Abstract
Noncoding, or intergenic, transcription by RNA polymerase II (RNAPII) is remarkably widespread in eukaryotic organisms, but the effects of such transcription remain poorly understood. Here we show that noncoding transcription plays a role in activation, but not repression, of the Saccharomyces cerevisiae PHO5 gene. Histone eviction from the PHO5 promoter during activation occurs with normal kinetics even in the absence of the PHO5 TATA box, showing that transcription of the gene itself is not required for promoter remodeling. Nevertheless, we find that mutations that impair transcript elongation by RNAPII affect the kinetics of histone eviction from the PHO5 promoter. Most dramatically, inactivation of RNAPII itself abolishes eviction completely. Under repressing conditions, an ≈2.4-kb noncoding exosome-degraded transcript is detected that originates near the PHO5 termination site and is transcribed in the antisense direction. Abrogation of this transcript delays chromatin remodeling and subsequent RNAPII recruitment to PHO5 upon activation. We propose that noncoding transcription through positioned nucleosomes can enhance chromatin plasticity so that chromatin remodeling and activation of traversed genes occur in a timely manner.
Keywords: elongation, intergenic transcription, RNA polymerase II
In addition to transcribing all protein-encoding genes, RNA polymerase II (RNAPII) also transcribes a large group of less known and poorly understood untranslated RNAs. Recent genome-wide studies in several species reveal that such noncoding transcription is much more extensive than previously thought and that it occurs across intergenic regions, introns, and exons (see, for example, refs. 1 and 2). Recently, genome-wide studies in yeast have identified many cases of intergenic transcripts associated with promoters (3–5), raising the question of whether and how intergenic transcription across a promoter is used as a means of regulating that gene's transcription.
During our studies on elongation and RNA processing factors in yeast, we discovered an intergenic transcript across the PHO5 promoter. This finding led us to investigate whether noncoding transcription might play a role in regulating this gene. PHO5 encodes an acid phosphatase that is regulated by phosphate availability (6). In high phosphate, four positioned nucleosomes are associated with the PHO5 promoter region (7). During phosphate starvation, the Pho4 activator translocates to the nucleus (8) and binds to PHO5 upstream activation sequences (UASp1 and UASp2) along with the Pho2 activator (9–11). This leads to eviction of the four positioned nucleosomes, making a 600-bp region effectively fully accessible (7, 12–14). Promoter remodeling is facilitated by, although not always absolutely dependent on, several transcription factor complexes including SAGA, Swi/Snf complex, INO80, and the Asf1 chaperone (14–18). High phosphate causes Pho4 accumulation in the cytoplasm, nucleosome reassembly on the promoter, and transcriptional repression of the gene.
Here we show that intergenic transcription plays a role in the kinetics of PHO5 promoter remodeling.
Results
An Intergenic Transcript Across the PHO5 Promoter.
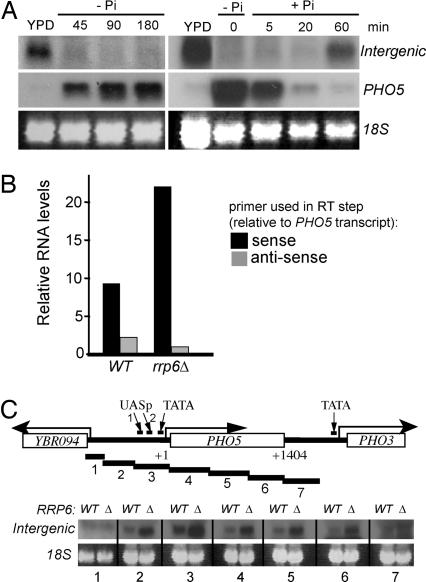
We initially noticed the appearance of an additional PHO5 RNA species during our characterization of transcription in rrp6 mutants (J.P.U., unpublished data), which lack a functional nuclear RNA exosome and accumulate intergenic transcripts (ref. 4 and references therein). However, detection of this transcript is possible even in wild-type cells in which noncoding RNAs are otherwise unstable. It is ≈2.4 kb in size and is observed only in cells grown in high-phosphate (repressing) conditions (Fig. 1A).
Fig. 1.
An intergenic transcript is detected across the repressed PHO5 promoter. (A) Northern blots of total RNA hybridized with PHO5 promoter-specific probe (intergenic) and an ORF probe (PHO5). (Left) PHO5 induction. (Right) PHO5 shutoff. 18S is shown as loading control. (B) Two-step reverse transcriptase, real-time PCR was performed on total RNA from wild-type (WT) or rrp6 cells grown in YPD using PHO5 promoter-specific primers (across UASp2; see C Upper). The initial reverse transcriptase step was performed with a primer that was either complementary to (antisense) or on the same strand as (sense) the PHO5 transcript. y axis indicates relative RNA levels (arbitrary units). The result shown is representative of two independent experiments. (C Upper) Schematic drawing of the PHO5 region (+1 indicates the first base of the ORF; +1,404 indicates the last) and the probes (numbered) used to detect the intergenic transcript. The positions of UASp1 and -2 and the TATA box are also indicated. (C Lower) Northern blots on wild-type and rrp6 RNA from cells grown in YPD using the probes indicated by numbers below. The intergenic transcript is detected with probes 2–6 (and stronger in rrp6).
Using strand- and promoter-specific RT-PCR on total RNA from wild-type and rrp6 cells (Fig. 1B), we deduced that intergenic transcription is antisense relative to PHO5 mRNA.
A series of probes across the PHO5 locus was hybridized with RNA isolated from wild type and rrp6, respectively. All probes that spanned the ≈2.4-kb region from 950 bp upstream of the PHO5 transcription start site to the 3′ end of the PHO5 ORF hybridized to the intergenic transcript, but the transcript was not detected with flanking probes 1 and 7 (Fig. 1C).
A strain carrying a temperature-sensitive allele of the largest subunit of RNAPII (rpb1-1) (19) was used to establish that the intergenic transcript is produced by RNAPII and that RNAPII can indeed be detected in the upstream PHO5 promoter by ChIP, even in repressing conditions [supporting information (SI) Fig. 6].
Together, the above results suggest that RNAPII actively transcribes across the uninduced PHO5 gene and its promoter, producing an unstable, noncoding, antisense RNA.
Intergenic Transcription Is Not Important for PHO5 Repression.
Because some previous studies of PHO5 chromatin structure and activation were conducted with versions of the PHO5 gene that did not contain the region in which the intergenic transcript originates (20, 21), an effect of intergenic transcription on the capability of the PHO5 gene to be turned on or off was not expected to be absolute. Indeed, it seemed reasonable to expect that the effect, if any, might be reminiscent of that observed upon mutation of the histone acetyltransferase GCN5, which affects the kinetics, but not the final level, of activated PHO5 transcription (15).
We first examined whether transcription in general is required to establish or maintain histones at the repressed PHO5 promoter. If not, a role for the intergenic transcript (representing only a fraction of general transcription) would be highly unlikely. For this purpose, wild-type and rpb1-1 cells were grown in high phosphate at 23°C and then shifted to 37°C. Comparison of histone H3 levels by ChIP assays revealed no significant difference in histone density at the promoter between wild type and rpb1-1 cells, indicating that ongoing transcription is not required to maintain histones at the promoter during repression (Fig. 2A). We also found that general RNAPII transcription is not required for establishment of repression (Fig. 2B). In this experiment, wild-type and rpb1-1 cells were grown at 23°C in phosphate-free medium and then shifted to 37°C for 30 min before adding phosphate. H3C ChIP analysis showed that, upon adding phosphate, histones were rapidly deposited onto the PHO5 promoter also in the transcription-defective rpb1-1 cells (Fig. 2B), indicating that transcription is not required for a normal rate of promoter closing either.
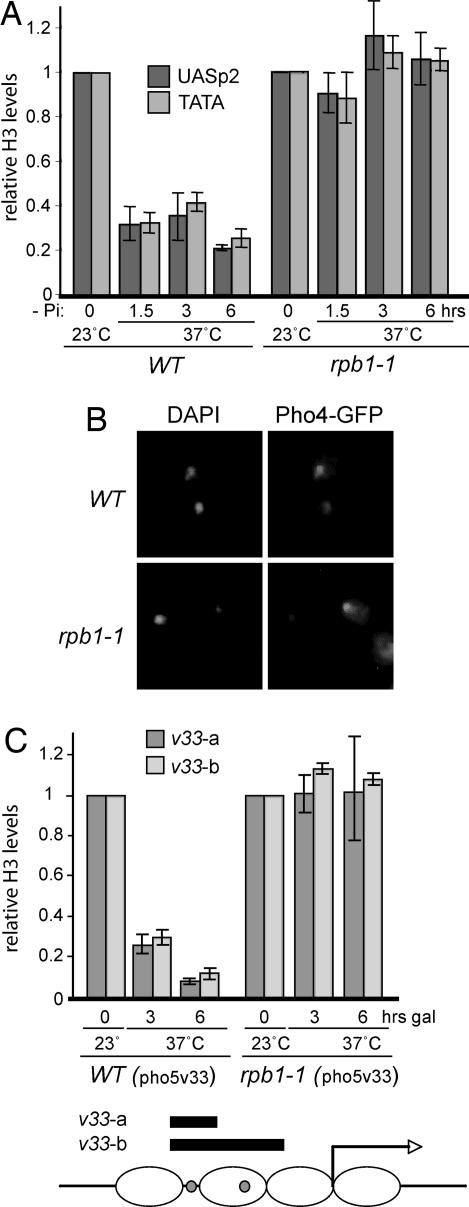
Fig. 2.
RNAPII activity is not required to maintain or establish repressed promoter chromatin. (A) Relative histone H3 occupancy (H3C ChIP) at the PHO5 promoter in WT and rpb1-1 cells grown in YPD at 23°C or 37°C, as indicated. y axis indicates relative histone H3 levels (arbitrary units). (B) Relative histone H3 density (H3C ChIP) at the PHO5 promoter in WT and rpb1-1 cells during the establishment of phosphate repression. Measurements from cells grown in high phosphate (YPD), in the absence of phosphate (−Pi, time = 0), and at different times after shifting to 37°C and into phosphate-containing medium (+Pi, different time points). Density in YPD was set to 1, and all other values are expressed relative to that.
Transcription Is Required for Normal Kinetics of Promoter Remodeling.
We next asked whether general transcription might somehow facilitate histone eviction from the PHO5 promoter during activation. Wild-type and rpb1-1 cells were grown at the permissive temperature and then shifted to 37°C for 30 min before phosphate starvation. By 6 h of induction, wild-type cells had ≈80% fewer histones at the promoter than in high phosphate (Fig. 3A). In sharp contrast, histones were not evicted at all upon phosphate starvation in the rpb1-1 strain, suggesting that transcription might indeed play a role in PHO5 promoter opening.
Fig. 3.
PHO5 promoter remodeling is abolished when RNAPII is inactivated. (A) Relative histone H3 density (H3C ChIP) in the PHO5 promoter in WT and rpb1-1 cells during phosphate starvation at 37°C. (B) WT and rpb1-1 cells expressing a Pho4-GFP fusion protein were grown in high phosphate and then transferred to medium lacking phosphate at 37°C. GFP and DAPI fluorescence are shown. (C) Relative histone H3 density (H3C ChIP) in the indicated regions of the galactose-regulated PHO5v33 promoter in WT and rpb1-1 cells during galactose induction at 37°C. In both A and C, density at time = 0 (23°C) was set to 1 and all other values are expressed relative to that. The schematic in C indicates location of PCR products used to measure histone H3 density in the PHO5v33 promoter. Small gray spheres indicate the position of Gal4 binding sites (replacing UASp1 and 2).
To check that the inability to remodel in rpb1-1 was not simply due to a defect in PHO signaling, we monitored Pho4 localization. Pho4 translocates to the nucleus upon successful signal transduction (8). Using GFP-tagged Pho4, we found that at 37°C the majority of Pho4 is in fact found in the nucleus by 1.5 h of phosphate starvation in both wild-type and rpb1-1 cells (Fig. 3B). Furthermore, the PHO induction cascade was bypassed altogether by using a PHO5 promoter derivative (PHO5v33), where the Pho4 binding sites are replaced by Gal4 binding sites (15). This promoter responds to the addition of galactose in a manner virtually identical to the response of the normal PHO5 promoter during phosphate starvation (ref. 15 and references therein). Also using this construct, histone eviction was negligible at the restrictive temperature in rpb1-1, whereas histone density was reduced to very low levels in wild type (Fig. 3C).
The TATA Box Is Not Required for Normal Kinetics of Promoter Remodeling.
Previous experiments by Hörz and coworkers (22) with promoter derivatives lacking a TATA box have argued against transcription of the PHO5 gene being an absolute requirement for its chromatin remodeling, but to address the possibility that it might affect the kinetics of chromatin remodeling we mutated the PHO5 TATA box at its native genomic site from TATATAA to CCTAGGA, asking whether this mutation affected histone eviction from the promoter during activation. RNAPII ChIP showed that, as expected, polymerase recruitment to the TATA-less promoter in response to activation was virtually abolished as a consequence of this mutation, but the kinetics of histone eviction during PHO5 induction was largely unaffected by this defect in polymerase recruitment (SI Fig. 7), confirming and extending the conclusion from previous studies of TATA-less PHO5 plasmid constructs (22): RNAPII recruitment to the PHO5 promoter is not required for normal histone eviction.
Defects in Transcriptional Elongation Affect the Kinetics of Promoter Remodeling at PHO5.
To further test the idea that intergenic transcription helps condition the PHO5 promoter for remodeling, we specifically targeted the elongating form of RNAPII using strains or conditions causing defects in this process, namely rpb2-10 (carrying an elongation-impairing mutation in the Rpb2 subunit) or dst1Δ (lacking the gene encoding TFIIS) strains and the elongation inhibitor 6-azauracil (6AU). Although, as expected, the results were much less dramatic than upon complete disruption of RNAPII transcription, the effect on PHO5 promoter histone eviction of these phenotypically innocuous mutations and 6AU was nevertheless significant and highly reproducible. H3C ChIP revealed that 6AU treatment led to a slower loss of histones (Fig. 4A). Significantly, RNAPII recruitment to the PHO5 TATA box was also both delayed and decreased under these elongation-prohibiting conditions, this effect being more dramatic in dst1Δ cells (Fig. 4B). Similar results were also obtained here by using the Gal-responsive PHO5 promoter derivative (PHO5v33) used in Fig. 3C (data not shown).
Fig. 4.
PHO5 promoter remodeling is delayed when RNAPII elongation is impaired. (A) Relative histone H3 density (H3C ChIP) in the indicated regions of the PHO5 promoter (UASp2 and TATA) in WT and dst1 cells during phosphate starvation, with or without 6AU treatment to inhibit RNAPII transcript elongation. (B) As in A, but RNAPII recruitment to the TATA box (4H8 ChIP). Density at time = 0 was set to 1, and other values are expressed relative to that.
Similarly, in the rpb2-10 strain (SI Fig. 8), an almost 2-fold-higher level of histones remained at the promoter (both at UASp2 and TATA) after 1.5 h of phosphate starvation compared with wild type. Only at later time points did the level of histone eviction reach wild-type levels, and there was an accompanying delay in transcriptional activation of PHO5, as revealed by delayed RNAPII recruitment to the PHO5 TATA box.
Together, these data further support the idea that efficient transcript elongation across the PHO5 gene under repressing conditions is required for its rapid activation.
Deletion of the 3′ End of the PHO5 ORF Affects Histone Eviction from the Promoter.
We finally sought to more specifically block intergenic transcription across the PHO5 promoter, to investigate whether it is relevant for PHO5 promoter remodeling. Because abrogation of RNAPII transcriptional initiation can be achieved only by removing the TATA box or initiator, we initially looked for potential TATA boxes near the end of the PHO5 ORF. No potential TATA boxes inside the PHO5 ORF were detected by sequence searching, but in the region immediately downstream from it, two fairly conserved cryptic sites were found. Three different approaches were then pursued to block intergenic transcription. First, we attempted to block usage of the potential TATA sites, either by inserting a URA3 marker right after the PHO5 STOP codon or by replacing the entire downstream region between PHO5 and PHO3 with a bidirectional terminator sequence (normally found between FBA1 and YKL061W), which has no discernable TATA sequences. Neither of these approaches halted intergenic transcription, nor did they affect PHO5 promoter activation (data not shown). This suggests that intergenic transcription is initiated by (non-TATA) sequence elements near the 3′ end of the PHO5 ORF itself. Second, we inserted the bidirectional terminator at a position 500 bp into the PHO5 ORF, hoping to thereby block intergenic transcription through the PHO5 promoter region. Interestingly, although this approach resulted in the disappearance of a stable, detectable intergenic transcript in wild-type cells, a slightly longer intergenic transcript could still be detected across the PHO5 promoter in rrp6 cells (SI Fig. 9), supporting the idea that RNAPII continued to transcribe through the PHO5 promoter region (SI Fig. 10). Accordingly, insertion of the terminator at this position also failed to affect PHO5 promoter activation (data not shown).
Given that the above approaches did not halt intergenic transcription, the region containing the intergenic transcription start site region was deleted by substitution with a URA3 marker (PHO5-3′Δ) orientated in the sense direction, in the hope that any potential transcription through the PHO5 promoter from the inserted marker gene itself, as well as from initiation region(s) downstream from the site of insertion, could be abrogated (the marker replaced PHO5 ORF sequence from +751 to +1,404; Fig. 5A Upper). The intergenic transcript was indeed absent from wild-type and, importantly, also from rrp6Δ cells carrying this modification (Fig. 5A Lower and data not shown), although RNAPII ChIP analysis suggested that transcription through the promoter had still not been reduced to background levels (SI Fig. 10), pointing to some “sporadic,” noncoding transcription still occurring across the locus.
Fig. 5.
Abolishing the intergenic transcript leads to slower PHO5 promoter remodeling and RNAPII recruitment to the PHO5 TATA box. (A Upper) Schematic of the PHO5 locus in WT and PHO5-3′Δ. URA3 replaces PHO5 +751–1,404 (to end of gene). (A Lower) Northern blot (using probe 3 from Fig. 1C) showing absence of intergenic transcription in the rrp6 version of the strain is shown. Note that only RRP6 cells were used in B and C and other experiments addressing the functional consequences of intergenic transcription. (B) Relative histone H3 occupancy (H3C ChIP) in the indicated regions of the PHO5 promoter in WT and PHO5-3′Δ cells during the course of phosphate starvation. (C) As in B, but RNAPII recruitment to the TATA box (4H8 ChIP). Density at time = 0 was set to 1, and other values are expressed relative to that. Histone density in the two strains at time = 0 was similar.
Nevertheless, this specific block clearly led to slower PHO5 promoter remodeling, with H3C ChIP revealing a significant delay in histone loss compared with wild-type cells (Fig. 5B). Thus, considering that four nucleosomes are being evicted from the PHO5 promoter during remodeling (13, 14), the wild-type cell population lost nucleosomes at a rate of ≈2.2 nucleosomes per hour, whereas cells lacking the intergenic transcript did so at a rate of ≈1.1 nucleosomes per hour. A corresponding delay in RNAPII recruitment to the PHO5 regulatory region upon activation was also observed in the absence of the intergenic transcript (Fig. 5C). Conversely, we obtained further evidence that the intergenic transcript does not play a role in maintaining a closed promoter and repressing PHO5 transcription under noninducing conditions (data not shown).
Taken together, these data support the idea that noncoding transcription through the PHO5 promoter affects the speed of histone remodeling during transcriptional activation, but not repression, of the gene.
Discussion
Here we identified an intergenic RNAPII-generated transcript that initiates in the region around the end of the PHO5 ORF and is transcribed in the antisense direction up to and across the PHO5 promoter. This transcript was also recently independently identified by whole-genome microarray studies (3–5). By employing various mutations (and the compound 6AU) affecting RNAPII transcript elongation and generation of the intergenic transcript, we uncovered evidence for the idea that the kinetics of activation, but not repression, of PHO5 is affected by intergenic transcription. Below we argue that transcription across the PHO5 promoter somehow contributes positively to chromatin plasticity, enabling rapid nucleosome disassembly upon activation.
Intergenic Transcription and Its Consequences.
Data published over the last few years indicate that intergenic transcription is much more widespread than had previously been expected. However, studies addressing the putative role(s) of such transcription are few (23–26). Previous data in yeast demonstrated a repressive role for intergenic transcription in the regulation of SER3, activated when serine is limiting (25). RNAPII generating this noncoding transcript (called SRG1) represses transcription by transcriptional interference, i.e., by inhibiting binding of activators to the SER3 UAS, and of TBP to its TATA box. The situation at PHO5 is fundamentally different from that at SER3/SRG1: in contrast to SRG1, the intergenic transcript across PHO5 is transcribed at low levels. Second, the SRG1 transcript initiates upstream of SER3, and on the same strand, whereas the PHO5 (antisense) intergenic transcript is initiated ≈1,400 bases downstream from the PHO5 TATA box. Transcription across the PHO5 promoter does not result in transcriptional interference, but instead seems to allow efficient histone eviction upon activation, promoting timely recruitment of RNAPII to the PHO5 TATA box and transcription of the gene.
Possible Mechanism Underlying the Effect of PHO5 Intergenic Transcription.
Intergenic transcription could in theory affect histone–DNA interactions at PHO5 in at least three different ways. First, the RNA transcript itself could facilitate more rapid activation, perhaps by acting as a histone acceptor/chaperone, as RNA has been shown to do in vitro (27). Second, physical movement of RNAPII through the promoter might increase the level of histone modification, such as acetylation and methylation, or increase insertion of the histone H2A variant Htz1. Finally, RNAPII movement through the region might cause temporary histone/nucleosome displacement, which could be required for or increase histone loss at the locus upon activation. Indeed, RNAPII-generated changes in chromatin integrity are well documented in a number of experimental systems (28–31).
If the presence of the RNA transcript itself were important, we reasoned that a higher level of the intergenic transcript near the PHO5 locus might facilitate promoter remodeling. The rrp6 stain allowed a test of these conditions because it has significantly higher levels of the intergenic transcript. In turn, nucleosomes might be evicted faster in rrp6 cells than in wild type. However, we found that histone eviction in rrp6 cells is similar to that of wild-type cells (data not shown). Likewise, expressing the intergenic transcript in trans from a plasmid failed to suppress the slower histone eviction and RNAPII TATA box recruitment observed in PHO5-3′Δ (data not shown). Finally, insertion of a terminator ≈500 bp into PHO5 did not affect the kinetics of PHO5 activation, although it resulted in a dramatic decrease of stable intergenic transcript. Together, these results argue against, but do not rule out, a positive role for the intergenic RNA transcript itself.
We also used the rpb1-1 and PHO5-3′Δ strains to test whether transcription across the PHO5 promoter affects the acetylation (H3-K9, H3-K27, H3-K18, H4-K5, and H4-K12) or methylation (H3-diMetK4, H3-diMetK36, and triMetK4) level of histones, or density of the histone variant Htz1, but ChIP assays failed to detect major changes in these histone characteristics (after normalizing to histone density) when transcription was inactivated (data not shown). Although these data in themselves do not rule out the possibility that RNAPII-mediated changes in histone modification play a role in PHO5 histone eviction, they do argue against this possibility. The PHO5-3′Δ mutation also does not result in a measurable change in the accessibility of promoter chromatin to restriction enzymes (data not shown), suggesting that, as expected, nucleosome positioning is not dramatically altered by intergenic transcription.
We also considered the possibility that the result we obtained with PHO5-3′Δ might be due to loss of putative promoter–terminator contacts (“gene looping”), rather than loss of intergenic transcription. However, slower PHO5 activation was not observed in strains where such looping would be disrupted because the PHO5 terminator was removed (but intergenic transcription still occurred) (data not shown), arguing against a role for gene looping.
A Model to Explain the Effect of Intergenic and Sporadic Transcription.
Considering the results described above, as well as previous data from others on the disruptive effect of RNAPII transcription through nucleosomes, we suggest that the actual movement of RNAPII through the PHO5 promoter affects nucleosome eviction by somehow increasing the local rate of nucleosome exchange/turnover. In support of this idea, more generally inhibiting or abrogating transcription (6AU and in particular rpb1-1) within and outside the PHO5 locus reduced nucleosome eviction to a much greater extent than specific abrogation of the intergenic transcript (PHO5-3′Δ). Indeed, it is possible that noncoding, sporadic transcription, i.e., random transcription originating from multiple spurious initiation sites and terminating at random, is as widespread as the intergenic transcription that gives rise to detectable transcripts. The concept of widespread, sporadic transcription may well be of substantial general importance in all eukaryotes, but it might play a particularly important role in maintaining chromatin plasticity in an active genome such as that of Saccharomyces cerevisiae.
The Kinetics of Gene Regulation.
It is obvious that the speed at which a cell responds to a stimulus by activating a set of genes and repressing another is of pivotal importance to the fate of that cell. However, this aspect of gene regulation is often not appreciated. Instead, the amplitude of regulation or the absolute levels of expression are generally seen as the hallmarks of a regulated gene. Here we have shown that the rate of activation of the PHO5 promoter is affected by intergenic transcription across it, whereas the final level of induction is not. Given that intergenic transcripts are often found to be expressed in a tissue- or time-dependent manner, the effect of noncoding transcription described here may be a general one, and one important consequence of it could be to increase the rate of chromatin remodeling and thereby the rate of gene induction, rather than to affect the final steady-state levels of expression. Experiments in higher cells suggest that intergenic transcription is generally extremely widespread. Our findings thus have important implications for the approaches that should be taken to study the effect of noncoding transcription also in metazoans.
Materials and Methods
Yeast Strains and Growth Conditions.
Strains used are listed in SI Text. For PHO5 activation, yeast strains were grown in yeast extract/peptone/dextrose (YPD; high-phosphate conditions) to mid-log phase, and then shifted to phosphate-free minimal media in a final concentration of typically 0.5 × 107 cells per milliliter. KH2PO4 was added to 10 mM final concentration to cultures in PHO5 shutoff experiments. For galactose induction (PHO5v33), strains were grown in yeast extract/peptone medium containing 2% raffinose. Galactose was added to 2% final concentration, and cells were diluted to 0.5 × 107 cells per milliliter. rpb1-1 and its wild-type counterpart were grown at 23°C as the permissive temperature and at 37°C as the restrictive temperature. Temperature shifts were performed 30 min before phosphate starvation, phosphate addition, or galactose induction. 6AU was added to a final concentration of 50 μg/ml, with cells made URA3+ by transformation with a CEN plasmid, as required.
ChIP Assays and Real-Time PCR Analysis.
ChIP assays were performed essentially as previously described (32). 4H8 (anti-Rpb1 antibody) was from Upstate Biotechnology (Lake Placid, NY), and the H3C antibody was a gift from Alain Verreault (Institute for Research in Immunology and Cancer, Montreal, QC, Canada). Coprecipitated DNA was analyzed in triplicate by quantitative PCR using the ABI Prism 7000 Sequence detection system (Applied Biosystems, Foster City, CA). Primer sequences are available upon request. Values were normalized to inputs. All values expressed in bar graphs represent means ± SEM of at least two independent experiments and three independent ChIP assays.
Northern Blots.
Total RNA was extracted by using the hot acid phenol method. RNA was separated on 1% formaldehyde agarose gels and blotted onto Nylon membranes. Blots were hybridized with PCR probes (primer sequences available upon request) labeled by random primer labeling. Details are available upon request.
RT-PCR Analysis.
Total RNA was extracted using RNeasy mini kits (Qiagen, Valencia, CA) followed by DNase treatment with the Turbo DNA-free kit (Ambion, Austin, TX). For asymmetric RT-PCR, 1 μg of RNA was incubated with one primer only, either sense or antisense, in a reaction using Multiscribe reverse transcriptase. Next, the DNA product was amplified by standard real-time PCRs using both sense and antisense primers and Absolute QPCR SYBR green reagents (Abgene, Rochester, NY). Values were normalized to those obtained in the FBA1 gene (primer sequences available upon request).
Microscopy.
Cells were prepared and visualized as previously described (15).
Acknowledgments
We thank Alain Verreault (Institute for Research in Immunology and Cancer), Rick Young (Whitehead Institute for Biomedical Research, Cambridge, MA), Kevin Struhl (Harvard Medical School, Boston, MA), Anita Corbett (Emory University, Atlanta, GA), Erin O'Shea (University of California, San Francisco, CA) and Wolfram Hörz (Universität München, Munich, Germany) for strains or plasmids; Arnold Kristjuhan and Jonathan Baxter for technical advice; and Philip Korber, Peter Verrijzer, Kristian Kvint, and Arnold Kristjuhan for discussions and comments on the manuscript. This work was supported by a grant from Cancer Research UK (to J.Q.S.).
Abbreviations
- RNAPII
RNA polymerase II
- 6AU
6-azauracil
- YPD
yeast extract/peptone/dextrose.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702431104/DC1.
References
- 1.Tupy JL, Bailey AM, Dailey G, Evans-Holm M, Siebel CW, Misra S, Celniker SE, Rubin GM. Proc Natl Acad Sci USA. 2005;102:5495–5500. doi: 10.1073/pnas.0501422102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, Gerstein M, Snyder M. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 3.Samanta MP, Tongprasit W, Sethi H, Chin CS, Stolc V. Proc Natl Acad Sci USA. 2006;103:4192–4197. doi: 10.1073/pnas.0507669103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis CA, Ares M., Jr Proc Natl Acad Sci USA. 2006;103:3262–3267. doi: 10.1073/pnas.0507783103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM. Proc Natl Acad Sci USA. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kramer RA, Andersen N. Proc Natl Acad Sci USA. 1980;77:6541–6545. doi: 10.1073/pnas.77.11.6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Almer A, Rudolph H, Hinnen A, Hörz W. EMBO J. 1986;5:2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Neill EM, Kaffman A, Jolly ER, O'Shea EK. Science. 1996;271:209–212. doi: 10.1126/science.271.5246.209. [DOI] [PubMed] [Google Scholar]
- 9.Barbaric S, Munsterkotter M, Goding C, Hörz W. Mol Cell Biol. 1998;18:2629–2639. doi: 10.1128/mcb.18.5.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fascher KD, Schmitz J, Hörz W. EMBO J. 1990;9:2523–2528. doi: 10.1002/j.1460-2075.1990.tb07432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komeili A, O'Shea EK. Science. 1999;284:977–980. doi: 10.1126/science.284.5416.977. [DOI] [PubMed] [Google Scholar]
- 12.Bergman LW, Kramer RA. J Biol Chem. 1983;258:7223–7227. [PubMed] [Google Scholar]
- 13.Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Mol Cell. 2003;11:1587–1598. doi: 10.1016/s1097-2765(03)00231-4. [DOI] [PubMed] [Google Scholar]
- 14.Reinke H, Hörz W. Mol Cell. 2003;11:1599–1607. doi: 10.1016/s1097-2765(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 15.Barbaric S, Walker J, Schmid A, Svejstrup JQ, Hörz W. EMBO J. 2001;20:4944–4951. doi: 10.1093/emboj/20.17.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steger DJ, Haswell ES, Miller AL, Wente SR, O'Shea EK. Science. 2003;299:114–116. doi: 10.1126/science.1078062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adkins MW, Howar SR, Tyler JK. Mol Cell. 2004;14:657–666. doi: 10.1016/j.molcel.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 18.Korber P, Barbaric S, Luckenbach T, Schmid A, Schermer UJ, Blaschke D, Hörz W. J Biol Chem. 2006;281:5539–5545. doi: 10.1074/jbc.M513340200. [DOI] [PubMed] [Google Scholar]
- 19.Nonet M, Scafe C, Sexton J, Young R. Mol Cell Biol. 1987;7:1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Straka C, Hörz W. EMBO J. 1991;10:361–368. doi: 10.1002/j.1460-2075.1991.tb07957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Mol Cell. 2004;14:667–673. doi: 10.1016/j.molcel.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 22.Fascher KD, Schmitz J, Hörz W. J Mol Biol. 1993;231:658–667. doi: 10.1006/jmbi.1993.1317. [DOI] [PubMed] [Google Scholar]
- 23.Ashe HL, Monks J, Wijgerde M, Fraser P, Proudfoot NJ. Genes Dev. 1997;11:2494–2509. doi: 10.1101/gad.11.19.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gribnau J, Diderich K, Pruzina S, Calzolari R, Fraser P. Mol Cell. 2000;5:377–386. doi: 10.1016/s1097-2765(00)80432-3. [DOI] [PubMed] [Google Scholar]
- 25.Martens JA, Laprade L, Winston F. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- 26.Schmitt S, Prestel M, Paro R. Genes Dev. 2005;19:697–708. doi: 10.1101/gad.326205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson T, Wiegand R, Brutlag D. Biochemistry. 1981;20:2594–2601. doi: 10.1021/bi00512a035. [DOI] [PubMed] [Google Scholar]
- 28.Kaplan CD, Laprade L, Winston F. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- 29.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 30.Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, Studitsky VM. Mol Cell. 2002;9:541–552. doi: 10.1016/s1097-2765(02)00472-0. [DOI] [PubMed] [Google Scholar]
- 31.Svejstrup JQ. Curr Opin Genet Dev. 2002;12:156–161. doi: 10.1016/s0959-437x(02)00281-2. [DOI] [PubMed] [Google Scholar]
- 32.Kristjuhan A, Walker J, Suka N, Grunstein M, Roberts D, Cairns BR, Svejstrup JQ. Mol Cell. 2002;10:925–933. doi: 10.1016/s1097-2765(02)00647-0. [DOI] [PMC free article] [PubMed] [Google Scholar]