatched DNA ends. (A) Purification of NHEJ proteins. Coomassie staining after SDS/PAGE shows our purified preparations of Ku, DNA-PKcs (Pk), Cernunnos (C), and XL. (B) Cernunnos stimulates the joining of noncohesive ends by Ku, DNA-PKcs, and XL. Linear DNA substrates were incubated with no protein (dark gray bars), Ku, DNA-PKcs and XL (light gray bars), or Ku, DNA-PKcs, XL, and Cernunnos (black bars). The orientation of DNA is depicted in the upper left corner. We tested compatible ends (rows 1–3) with cohesive 5′ overhangs (EcoRI–EcoRI), cohesive 3′ overhangs (KpnI–KpnI), or blunt ends (EcoRV–EcoRV). We also tested eight combinations of mismatched ends (rows 4–11). Note that SacII creates a 2-nt 3′ overhang. All other 3′ and 5′ overhangs were 4 nt. We measured joining efficiency by quantitative PCR and expressed efficiency as the percentage of input DNA ends joined (18). Because of large differences among experiments, we plotted joining efficiency on a logarithmic scale. Concentrations of Ku, DNA-PKcs, XL, and Cernunnos were 5, 5, 0.5, and 2.5 nM, respectively, here and elsewhere unless otherwise noted. (Insets) The overhangs from the KpnI–SacI, Acc65I–EcoRI, and EcoRI–BamHI ends.
Next, we tested whether Cernunnos might affect the joining of noncohesive DNA ends. The addition of Cernunnos to Ku, DNA-PKcs, and XL stimulated the joining of two blunt ends (EcoRV–EcoRV) by 6-fold (Fig. 1B, row 3). In the absence of Cernunnos, Ku, DNA-PKcs, and XL joined blunt ends paired with either 5′ or 3′ overhangs with low and approximately equal efficiencies (Fig. 1B, rows 4–7). However, the addition of Cernunnos stimulated joining 30- and 55-fold for the blunt-3′ ends, EcoRV–KpnI and EcoRV–SacII, and 8- and 9-fold for the blunt-5′ ends, EcoRV–EcoRI and EcoRV–BamHI. Thus, Cernunnos had a greater effect on blunt-3′ ends than on blunt-5′ ends.
Cernunnos also stimulated joining of ends with mismatched overhangs by 40- to 150-fold (Fig. 1B, rows 8–11). Ku, DNA-PKcs, XL, and Cernunnos joined mismatched 5′ overhangs (Acc65I–EcoRI, 0.0039%) with 23-fold lower efficiency than mismatched 3′ overhangs (KpnI–SacI, 0.091%). However, Cernunnos facilitated more efficient joining when the 5′ overhangs contained partially complementary sequences (BamHI–EcoRI, 0.25%).
In summary, Cernunnos exhibited a preference for 3′ overhangs over 5′ overhangs. This preference also occurred when we omitted DNA-PKcs from the joining reaction [supporting information (SI) Fig. 6]. The effect of Cernunnos appeared to be specific, because Cernunnos had no effect on ligation of blunt ends to 3′ overhangs by bacteriophage T4 DNA ligase (SI Fig. 7).
Cernunnos Requires Ku to Promote Mismatched End Joining by XL.
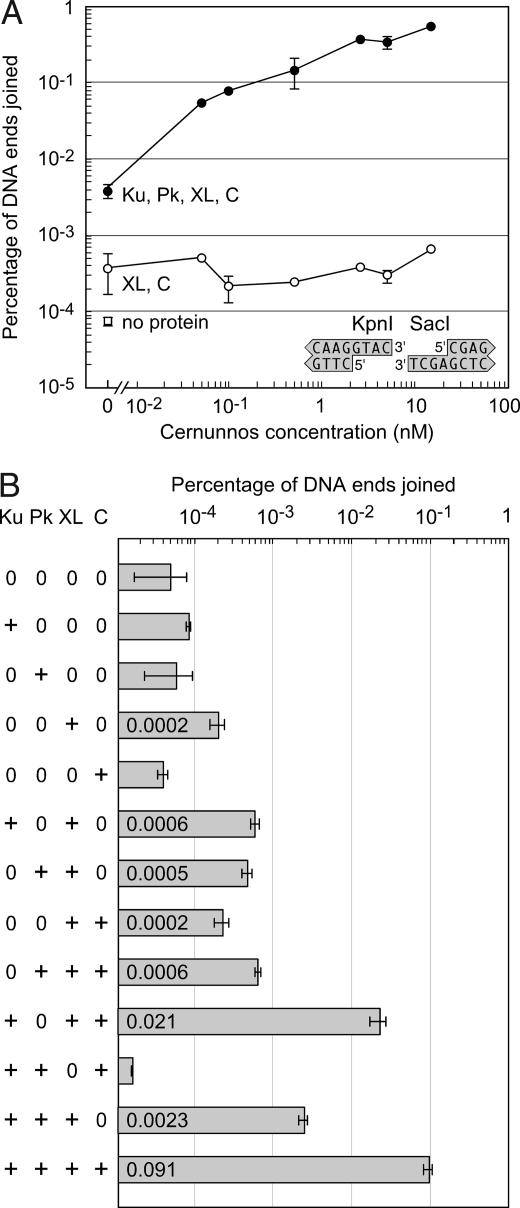
We were particularly interested in the joining of mismatched 3′ overhangs because they are produced during V(D)J recombination in vivo (15) and because Cernunnos stimulated their joining by 40-fold in vitro (Fig. 1B, row 9). Interestingly, purified XL joined KpnI–SacI ends with low but detectable efficiency, although the addition of Cernunnos failed to stimulate joining by XL alone (Fig. 2A). However, even a substoichiometric concentration of Cernunnos (0.05 nM) stimulated joining by XL (0.5 nM) in the presence of Ku (5 nM) and DNA-PKcs (5 nM). A stoichiometric excess of Cernunnos (15 nM) stimulated joining 200-fold, up to a level of nearly 1%.
Fig. 2.
Ku, DNA-PKcs, XL, and Cernunnos act in concert to join mismatched 3′ overhangs. (A) Cernunnos stimulates the joining of mismatched ends with Ku, DNA-PKcs, and XL but not with XL alone. DNA substrates with mismatched 3′–3′ overhangs (KpnI–SacI) were incubated with XL alone or XL, Ku, and DNA-PKcs (Pk). Concentrations of Ku, DNA-PKcs, and XL were 5, 5, and 0.5 nM, respectively. Cernunnos (C) was added at concentrations of 0.05, 0.1, 0.5, 2.5, 5, and 15 nM. The point in the lower left shows the background PCR signal in the absence of protein. Error bars represent the variation in duplicate measurements. (B) Cernunnos requires Ku to promote mismatched end joining by XL. DNA substrates with mismatched 3′ overhangs (KpnI–SacI) were incubated with different combinations of the core NHEJ proteins Ku, DNA-PKcs, XL, and Cernunnos (2.5 nM).
To determine which proteins were required for joining mismatched ends, we incubated the DNA with Ku, DNA-PKcs, XL, and Cernunnos in different combinations (Fig. 2B). As single proteins, only XL produced detectable joining of the KpnI–SacI ends. The addition of Ku and DNA-PKcs to XL increased joining efficiency 12-fold. Starting from the reaction with all four protein preparations, we omitted each protein one at a time. The omission of DNA-PKcs reduced joining only 4-fold, but the omission of Ku or Cernunnos reduced joining 150-fold or 40-fold, respectively. Note that the omission of Cernunnos reduced joining 200-fold when the concentration of Cernunnos was 15 nM rather than 2.5 nM (Fig. 2A). Significantly, the omission of XL eliminated joining activity completely. Thus, Ku, XL, and Cernunnos were required for efficient joining of mismatched ends. These data suggest that Cernunnos and Ku stimulated a latent joining activity for mismatched ends contained in Ligase IV.
Cernunnos Promotes the Preservation of 3′ Overhangs.
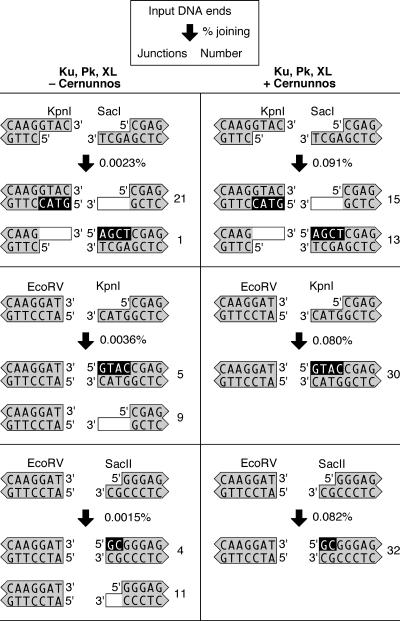
To further characterize the joining of mismatched KpnI–SacI ends, we sequenced the junctions created by Ku, DNA-PKcs, and XL with or without the addition of Cernunnos. Surprisingly, the sequence from one 3′ overhang was retained in every junction, whereas the sequence of the other 3′ overhang was lost (Fig. 3). In the absence of Cernunnos, 21 of the 22 sequenced junctions retained the 3′ KpnI overhang, whereas only 1 junction retained the SacI overhang. This preference occurred only in the absence of Cernunnos and remains unexplained. However, in the presence of Cernunnos, the 3′ overhangs were retained with equal probability.
Fig. 3.
Cernunnos promotes the retention of overhanging ends. We incubated DNA substrates with Ku, DNA-PKcs, and XL with or without the addition of Cernunnos. The DNA substrates had mismatched 3′–3′ ends (KpnI–SacI), blunt-3′ ends with 4-nt overhangs (EcoRV–KpnI), or blunt-3′ ends with 2-nt overhangs (EcoRV–SacII). We used quantitative PCR to measure the percentage of DNA ends joined and sequenced products from each reaction to characterize the junctions. The key shown at the top indicates how the data are presented for each reaction. The junctions recovered from PCR amplification are depicted to show nucleotide addition (black background) and nucleotide deletion (white background).
We also characterized the joining of blunt ends to 3′ overhangs of 4 nt (EcoRV–KpnI) or 2 nt (EcoRV–SacII). In the absence of Cernunnos, most junctions (20 of 29) lost the 3′ overhang, and only a minority (9 of 29) retained the 3′ overhang (Fig. 3). In the presence of Cernunnos, 62 of 62 sequenced junctions retained the 3′ overhang (Fig. 3). Cernunnos exhibited the same effect when added to Ku and XL in the absence of DNA-PKcs (SI Fig. 8). Thus, the addition of Cernunnos promoted retention of sequences from the 3′ overhangs.
Finally, we characterized the joining of blunt ends to 5′ overhangs (SI Fig. 9). Cernunnos exhibited only a moderate effect in promoting retention of the 5′ overhang for EcoRV–BamHI ends, and no effect on retention of the 5′ overhang for EcoRV–EcoRI ends.
Ku, XL, and Cernunnos Ligate One of the Two Strands from Mismatched DNA Ends.
In an effort to account for the retention or deletion of 3′ overhangs, we examined our purified protein preparations for polymerase or nuclease activities. Polymerase activity was absent because the joining reactions did not include dNTPs and because joining efficiency was unaffected by addition of dNTPs (data not shown). Nuclease activity also was absent because Cernunnos strongly stimulated joining of blunt-3′ ends with retention of 3′ overhanging sequences (Fig. 3 and SI Fig. 8). Furthermore, Cernunnos, Ku, XL, and DNA-PKcs failed to degrade radiolabeled single-strand DNA substrates (SI Fig. 10) and did not delete any nucleotides in 112 junctions from pairs of blunt ends (EcoRV–EcoRV) and cohesive ends (EcoRI–EcoRI and KpnI–KpnI) (SI Fig. 11).
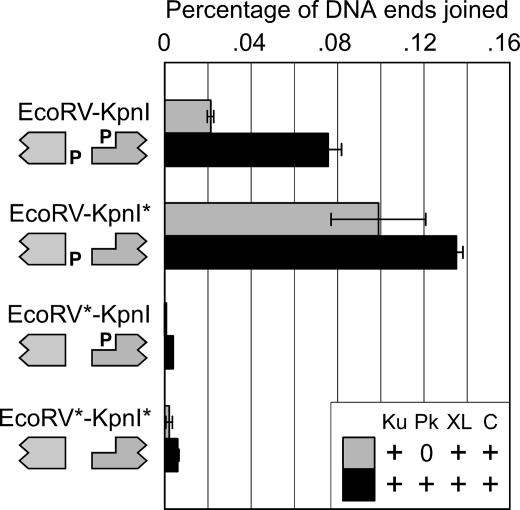
Taken together, our data suggest that Cernunnos and Ku stimulated XL to ligate one of the two strands from mismatched DNA ends. The deletion or retention of 3′ overhangs would depend on which strand was ligated. If Ligase IV catalyzes ligation of only one strand from mismatched ends, the 5′ phosphate group of one DNA substrate should be dispensable for ligation. To test this hypothesis, we removed 5′ phosphates from the DNA substrates with 3′ overhanging KpnI ends or blunt EcoRV ends and incubated the DNA with Ku, XL, and Cernunnos with or without DNA-PKcs (Fig. 4). When we removed the recessed 5′ phosphate on the KpnI end, joining actually increased because of decreased competition from intramolecular circularization of the KpnI substrate. This result demonstrated that the 5′ phosphate on the KpnI end was dispensable for joining. By contrast, when we removed the 5′ phosphate from the blunt EcoRV end, joining decreased to the levels observed when both DNA ends lacked 5′ phosphates. Taken together with the junction sequences (Fig. 3), these data indicated that Cernunnos stimulated the ligation of one strand from each end, joining the overhanging 3′ hydroxyl on the KpnI end to the 5′ phosphate on the blunt EcoRV end.
Fig. 4.
Ku, XL, and Cernunnos join mismatched ends with the 5′ phosphate from only one end. To determine how Ku, XL, and Cernunnos joined mismatched ends, we treated the EcoRV and KpnI substrates with DNA phosphatase (Antarctic phosphatase; New England Biolabs). EcoRV* and KpnI* denote dephosphorylated DNA molecules. We confirmed the successful removal of the 5′ phosphates by incubating each DNA preparation with T4 ligase and were able to demonstrate a loss of >90% of the ligation products by using agarose gel electrophoresis (data not shown). We incubated mismatched DNA substrates (EcoRV–KpnI, EcoRV–KpnI*, EcoRV*–KpnI, or EcoRV*–KpnI*) with Ku, XL (1 nM), and Cernunnos (2 nM), with or without DNA-PKcs; amplified the junctions by quantitative PCR; and plotted joining efficiency on a linear scale.
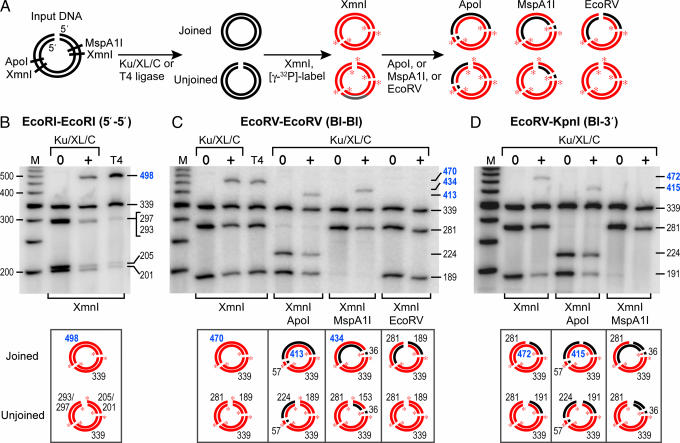
We wanted to obtain direct evidence for what we shall refer to as mismatched end (MEnd) DNA ligase activity. To avoid PCR amplification, we designed a gel-based assay with radiolabeled DNA. To unambiguously identify the joined products and improve joining efficiency, we assayed intramolecular joining of linear DNA. Each DNA molecule contained cohesive EcoRI–EcoRI ends, blunt EcoRV–EcoRV ends, or mismatched blunt-3′ EcoRV–KpnI ends (Fig. 5).
Fig. 5.
Ku, XL, and Cernunnos join mismatched ends by ligation of one strand. (A) Schematic of the gel-based assay for joining. We incubated Ku, XL, and Cernunnos (Ku/XL/C) or T4 ligase (T4) with a linear DNA substrate containing EcoRI–EcoRI, EcoRV–EcoRV, or EcoRV–KpnI ends; digested the DNA products with XmnI; and used T4 polynucleotide kinase to radiolabel the DNA. Concentrations of XL and Cernunnos were 1 nM and 5 nM, respectively. For EcoRV–EcoRV and EcoRV–KpnI ends, we performed a second digest with ApoI or MspA1I to remove the radiolabel from one DNA strand. To confirm that ligation created a new phosphodiester bond, we digested the products of the EcoRV–EcoRV joining reaction with EcoRV. Radiolabeled DNA strands are red with red asterisks at the 5′ end, and unlabeled DNA strands are black. DNA products were resolved with denaturing gel electrophoresis. (B) Ku, XL, and Cernunnos joined cohesive EcoRI–EcoRI ends. Ku, XL, and Cernunnos (lane 3) and T4 ligase (lane 4) produced a higher molecular weight band of 498 nt. The molecular weight markers consisted of a radiolabeled 50-bp ladder (lane 1). Joining efficiencies for Ku/XL/C and T4 (calculated from the ratio of intensities in the 498-nt band to the 339-nt band) were 39% and 89%, respectively. Sizes of the ligated higher molecular weight bands in B–D appear in blue typeface. (C) Ku, XL, and Cernunnos joined blunt EcoRV–EcoRV ends. Ku, XL, and Cernunnos (lane 3) and T4 ligase (lane 4) produced a higher molecular weight band of 470 nt, with joining efficiencies of 20% and 14%, respectively. ApoI digestion converted the 470-nt band to 413-nt and 57-nt bands and converted the 281-nt band to 224-nt and 57-nt bands (lane 6). MspA1I digestion converted the 470-nt band to 434-nt and 36-nt bands and the 189-nt band to 153-nt and 36-nt bands (lane 8). Intensities of the 413-nt and 434-nt bands were reduced to 50% of the 470-nt band because the second digestion removed the radiolabel from one strand. EcoRV digestion eliminated the 470-nt band (lane 10), demonstrating that the DNA junction contained new phosphodiester bonds. The 153-, 57-, and 36-nt bands are not shown. (D) Ku, XL, and Cernunnos joined only one strand from mismatched EcoRV–KpnI ends. Ku, XL, and Cernunnos (lane 3) produced a higher molecular weight band of 472 nt, with a joining efficiency of 5%. ApoI digestion converted the 472-nt band to 415-nt and 57-nt bands and the 281-nt band to 224-nt and 57-nt bands (lane 5). The intensities of the 472-nt and 415-nt bands were equivalent (lanes 3 and 5). MspA1I digestion converted the 472-nt band to a 36-nt band and converted the 191-nt band to a 36-nt band (lane 7), demonstrating the ligation of only one strand.
We incubated the DNA with Ku, XL, and Cernunnos. To detect the joined products by denaturing gel electrophoresis, we cleaved the DNA products with XmnI and labeled the ends with [γ-32P]ATP and T4 polynucleotide kinase (Fig. 5A). This procedure labeled the 5′ ends of DNA strands terminating at XmnI, EcoRI, and EcoRV sites. We measured joining by appearance of a higher molecular weight DNA fragment of the appropriate size. To determine whether joining occurred for a specific strand, we removed the radiolabel on one strand or the other by cleaving the DNA with ApoI or MspA1I.
For the cohesive 5′ overhanging EcoRI–EcoRI ends (Fig. 5B), Ku, XL, and Cernunnos joined 39% of the input DNA. By contrast, T4 ligase joined 89% of the input DNA. For blunt EcoRV–EcoRV ends (Fig. 5C), Ku, XL, and Cernunnos joined 20%, and T4 ligase joined 14% of the input DNA. When we removed the radiolabel from one strand by cleaving the DNA with ApoI or MspA1I, 50% of the signal from the higher molecular weight DNA disappeared (Fig. 5C). Thus, the assay was able to discriminate the joining of one strand vs. the other. The junction formed by joining of the blunt EcoRV ends was susceptible to cleavage by the phosphodiesterase activity of EcoRV, indicating that Cernunnos stimulated a ligation event that created a bona fide phosphodiester bond.
For blunt-3′ EcoRV–KpnI ends, Ku, XL, and Cernunnos joined 5% of the input DNA (Fig. 5D), which was higher than the result from quantitative PCR of the same DNA products, which detected ligation of 1.6% of the input DNA (data not shown). When we removed the radiolabel from the strand containing the 3′ overhang by MspA1I cleavage, 100% of the signal from the higher molecular weight DNA disappeared. By contrast, removal of the radiolabel from the other strand by ApoI reduced the size of the higher molecular weight fragment by 57 nt without diminishing the signal. This result provided direct evidence for single-strand ligation of the hydroxyl group of the 3′ overhang to the 5′ phosphate of the blunt end. Remarkably, Ku, XL, and Cernunnos ligated blunt-3′ ends with an efficiency of 5%, even in the absence of DNA-PKcs. For comparison, T4 ligase ligated blunt ends with an efficiency of 14%.
Discussion
V(D)J recombination often generates ends with 3′ overhangs. RAG1 and RAG2 initiate recombination by cleaving the immunoglobulin gene to create hairpin ends (16). Artemis then opens the hairpins to leave 3′ overhangs (8, 15). TdT extends the 3′ overhang further by nontemplated nucleotide addition to generate additional immunoglobulin diversity.
To preserve the sequence of the 3′ overhang, NHEJ must use an unconventional enzymatic activity. The problem is that DNA polymerases require a primer strand and always replicate DNA in the 5′ to 3′ direction. To address this problem, Nick McElhinny et al. (10) reported that pol mu has the unusual ability to fill in a 2-nt 3′ overhang by priming from the opposing DNA end. However, there are no data showing that pol mu can fill in longer 3′ overhangs, such as those created during V(D)J recombination.
MEnd ligase activity may explain how V(D)J recombination preserves 3′ overhang sequences in vivo. We propose that Ku, XL, and Cernunnos ligate the 3′ overhanging hydroxyl group to the 5′ phosphate of the opposing end. Thus, ligation creates a continuous single strand, which serves as a template for DNA polymerase. Consistent with this proposal, Cernunnos-defective cells join coding ends with decreased efficiency and with abnormally large deletions (13, 14, 17).
Previously, we showed that human cell extracts recapitulated NHEJ as observed in vivo (18). Extracts join blunt-3′ ends with high efficiency and use polymerase activity to preserve the sequence from the 3′ overhang. MEnd ligase activity may explain this phenomenon. In fact, polymerase activity during NHEJ in extracts exhibited absolute dependence on XL (19), which is an essential component of MEnd ligase activity.
Cernunnos stimulates XL to ligate mismatched DNA ends by a mechanism that remains unknown. We speculate that Cernunnos, together with Ku and XRCC4, might align mismatched ends to promote ligation by Ligase IV. Consistent with our results, Hentges et al. recently reported that Cernunnos binds to DNA and stimulates XL to join PvuI–PvuI ends with 2-nt 3′ overhangs (20). In this paper, we showed that Cernunnos and Ku stimulated XL to join blunt ends and several types of mismatched ends that are noncomplementary or partially complementary (Fig. 1). However, Cernunnos failed to stimulate joining of ends with cohesive 4-nt overhangs. Thus, Cernunnos may be dispensable for cohesive end joining with 4-nt overhangs but may provide the necessary alignment for XL to join blunt ends, ends that are less cohesive, or ends with mismatched overhangs.
Ma et al. (21) reported a single-strand ligase activity for XL alone. However, joining required alignment of the ends by at least two complementary base pairs and occurred in the absence of Ku or Cernunnos. In our experiments, Cernunnos and Ku stimulated XL to ligate mismatched ends by >100-fold (Fig. 2B), even when the ends lacked complementary base pairing. Thus, Ku, XL, and Cernunnos possess a novel MEnd ligase activity.
In conclusion, our data suggest that MEnd ligase activity contributes to the preservation of DNA sequence during NHEJ. For V(D)J recombination in vivo and for cell extracts in vitro, NHEJ preserves the sequence of 3′ overhangs. In these cases, the MEnd ligase activity may preserve sequence by direct ligation of the 3′ overhang to the 5′ phosphate of the opposing end. Finally, MEnd ligase activity may have applications for recombinant DNA technology. For example, sheared genomic DNA and PCR-amplified DNA contain 3′ overhangs generated by the shearing process or by the weak TdT activity of Taq polymerase, respectively. MEnd ligase can ligate such DNA molecules directly into a blunt-ended cloning vector.
Materials and Methods
Expression Constructs and Protein Purification.
We used PCR to amplify the human Cernunnos ORF from a human cDNA clone (no. MGC-32656; American Type Culture Collection, Manassas, VA); subcloned it into pET101/D-TOPO (Invitrogen, Carlsbad, CA), which added a His tag to the C terminus of the protein; and expressed recombinant Cernunnos in bacteria. We purified Cernunnos from the bacterial lysate to apparent homogeneity on Ni-NTA beads (Qiagen, Valencia, CA) followed by Superdex-200 (26/60) and Hi-trap heparin columns (Amersham Pharmacia Biotech, Piscataway, NJ).
We purified XL as described (19). In brief, we infected Sf9 insect cells with baculoviruses containing XRCC4 and Ligase IV-His (a gift from William Dynan, Medical College of Georgia, Augusta, GA), lysed the cells, and purified the proteins on Ni-NTA beads (Qiagen), followed by Superdex-200 (26/60) and Mono Q (5/50 GL) columns (Amersham Pharmacia Biotech).
We purified Ku from the lysate of Sf9 cells coinfected with baculoviruses containing Ku70-His and Ku86 (a gift from Beverly Mitchell, Stanford University, Stanford, CA) by binding Ku to Ni-NTA beads (Qiagen) and by gel filtration on a Superose-12 (HR 10/30) column (Amersham Pharmacia Biotech).
We purified DNA-PKcs from HeLa cells, which were provided as a cell pellet by the National Cell Culture Center. We prepared extracts with minor modifications of the protocol by Dvir et al. (22) and purified DNA-PKcs by using methods from Ding et al. (23). Peak fractions containing Cernunnos, XL, Ku, and DNA-PKcs were flash-frozen and stored at −80°C.
Preparation of DNA Substrates.
We prepared DNA substrates for the PCR-based assay as previously described (18, 24) and prepared DNA substrates for the gel-based assay by PCR amplification of a 1-kb DNA fragment. Digestion of the PCR product with EcoRI or EcoRV or double digestion with EcoRV and KpnI released a DNA fragment with EcoRI–EcoRI, EcoRV–EcoRV, or EcoRV–KpnI ends, which was resolved by agarose gel electrophoresis and purified from the gel.
End-Joining Reactions.
We performed end-joining reactions as described in SI Materials and Methods, purified the DNA products on a QIAquick column (Qiagen), measured joining efficiencies by quantitative PCR, and sequenced the DNA junctions as previously described (18).
For the gel-based assay, we performed end-joining reactions by incubating 0.4 pmol of the DNA substrate with no protein; 2 pmol Ku, 0.4 pmol XL, and 2 pmol Cernunnos or 1 unit of T4 DNA ligase (USB) in NHEJ buffer in a total volume of 200 μl. After incubation, we purified the DNA products on a QIAquick column (Qiagen), digested the DNA with XmnI at 37°C for 1 h, dephosphorylated the DNA with 1 unit Antarctic phosphatase (New England Biolabs, Beverly, MA) at 37°C for 15 min, and then labeled the DNA with [γ-32P]ATP and T4 polynucleotide kinase (New England Biolabs). We purified the DNA by phenol/chloroform extraction and ethanol-precipitation; digested the DNA further with ApoI, MspA1I, or EcoRV; resolved the products by electrophoresis in a TBE-urea 5% polyacrylamide gel (Bio-Rad, Hercules, CA); and quantified joining by PhosphorImager analysis (Molecular Dynamics). For detailed descriptions of all methods, see SI Materials and Methods.
Acknowledgments
We thank Dr. William Dynan for XRCC4 and Ligase IV baculoviruses and for advice on DNA-PKcs purification, Dr. Beverly Mitchell for Ku70 and Ku86 baculoviruses, and Drs. Dale Ramsden and Joe Budman for helpful discussions. This work was supported by a National Science Fellowship (to S.A.K.), National Institutes of Health Grant RO1 GM58120 (to G.C.), and a Translational Cancer Research Award from the Stanford Comprehensive Cancer Center (to G.C.).
Abbreviations
- NHEJ
nonhomologous end-joining
- DNA-PKcs
DNA-dependent protein kinase catalytic subunit
- XL
XRCC4/Ligase IV
- XLF
XRCC4-like factor
- MEnd
mismatched end.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702620104/DC1.
References
- 1.Lieber MR, Ma Y, Pannicke U, Schwarz K. DNA Repair. 2004;3:817–826. doi: 10.1016/j.dnarep.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 2.Smider V, Rathmell WK, Lieber M, Chu G. Science. 1994;266:288–291. doi: 10.1126/science.7939667. [DOI] [PubMed] [Google Scholar]
- 3.Taccioli G, Gottlieb T, Blunt T, Priestly A, Demengeot J, Mizuta R, Lehmann A, Alt F, Jackson S, Jeggo P. Science. 1994;265:1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- 4.Hammarsten O, Chu G. Proc Natl Acad Sci USA. 1998;95:525–530. doi: 10.1073/pnas.95.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeFazio L, Stansel R, Griffith J, Chu G. EMBO J. 2002;21:3192–3200. doi: 10.1093/emboj/cdf299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grawunder U, Wilm M, Xiantuo W, Kulezla P, Wilson TE, Mann M, Lieber MR. Nature. 1997;388:492–494. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 7.Goodarzi AA, Yu Y, Riballo E, Douglas P, Walker SA, Ye R, Harer C, Marchetti C, Morrice N, Jeggo PA, et al. EMBO J. 2006;25:3880–3889. doi: 10.1038/sj.emboj.7601255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma Y, Pannicke U, Schwarz K, Lieber MR. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 9.Niewolik D, Pannicke U, Lu H, Ma Y, Wang LC, Kulesza P, Zandi E, Lieber MR, Schwarz K. J Biol Chem. 2006;281:33900–33909. doi: 10.1074/jbc.M606023200. [DOI] [PubMed] [Google Scholar]
- 10.Nick McElhinny SA, Havener JM, Garcia-Diaz M, Juarez R, Bebenek K, Kee BL, Blanco L, Kunkel TA, Ramsden DA. Mol Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 11.Bertocci B, De Smet A, Berek C, Weill JC, Reynaud CA. Immunity. 2003;19:203–211. doi: 10.1016/s1074-7613(03)00203-6. [DOI] [PubMed] [Google Scholar]
- 12.Bertocci B, De Smet A, Weill JC, Reynaud CA. Immunity. 2006;25:31–41. doi: 10.1016/j.immuni.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 13.Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche MC, Sanal O, Plebani A, Stephan JL, Hufnagel M, le Deist F, et al. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 14.Ahnesorg P, Smith P, Jackson SP. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 15.Schlissel M. Mol Cell Biol. 1998;18:2029–2037. doi: 10.1128/mcb.18.4.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Gent D, Ramsden D, Gellert M. Cell. 1996;85:107–113. doi: 10.1016/s0092-8674(00)81086-7. [DOI] [PubMed] [Google Scholar]
- 17.Dai Y, Kysela B, Hanakahi LA, Manolis K, Riballo E, Stumm M, Harville TO, West SC, Oettinger MA, Jeggo PA. Proc Natl Acad Sci USA. 2003;100:2462–2467. doi: 10.1073/pnas.0437964100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Budman J, Chu G. EMBO J. 2005;24:849–860. doi: 10.1038/sj.emboj.7600563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Budman J, Kim SA, Chu G. J Biol Chem. 2007;282:11950–11959. doi: 10.1074/jbc.M610058200. [DOI] [PubMed] [Google Scholar]
- 20.Hentges P, Ahnesorg P, Pitcher RS, Bruce CK, Kysela B, Green AJ, Bianchi J, Wilson TE, Jackson SP, Doherty AJ. J Biol Chem. 2006;281:37517–37526. doi: 10.1074/jbc.M608727200. [DOI] [PubMed] [Google Scholar]
- 21.Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, Koiwai O, Hsieh CL, Schwarz K, Lieber MR. Mol Cell. 2004;16:701–713. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 22.Dvir A, Stein LY, Calore BL, Dynan WS. J Biol Chem. 1993;268:10440–10447. [PubMed] [Google Scholar]
- 23.Ding Q, Reddy YV, Wang W, Woods T, Douglas P, Ramsden DA, Lees-Miller SP, Meek K. Mol Cell Biol. 2003;23:5836–5848. doi: 10.1128/MCB.23.16.5836-5848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Budman J, Chu G. Methods Enzymol. 2006;408:430–444. doi: 10.1016/S0076-6879(06)08027-X. [DOI] [PubMed] [Google Scholar]