Abstract
Fabry disease is a disorder of α-d-galactosyl-containing glycolipids resulting from a deficiency of α-galactosidase A. Patients have a poorly understood vascular dysregulation. We hypothesized that disease-related perturbation by using enzyme replacement therapy in the murine model of Fabry disease would provide insight into abnormal biological processes in Fabry disease. Gene expression analyses of the heart, aorta, and liver of male α-galactosidase A knockout mice 28 weeks of age were compared with that of WT mice. Microarray analyses were performed before and after six weekly injections of α-galactosidase A. Alteration of Rpgrip1 ranked highest statistically in all three organs when knockout mice were compared with WT, and its splice variants responded in a unique way to α-galactosidase A. Enzyme replacement therapy tended to not only normalize gene expression, e.g., reduce the overexpression of securin, but also specifically modified gene expression in each tissue examined. Following multiple comparison analysis, gene expression correlation graphs were constructed, and a priori hypotheses were examined by using structural equation modeling. This systems biology approach demonstrated multiple and complex parallel cellular abnormalities in Fabry disease. These abnormalities form the basis for informed, in a Bayesian sense, sequential, hypothesis-driven research that can be subsequently tested experimentally.
Keywords: glycolipids, growth factor, lysosomal, reactive oxygen species
Fabry disease is an X-linked metabolic disorder due to deficiency of the lysosomal enzyme α-galactosidase A, resulting in reduced and altered catabolism of α-d-galactosyl-containing compounds (1). Glycosphingolipids, particularly globotriaosylceramide (Gb3), accumulate in endothelial cells and other tissues (2–4). It is thought that the deposition of such lipids contributes directly to endothelial dysfunction. The precise relationship between abnormal sphingolipid metabolism and impaired endothelial function is not clear (5). Most of the burden of disease results from a vasculopathy associated with progressive damage of the heart and kidney as well as cerebrovascular strokes. Evidence suggests that the vasculopathy of Fabry disease has significant functional components and is not merely an obstructive vasculopathy. The vasculopathy may result from an impairment of the nitric oxide (NO) pathway probably due to excess superoxide production, peroxynitrite, and other reactive nitrogen species capable of causing oxidative damage. Abnormalities of this nature are present in the cerebral circulation and dermal vasculature of patients with Fabry disease (6).
One consequence of NO pathway dysregulation may be cerebral hyperperfusion resulting in altered vascular reactivity and elongation of cerebral blood vessels predominantly in the vertebrobasilar circulation (5). Evidence for excess formation of superoxide or reactive oxygen species (ROS) was provided recently by a magnetic resonance imaging and arterial spin labeling study (7). Elevated vascular ROS formation, together with increased myeloperoxidase, has been associated with accelerated atherosclerosis (7). The abnormalities in cerebral blood flow, including cerebral hyperperfusion and delayed vascular reactivity to acetazolamide, were reversed by enzyme replacement therapy (ERT) (7–9). Vascular NO dysregulation, increased nitrotyrosine staining, and accelerated atherosclerosis were recently confirmed in the murine model of Fabry disease (10).
The murine model of Fabry disease developed at the National Institutes of Health has been well characterized. Histological evidence suggests many similarities with human Fabry disease, although some discordance is present (4, 11–13). One significant advantage of the murine model is the ability to investigate Fabry tissues such as the aorta and heart, which are generally inaccessible in clinical studies. Gene expression analysis using microarrays allows a near complete description of biological processes within a tissue. We performed gene expression analysis on the heart and aorta of the knockout (KO) model of Fabry disease. The liver was also examined as a tissue that is not critically involved in Fabry disease. The goal of the current work was to develop a systems biology approach to Fabry disease by using ERT with agalsidase-α as a disease-related perturbation, allowing mapping of the significantly associated gene pathways suitable for development of hypothesis-driven research.
Results
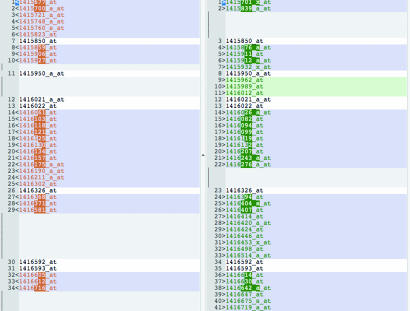
Gene-wise comparison using a robust general linear model estimation of the aorta, heart, and liver was performed for the following states: untreated Fabry mice, Fabry mice that received ERT (Fabry-ERT), and wild-type (WT) mice. These findings are presented and tabulated in supporting information (SI) Tables 1–9. Results for Holm correction [family-wise error (FWE) correction] are presented before results for a false discovery rate (FDR) multiple comparison correction. For aortic tissue, contrasts between Fabry-ERT and Fabry after FWE correction gave 10 significant differentially expressed genes, Fabry-ERT vs. WT gave 65 significant differentially expressed genes, and Fabry and WT gave 30 significant differentially expressed genes. FDR correction gave aortic Fabry-ERT and Fabry (n = 180), Fabry-ERT and WT (n = 751), and Fabry and WT (n = 551) significant differentially expressed genes. SI Fig. 5 is a heat plot illustrating the significant differential gene expression of Fabry aortic tissue with and without ERT. The red areas indicate up-regulation of gene expression, and the green areas indicate down-regulation. ERT caused a predominantly up-regulatory response in aortic tissue gene expression. For cardiac tissue, contrasts between Fabry-ERT and Fabry after FWE correction revealed seven significant differentially expressed genes, contrasts between Fabry-ERT and WT showed 52 significant differentially expressed genes, and contrasts between Fabry and WT gave 69 significant differentially expressed genes. FDR correction comparing cardiac Fabry-ERT and Fabry showed 129 differentially expressed genes, 386 genes between Fabry-ERT and WT, and 334 genes between Fabry and WT. For liver tissue, contrasts between Fabry-ERT and Fabry after FWE correction gave 11 significant differentially expressed genes, Fabry-ERT and WT gave 41 significant differentially expressed genes, and Fabry and WT revealed 31 significant differentially expressed genes. FDR correction for the liver between Fabry-ERT and Fabry gave 41 differentially expressed genes, Fabry-ERT and WT 401 differentially expressed genes, and Fabry and WT 186 differentially expressed genes. Upon comparison of the significant differentially expressed genes between not only the Fabry vs. WT, but also the Fabry-ERT vs. WT, we found 61 shared genes in the aorta (Fig. 1), 139 genes in the heart, and 85 genes in the liver. ERT did not completely normalize the gene expression profile; in fact, it actually induced novel sets of genes in each tissue examined.
Fig. 1.
Genes that are specifically modified by ERT. Comparative list of genes significantly expressed in the aorta of Fabry-ERT mouse vs. WT (Left) and Fabry vs. WT (Right). Genes in both lists have a white background, and unique genes for either list are in color.
To address the question of the biological relevance of these changes, we used two approaches. The first is a more traditional approach that searches for similarities and differences in the gene listings representing changes between the genes ranked by their statistical P value. The second was a systems biology approach looking for networks of genes in specific pathways that change expression due to Fabry disease and, more specifically, disease-related perturbations in tissues following ERT.
The first approach highlights the similarities between the aorta and heart lists, probably representing similar embryological origins of these tissues, whereas the liver list is different. In general, several genes, including Rpgrip1, Pttg1, Cap1, Syndecan 4, and Comt 1, appeared in all three tissues. The majority of genes in nontreated Fabry KO mice expression lists also occurred in agalsidase-α-treated KO gene expression lists (≈80%). In this regard, the gene expression lists comparing ERT-treated and nontreated Fabry KO mice are less informative probably due to the partial treatment effect of ERT. Pttg1 (securin) and syndecan 4 appear in the liver (Fabry-ERT contrasting with untreated Fabry mice), but not in a statistically significant manner. In general, despite the reduction in storage material observed in Fabry tissue after ERT, the mouse array experiment reveals at best a trend toward partial restoration of normal tissue function.
Rpgrip1 Gene Expression in the Fabry KO Mouse.
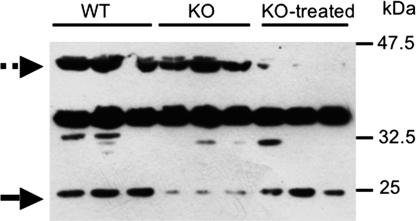
The expression of Rpgrip1 ranked at the top of all mutant lists in the three tissues tested regardless of treatment (see SI Tables 1, 3, 4, 6, 7, and 9). This gene is known to generate multiple splice variants and protein isoforms that are differentially expressed in various tissues and subcellular compartments in the mouse and human (14–16). In addition, some isoforms of Rpgrip1 undergo limited proteolytic processing (17). The Affymetrix (Santa Clara, CA) mouse gene expression chip contains four different probes for the Rpgrip1 gene, but only one (1421144_at) is significantly overexpressed in the Fabry mouse. This probe corresponds to the terminal 3′ region of mRNA for Rpgrip1. We tested the expression of the protein in tissue from WT, Fabry, and agalsidase-α-treated Fabry mice. A novel 25-kDa Rpgrip1 isoform, or a limited proteolytic product thereof, was missing in the liver of nontreated Fabry mice. This form appeared to be partially restored in tissue from Fabry-ERT mice, whereas another isoform of ≈35 kDa was repressed only in Fabry-ERT mice (Fig. 2). The apparent discrepancy between mRNA and protein expression may be explained by assuming an inhibitory role of the overexpressed form on other variants of the gene either at the mRNA or protein level.
Fig. 2.
Western blot for RPGRIP1γ in liver tissue. In Fabry mice, the expression of an immunoreactive RPGRIP1γ-related isoform is suppressed, whereas upon ERT its expression is induced again (arrow). Conversely, another RPGRIP1γ-related isoform seems to be suppressed upon treatment only (dashed arrow).
Securin (Pttg1) Epression in the Fabry KO Mouse.
As indicated in the gene list, the gene for securin was among the most highly overexpressed in the Fabry KO mouse. The expression of securin protein was also tested in Fabry mice. The level of protein expression was increased dramatically in both the heart (10-fold) and liver (16.8-fold), consistent with the microarray results. Interestingly, the level of expression decreased in both tissues with agalsidase-α treatment to 4.4- and 5.3-fold, respectively. These results indicate that, in this case, determining protein expression may offer a better marker to evaluate the ability of ERT to alleviate the burden of the disease in treated animals than measuring mRNA expression.
Structural Equation Modeling (SEM).
To develop SEM, assumptions of data normality, with an emphasis on kurtosis (18) of differential gene expression, were examined in the case of the FDR-corrected, ERT-treated aortic tissue, compared with nontreated Fabry KO aortic tissue. SI Fig. 6A is a histogram plot of the data set with a skew calculated at 0.43 and a kurtosis of 2.9. By D′Agostino skew test (skew = 0.43, Z = 5.2, P = 1.759e-07), the data are significantly skewed. By Anscombe–Glynn test for kurtosis, the data are not abnormally kurtotic (kurtosis = 2.9, Z = −0.66, P = 0.5; in an alternative hypothesis, kurtosis is not equal to 3). Despite the significant right skew of the data, SI Fig. 6B shows the Mahalanobis distance for the aortic ERT-Fabry vs. Fabry data set with relatively few outliers. These indices indicate the acceptable normality and kurtosis of the differential gene expression set used in path analysis and SEM. It has been established that the maximum likelihood estimator (MLE) properties are maintained for nonnormal data as long as there is not excess multivariate kurtosis.
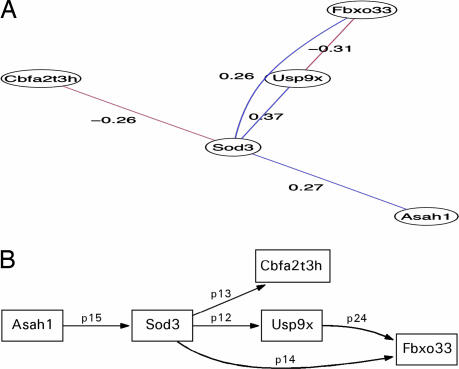
In the development of the subsequent systems biology models, particular attention was paid to the effect of ERT on Fabry vasculopathy by comparing the differential gene expression between the ERT-Fabry aortic tissue group and the nontreated Fabry aortic tissue group. Complete correlation graphs derived for the aortic tissue are shown in SI Fig. 7. Subsequent systems biology approaches develop an SEM concentrating on the up-regulated Sod3 gene following ERT because this is known to regulate vessel wall ROS (19). Prior clinical investigation indicates the involvement of ROS in Fabry vasculopathy (6, 7). This suggests that the model is an effective realization of vessel wall ROS involvement and allows the formation of a priori SEM related to the abnormal glycolipid disorder in Fabry (glycolipidopathy) and ERT. The model was a priori-derived from the correlation graph in Fig. 3a. An initial simplified structural model seen in Fig. 3b was examined and fitted by an MLE, giving a model fit of χ2 = 4.74, df = 5 (model overidentified), Pr (>χ2) = 0.45, null model χ2 = 11.21, df = 10, goodness-of-fit index (GFI) = 0.88, adjusted goodness-of-fit index (AGFI) = 0.65, Bentler–Bonnett normed fit index (NFI) = 0.58, root mean-squared error of approximation (RMSEA) index = 0.0, 90% confidence intervals (CI): (0, 0.41), Bayesian information criterion (BIC) = −7.68, with negative value signifying a fit superior to saturated model. Although none of the parameters was statistically significant, directionality (correlation graph value) was appropriately preserved with standardized parameter values in the small to medium effect range (small <0.1, medium 0.1–0.5). A trend significance value was found between Usp9x and Fbxo33 (P = 0.09). The standardized residuals ranged from −0.83 to 0.56, with small modification indices suggesting little utility in parameter refit and relative variable independence. The model parameters were used as initial values in confirmatory factor analysis (CFA), where glycolipidopathy and subsequently ERT were independently fitted to the path model with MLE convergence, although the BIC was reduced to 1.99 (glycolipidopathy) and −1.73 (ERT). These CFA models were subsequently combined into a model with structural and measurement components representing a hypothesized cellular abnormality of Fabry vascular tissue with up-regulation of growth factor genes such as Cbfa2t3h and Fbxo33, with ERT acting directly on glycolipidopathy as a latent variable (see SI Fig. 8B). These paths were chosen on the basis of the directed correlation in SI Fig. 8A, indicating that cellular growth dysregulation genes are up-regulated in the untreated Fabry aortic tissue. Sod3 is included in the path from the glycolipidopathy latent variable. The model was overidentified with the following fit parameters: Model χ2 = 7.7, df = 3, Pr(>χ2) = 0.053, GFI = 0.84, AGFI = 0.21, NFI = 0.31, RMSEA index = 0.38, 90% CI: (0, 0.72), BIC = 0.203. This model was trimmed (SI Fig. 8B) to achieve better global fit statistics by removing paths with high standard residuals so that Sod3 had a path to the ubiquitin genes Usp9x, Cbfa2t3h, and Fbxo33, whereas the effect of the glycolipidopathy is restricted to Cbfa2t3h and Fbxo33, with ERT acting as before directly on the glycolipidopathy latent variable. The fit was improved by this more parsimonious model as indicated by the global indices and the decrease in the BIC: Model χ2 = 7.6, df = 4 Pr(>χ2) = 0.10, GFI = 0.84, AGFI = 0.41, RMSEA index = 0.29, 90% CI: (0, 0.59597), BIC = −2.28. The χ2 value indicates that the model is acceptable, whereas the RMSEA is less than optimal despite the effective reduction in BIC. The standardized path coefficients for this model are as follows: p14 = 0.26, Sod3 → Fbxo33, p13 = −0.26, Sod3 → Cbfa2t3h, p12 = 0.37, Sod3 → Usp9x, lg3 = 0.27, glycolipidopathy → Cbfa2t3h, lg4 = 0.62, glycolipidopathy → Fbxo33, latent1 = 0.1, ERT → glycolipidopathy. SI Table 10 contains the unstandardized parameters and the estimated parameter significance.
Fig. 3.
SEM analysis of superoxide dismutase in aorta of Fabry mice that received ERT. (A) Graphical Gaussian model of Sod3-related gene expression in aortic tissue of the Fabry KO mouse after disease-related perturbation with agalsidase-α. (B) Path diagram of simplified Sod3 model based on the graphical Gaussian network illustrated in A (χ2 = 4.7, df = 5, P = 0.44).
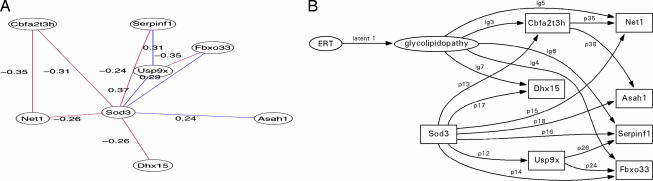
Nonuniqueness of SEM analysis is inherent in the technique. For this reason, a more complex model was analyzed. It is seen that the previously analyzed model is a subgraph of this more complex model (Fig. 4b) based on the correlational graph in Fig. 4a. The strategy previously adopted for the simplified model was developed, in that the initial model was analyzed and used to provide parameter start values for the subsequent structural equation model: Model χ2 = 8.9, df = 17, Pr(>χ2) = 0.94, null model χ2 = 16.476, df = 28, GFI = 0.88, AGFI = 0.74, NFI = 0.4604, RMSEA index = 0.0, 90% CI: (0, 0.047356), BIC = −33.356, normalized residuals min. −9.47e-01, 1st Qu. −5.35e-02, median 0.00e + 00, mean −1.83e-02, 3rd Qu. 1.59e-16, max. 1.05e + 00. The complex model containing both manifest and latent variables was formulated analogously to the more simple model by allowing the glycolipidopathy latent variable to have paths directly to cellular growth factors, whereas the ERT effect is mediated through the glycolipidopathy variable as before. The BIC is significantly decreased after addition of the latent variables over the measurement model; however, the global fit indices show a good overall model fit: Model χ2 = 3.15, df = 11, Pr(>χ2) = 0.99, null model χ2 = 16.476, df = 28, GFI = 0.93, AGFI = 0.80, NFI = 0.806, RMSEA index = 0.0, 90% CI: (0, 0), BIC = −24.181, normalized residuals min. −6.22e-01 1st Qu. −4.22e-02, median −1.12e-07, mean 2.51e-02, 3rd Qu. 5.73e-02, max. 1.05e + 00. SI Tables 11 and 12 contain the nonstandardized parameters and the estimated parameter significance, together with the standardized parameters. It is noted that the glycolipidopathy path parameter to Serpinf1 and the Usp9x path parameter to Serpinf1 are statistically significant.
Fig. 4.
SEM analysis of superoxide dismutase in ERT Fabry aorta using a more complex instantiation of the differential gene expression relations. (A) Complex graphical Gaussian model (correlational graph network) of Sod3-related gene expression in aortic tissue of the Fabry KO mouse after disease-related perturbation with agalsidase-α. (B) Corresponding Sod3 SEM showing the latent variables for ERT and glycolipidopathy based on the graphical Gaussian model illustrated in A (χ2 = 3.1, df = 11, P = 0.99)
Discussion
In this investigation, we first attempted to validate the results of the microarrays and demonstrate that the changes are valid at a protein level. We then investigated the manner by which these findings may help explain the pathogenesis of Fabry disease. The present work determined significant differences among treated, untreated Fabry mice, and WT mice. The exact number of gene differences varied for the tissues examined (aorta, heart, and liver) and comprised both up- and down-regulated gene expression levels. It appeared that the predominant effect of the Fabry phenotype was determined by excessive expression of regulatory growth genes in a tissue-dependent manner. This is seen, for example, in the SEM analysis with the association of Net 1 and Cbfa2t3h in the Fabry KO mouse aortic tissue. ERT appeared to induce the expression of counterregulatory and adaptive genes. The extent of significant differential gene expression in each tissue was determined by the application of MCC. Such correction was an absolute requirement due to the large number of highly correlated genes sampled simultaneously from single independent murine specimens. Two types of corrections were performed and recorded in the SI Tables 1–9: an FWE or Holm correction that is a relatively stringent procedure compared with the FDR technique. Both techniques are valid, but it is often considered that the FWE correction is probably too stringent.
The evaluation of the protein expression of two of the most highly ranked genes in all lists confirmed the changes in mRNA expression. The increase in expression of Rpgrip1 throughout the tissues studied boosts our confidence in the validity of the techniques used in this study. It points to a significant pathology in the Fabry KO mouse resulting in major alterations of gene expression to adapt to the mutation. The apparent reduction in expression of one form of the protein provides an example of how difficult it is to interpret gene expression data. However, this change illustrates our general finding that ERT specifically modulated a series of genes. Rpgrip1 was discovered in the retina (16), but multiple isoforms derived from alternative splicing are found across tissues and/or with tissue-selective expression (14, 16). RPGRIP1γ is unique among all Rpgrip1 isoforms because it colocalizes to a subpopulation of lysosomes in the retina and cell culture (16, 36), pointing to a possible interaction of Rpgrip1γ, a related isoform, or a processed proteolytic product, with α-galactosidase A. Changes in Rpgrip1 expression seem to be related to tissue growth factors, as is the trend in many other genes that show up in the Fabry mouse. The change in securin levels due to ERT may be used as a target to evaluate treatment in Fabry disease. The obvious limit in evaluating this information is the fact that there is no known association between these genes and Fabry disease. Another approach based on what is already known about Fabry disease may be more useful. Such an approach can identify targets in genes that may have changed less dramatically or in a more tissue-specific manner, but are more relevant to understanding the pathophysiology of the disease.
For consideration of a systems biology approach where process parallelism is implicit, the less stringent FDR approach is probably preferable because it allows the potential weaker causal associations to interact. We focused on the aortic tissue because of the significance of Fabry vasculopathy and evidence for an altered tissue NO and superoxide regulation (7). Of particular interest was the effect of ERT on the aortic tissue. For this reason, the 180 FDR-corrected gene expression values were chosen as the basis for the subsequent systems biology analysis, with a view to retaining tissue pathway parallelism as much as possible while removing significant sources of type I error.
A first step in the construction of potential gene expression pathways was the calculation of correlation graphs between the various gene nodes, followed again by multiple comparison correction (MCC) to prevent the construction of spurious paths. The resultant sparse network of graphical Gaussian models contains a series of biologically testable statistical hypotheses about the differences between ERT-treated and untreated Fabry aortic tissue. The derivation of the covariance matrix in a microarray context is ill-posed. However, a satisfactory approach based on Bayesian inference was adopted here, resulting in the reported sparse graphical Gaussian networks (20). To test hypotheses about these correlation graphs, we adopted an SEM approach focusing in particular on the adjacency graph of Sod3. Up-regulation of this gene following ERT is significant as an a priori hypothesis about the mechanism of dysfunction in Fabry vasculopathy relating to dysregulation of the NO and superoxide pathways. The implication of SEM for defining and testing causal networks depends on the strength of the a priori conditions on which the SEM analysis is based. What is apparent is the ability of SEM to handle parallel or conjunctional primitive events in the overall observed causal effect, such as relating interdependent gene expression values. This analysis allows the investigator to generate simplified and sequential hypothesis-driven investigations from groups of causal primitives while increasing understanding of a disease state at a systems biology level. The relationship of SEM to causality has a formal theoretical basis (21).
Sod3 is the known vascular extracellular isoform of superoxide dismutase (SOD), with the major source of synthesis considered to be smooth muscle cells. It is hypothesized that the major function of Sod3 is to protect NO as it diffuses from the endothelium to its major target, smooth muscle soluble guanylate cyclase, resulting in arterial wall relaxation (19). The up-regulation of Sod3 following ERT is likely an adaptive response to restoring vascular health in the Fabry aorta. Other genes present in the Sod3 subgraph are Cbfa2t3h, a transcription factor (22); Dhx15, which forms part of a putative family of RNA helicases (23); Fbxo33, which forms part of the F-box protein family components of modular E3 ubiquitin protein ligases (24); Asah1, which is the gene for acid ceramidase, the enzyme that is deficient in Farber Disease (25); and Serpinf1 (α-2-antiplasmin), which is part of the serine protease inhibitor family that has broad functions, including antiangiogenesis, but is also known to decrease the activity of free plasmin (26) while at a cellular level (e.g., in the brain) it is known to be neuroprotective (27). We recently found a generalized perturbation of α-2-antiplasmin and plasminogen in children with Fabry disease by using a similar disease-related perturbation (28). Usp9x is ubiquitin-specific protease 9 or deubiquitinating enzyme 9, resulting in the removal of ubiqitin moiety from ubiquitin-modified proteins, reversing protein targeting to the 26S proteosome (29, 30).
A potential interpretation for the simpler SEM is that the Fabry aortic phenotype results in enhanced cellular growth factor function with ERT at least initiating counterregulatory mechanisms such as enhanced Sod3 expression and deubiquitination of proteins destined for cellular proteolysis. The association of Asah1 may represent activation of a secondary metabolic pathway, again in a counterregulatory fashion. SEM mediates the effect of ERT through the glycolipidopathy latent variable to represent the clinical context of treatment of patients with Fabry disease in whom ERT is infused biweekly at a fixed dose. It is apparent that ERT has some effect on Fabry disease, and the present experimental design enables this effect to be evaluated at the genomic expression level. The more complex model provides global fit and BIC statistics that are substantially better than the simpler model, indicating a preference for the complex model on the basis of parsimony alone. On a statistical note, the small sample size in this work will have an effect on estimation of SEs. One way to overcome this problem is to use bootstrapping. Two paths are statistically significant: glycolipidopathy → Serpinf1, and Usp9x → Serpinf1. Although the sign of the Usp9x → Serpinf1 is similar to the graphical Gaussian model, the large negative of glycolipidopathy → Serpinf1 suggests a negative interaction with glycolipidopathy. A treatment effect was also seen by the up-regulation of Serpinf1 with ERT.
Further exploration of the Fabry phenotype from the current experimental data is ongoing. The applicability of the techniques developed in this article could serve equally well in the study of any disease state.
Methods
Male α-galactosidase A KO mice (Fabry mice) on a mixed C57BL/6 × 129/SvJ background (31), ≈28 weeks of age at the start of each study, were used for these studies. All Fabry animals and WT controls were progeny of the original colony of C57BL/6 × 129/SvJ mice. Mice were housed in a pathogen-free facility where standard rodent diet (Zeigler, Gardners, PA) and water were available ad libitum. All procedures involving animals were performed in compliance with the Guidelines of the American Association for the Accreditation of Laboratory Animal Care on Use of Animals in Research at the National Institutes of Health. Both WT and Fabry mice were genotyped by a PCR-based assay as previously described (32). Groups of Fabry mice received six weekly injections (100 μl) in the tail vain of α-galactosidase A (agalsidase-α; Shire Human Genetic Therapy, Cambridge, MA) diluted in normal saline at a dose of 1.0 mg/kg of body weight for a total of six injections. One week after the last injection, the thoracic aorta, liver, heart, and kidneys were harvested. Portions of these organs were either snap-frozen on dry ice and stored at −80°C before enzyme and lipid analyses or flash-frozen in liquid nitrogen and stored in the liquid phase of liquid nitrogen before RNA extraction. The thoracic aortas were flash-frozen in liquid nitrogen and stored in the liquid phase of liquid nitrogen before RNA extraction.
Total RNA Isolation.
Details of RNA isolation and Affymetrix microarray methods are available in SI Methods.
Data Normalization and Analyses.
A full description of data normalization and analyses is available in SI Methods. We followed the technique of Irizarry et al. (33) for analyzing gene expression data. MCCs used either an FWE correction technique (Holm) or an FDR correction technique (Hochberg–Benjamini) (34, 35). Two list types of significantly differentially expressed genes were presented, the FWE-corrected gene expression, which was likely to be too stringent, and the FDR corrected list, which may have been overinclusive.
Protein Expression Determination.
Aliquots of 10–30 mg of tissue samples stored in liquid nitrogen were homogenized in NuPAGE lithium dodecyl sulfate buffer and separated on 4–12% NuPAGE gels (Invitrogen, Carlsbad, CA). Rpgrip1 analysis was performed with tissue extracts prepared in Nonidet P-40 lysis buffer. Proteins were transferred and incubated with an antibody against Rpgrip1γ (36) and anti-securin antibody (a kind gift from Hui Zuo, University of Texas Southwestern Medical Center at Dallas). Densitometry was performed by using Alpha-Innotech (San Leandro, CA) software.
Statistical Analysis.
To develop SEM for selected metabolic pathway abnormalities in Fabry vasculopathy by using a systems biology approach, we examined significant gene expression in the KO aortic tissue contrasted with the KO aortic tissue from mice that received ERT (37, 38). SEM allowed construction of an a priori hypothesis related to excessive ROS noted in previous clinical work (6, 7). We were specifically interested in examining the significantly expressed superoxide dismutase 3 gene (Sod3). We assumed that weak causality exists (i.e., the differential gene expression associations exist, although not necessarily uniquely) among the FDR-corrected gene expression list and that, in a statistical sense, this may have been approached by deriving the covariance matrix between all significantly expressed genes resulting in a 180 × 180 square matrix. Derivation of a covariance matrix from high-dimensional correlated gene expression data was performed by using an empirical Bayesian technique, allowing derivation of an appropriately estimated correlation matrix followed by application of FDR correction on the connecting edges of the graphical Gaussian model (20). The overall distributional properties of the differential gene expression data used to generate the covariance matrix is seen in SI Fig. 6, with prior distribution estimates of kurtosis indicating satisfactory compliance with the multivariate kurtosis requirement for SEM. Subgraphs were formed by selecting particular genes (e.g., Sod3) and forming adjacency matrices from the selected gene nodes. Subgraph covariance matrices were then derived for the subgraph from the previously determined covariance matrix. SEM and derivation of the normalized gene expression data, together with the covariance matrix, were performed in the statistical programming language R (37, 39–42). For SEM-related analyses, the MLE was calculated by the SEM R package. Standardized SEM parameter values <0.1 were considered small, values of 0.1–0.5 were medium effects, with values >0.5 representing large effects (37, 39). The structural models can be assessed by global indices such as χ2 > 0.05, GFI, AGFI, and the Bentler–Bonnett NFI, where values closer to 1 suggested a better fit and values closer to 0 suggest a poorer fit. These global fit indices tended to be affected by sample size, with a larger n resulting in a better fit. Some relative fit indices are sample size-independent, such as Bollen's incremental fit index (IFI), but these indices are not widely available (43). The RMSEA is an absolute measure of global fit that asymptotically follows a noncentral χ2 distribution with adjustment for the sample size. RMSEA values <0.05 indicate a good fit of the data. The BIC, calculated as BIC = −2.Ln(L) + k.Ln(n), where n = the sample size, k = the number of parameters, and L is the maximized value of the likelihood function for the estimated model, was used to compare overidentified models, with more negative values indicating that the model has greater support than a just-identified model (no degrees of freedom). The BIC is a decreasing function of the residual sum of squares and an increasing function of the number of parameters, k. The BIC penalizes the number of free parameters (k) more strongly than the Akaike information criterion. A value of standardized residuals >0.1 suggested that the corresponding cor-relation was not well explained. The reporting of the SEM analysis conformed with current standards (37, 38, 44, 45).
Acknowledgments
This work was supported by the Intramural Program of the National Institute of Neurological Disorders and Strokes and National Institutes of Health Grants EY 11993 and 2P30-EY 005722-21 (to P.A.F.). P.A.F. is the Jules & Doris Stein Research to Prevent Blindness Professor.
Abbreviations
- AGFI
adjusted goodness-of-fit index
- BIC
Bayesian information criterion
- CFA
confirmatory factor analysis
- CI
confidence intervals
- ERT
enzyme replacement therapy
- FDR
false discovery rate
- FWE
family-wise error
- GFI
goodness-of-fit index
- KO
knockout
- MCC
multiple comparison correction
- ML
maximum likelihood
- MLE
maximum likelihood estimator
- NFI
normed fit index
- RMSEA
root mean square error of approximation
- ROS
reactive oxygen species
- SEM
structural equation modeling.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701991104/DC1.
References
- 1.Brady R, Gal AE, Bradley RM, Martensson E, Warshaw AL, Laster L. N Engl J Med. 1967;276:1163–1167. doi: 10.1056/NEJM196705252762101. [DOI] [PubMed] [Google Scholar]
- 2.Desnick R, Ioannou YA, Eng CM. In: The Metabolic and Molecular Bases of Inherited Disease. 8th Ed. Charles R, Scriver, Arthur L, Beaudet WS, Sly DV, editors. New York: McGraw–Hill; 2001. pp. 3733–3774. [Google Scholar]
- 3.Mitsias P, Levine SR. Ann Neurol. 1996;40:8–17. doi: 10.1002/ana.410400105. [DOI] [PubMed] [Google Scholar]
- 4.Moore D, Ye F, Schiffmann R, Butman JA. AJNR. 2003;24:1096–1101. [PMC free article] [PubMed] [Google Scholar]
- 5.Moore D, Altarescu G, Ling GSF, Jeffries N, Frei K, Weibel T, Charria-Ortiz G, Ferri R, Brady RO, Schiffmann R. Stroke. 2002;33:525–531. doi: 10.1161/hs0202.102601. [DOI] [PubMed] [Google Scholar]
- 6.Moore DF, Scott LJC, Gladwin MT, Altarescu G, Kaneski C, Suzuki K, Pease-Fye M, Ferri R, Brady RO, Herscovitch P, Schiffmann R. Circulation. 2001;104:1506–1512. doi: 10.1161/hc3801.096352. [DOI] [PubMed] [Google Scholar]
- 7.Moore DF, Ye F, Brennan ML, Gupta S, Barshop BA, Steiner RD, Rhead WJ, Brady RO, Hazen SL, Schiffmann R. J Magn Reson Imaging. 2004;20:674–683. doi: 10.1002/jmri.20162. [DOI] [PubMed] [Google Scholar]
- 8.Moore DF, Pursley R, Altarescu G, Schiffmann R, Dimitriadis E. In: Thomas G, Long R, editors. 14th IEEE Conference on Computer-Based Medical Systems; Bethesda, MD: IEEE; 2001. pp. 216–221. [Google Scholar]
- 9.Schiffmann R, Kopp J, Austin H, Sabnis, Sharda, Moore DF, Weibel T, Balow J, Brady RO. J Am Med Assoc. 2001;285:2743–2749. doi: 10.1001/jama.285.21.2743. [DOI] [PubMed] [Google Scholar]
- 10.Bodary PF, Shen Y, Vargas FB, Bi X, Ostenso KA, Gu S, Shayman JA, Eitzman DT. Circulation. 2005;111:629–632. doi: 10.1161/01.CIR.0000154550.15963.80. [DOI] [PubMed] [Google Scholar]
- 11.Ohshima T, Schiffmann R, Murray GJ, Kopp J, Quirk JM, Stahl S, Chan CC, Zerfas P, Tao-Cheng JH, Ward JM, Brady RO, Kulkarni AB. Proc Natl Acad Sci USA. 1999;96:6423–6427. doi: 10.1073/pnas.96.11.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen Y, Bodary PF, Vargas FB, Homeister JW, Gordon D, Osteno KA, Shayman JA, Eitzman DT. Stroke. 2006;37:1106–1108. doi: 10.1161/01.STR.0000206442.86238.39. [DOI] [PubMed] [Google Scholar]
- 13.Eitzman D, Bodary PF, Shen KY, Khairallah CG, Wild SR, Abe A, Shaffer-Hartman J, Shayman JA. J Am Soc Nephrol. 2003;14:298–302. doi: 10.1097/01.asn.0000043901.45141.d4. [DOI] [PubMed] [Google Scholar]
- 14.Roepman R, Bernoud-Hubac N, Schick DE, Maugeri A, Berger W, Ropers HH, Cremers FP, Ferreira PA. Hum Mol Genet. 2000;9:2095–2105. doi: 10.1093/hmg/9.14.2095. [DOI] [PubMed] [Google Scholar]
- 15.Castagnet P, Mavlyutov T, Cai Y, Zhong F, Ferreira P. Hum Mol Genet. 2003;12:1847–1863. doi: 10.1093/hmg/ddg202. [DOI] [PubMed] [Google Scholar]
- 16.Ferreira PA. Hum Mol Genet. 2005;14:R259–R267. doi: 10.1093/hmg/ddi272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu X, Guruju M, Oswald J, Ferreira PA. Hum Mol Genet. 2005;14:1327–1340. doi: 10.1093/hmg/ddi143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Browne MW. Br J Math Stat Psychol. 1984;37:62–83. doi: 10.1111/j.2044-8317.1984.tb00789.x. [DOI] [PubMed] [Google Scholar]
- 19.Faraci F, Didion SP. Arterioscler Throm Vasc Biol. 2004;24:1367–1373. doi: 10.1161/01.ATV.0000133604.20182.cf. [DOI] [PubMed] [Google Scholar]
- 20.Schafer J, Stimmer K. Bioinformatics. 2004;1:1–14. [Google Scholar]
- 21.Halpern J, Pearl J. Proceedings of 17th Conference on Unceratinty in Artificial Intelligence; San Francisco, CA: Morgan Kauffmann; 2001. pp. 194–202. [Google Scholar]
- 22.Ito Y. Oncogene. 2004;23:4196–4208. [Google Scholar]
- 23.Abdelhaleem M, Maltais L, Wain H. Genomics. 2003;81:618–622. doi: 10.1016/s0888-7543(03)00049-1. [DOI] [PubMed] [Google Scholar]
- 24.Winston J, Koepp DM, Zhu C, Elledge SJ, Harper JW. Curr Biol. 1999;9:1180–1182. doi: 10.1016/S0960-9822(00)80021-4. [DOI] [PubMed] [Google Scholar]
- 25.Moser H. In: The Molecular and Genetic Basis of Neurologic and Psychiatric Disease. Rosenberg R, Prusiner SB, DiMauro S, Barchi RL, Nestler EJ, editors. Boston: Butterworth–Heinemann; 2003. pp. 299–304. [Google Scholar]
- 26.Coughlin P. FEBS J. 2005;272:4852–4857. doi: 10.1111/j.1742-4658.2005.04881.x. [DOI] [PubMed] [Google Scholar]
- 27.Yepes M, Lawrence DA. Thromb Haemost. 2004;91:457–464. doi: 10.1160/TH03-12-0766. [DOI] [PubMed] [Google Scholar]
- 28.Moore DF, Krokhin OV, Beavis RC, Ries M, Robinson C, Goldin E, Brady RO, Wilkins JA, Schiffmann R. Proc Natl Acad Sci USA. 2007;104:2873–2878. doi: 10.1073/pnas.0611315104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li M, Chen D, Shiloh A, Luo J, Nikolaev A, Qin J, Gu W. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 30.Hershko A. Curr Opin Cell Biol. 1997;9:788–799. doi: 10.1016/s0955-0674(97)80079-8. [DOI] [PubMed] [Google Scholar]
- 31.Ohshima T, Murray GJ, Swaim WD, Longenecker G, Quirk JM, Cardarelli CO, Sugimoto Y, Pastan I, Gottesman MM, Brady RO, Kulkarni AB. Proc Natl Acad Sci USA. 1997;94:2540–2544. doi: 10.1073/pnas.94.6.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gelderman M, Oliver KL, Yazdani AT, Murray GJ, Miller GF, Cameron TI, Garger SJ, Turpen TH, Holtz RB, Brady RO. Preclinica. 2004;2:67–74. [Google Scholar]
- 33.Irizarry R, Gautier L, Cope LM. In: The Analysis of Gene Expression Data. Parmigiani G, Garreyy ES, Irizarry RA, Zeger SL, editors. New York: Springer; 2003. [Google Scholar]
- 34.Hochberg Y, Benjamini Y. Stat Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 35.Holm S. Scand J Stat. 1979;6:65–70. [Google Scholar]
- 36.Lu X, Ferreira PA. Invest Ophthalmol Vis Sci. 2005;46:1882–1890. doi: 10.1167/iovs.04-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kline R. Principles and Practice of Structural Equation Modeling. New York: Guilford; 2005. [Google Scholar]
- 38.Bollen KA. Structural Equations with Latent Variable. New York: Wiley Interscience; 1989. [Google Scholar]
- 39.Fox J. An R, S-Plus Companion to Applied Regression. Thousand Oaks, CA: Sage; 2002. [Google Scholar]
- 40.Loehlin J. Latent Variable Models. Mahwah, NJ: Lawrence Erlbaum Associates; 2004. [Google Scholar]
- 41.Team TRD. The R Reference Manual–Base Package Volume 1 and 2. Bristol, UK: Network Theory Limited; 2004. [Google Scholar]
- 42.Bentler P. EQS Structural Equation Program Manual. Encino, CA: Multivariate Software; 1995. [Google Scholar]
- 43.Bollen KA. Psychological Bulletin. 1990;107:256–259. [Google Scholar]
- 44.Raykov T, Tomer A, Nesselroade JR. Psychol Aging. 1991;6:499–503. doi: 10.1037//0882-7974.6.4.499. [DOI] [PubMed] [Google Scholar]
- 45.McDonald RP, Ho MH. Psychol Methods. 2002;7:64–82. doi: 10.1037/1082-989x.7.1.64. [DOI] [PubMed] [Google Scholar]