Abstract
The RNA polymerase subunit RPB10 displays a high level of conservation across archaea and eukarya and is required for cell viability in yeast. Structure determination of this RNA polymerase subunit from Methanobacterium thermoautotrophicum reveals a topology, which we term a zinc-bundle, consisting of three α-helices stabilized by a zinc ion. The metal ion is bound within an atypical CX2CXnCC sequence motif and serves to bridge an N-terminal loop with helix 3. This represents an example of two adjacent zinc-binding Cys residues within an α-helix conformation. Conserved surface features of RPB10 include discrete regions of neutral, acidic, and basic residues, the latter being located around the zinc-binding site. One or more of these regions may contribute to the role of this subunit as a scaffold protein within the polymerase holoenzyme.
Understanding the mechanism and regulation of transcription is a long-standing challenge in the field of molecular biology. The central players in this process, multisubunit RNA polymerases (RNAPs), are conserved throughout the eubacterial, archaeal, and eukaryal kingdoms. In detail, archaeal RNAP most closely resembles eukaryal RNA polymerase II (RNAPII), the enzyme responsible for transcribing all protein-encoding genes in higher organisms. The similarity in subunit composition between archaeal RNAP and RNAPII extends to function, as both polymerases use the basal transcription factors TBP/TFB and TFIIB and recognize similar promoter sequences within histone-mediated DNA packaging (1). Given the availability of several archaeal genome sequences, the enhanced stability of many archaeal proteins compared with their eukaryal counterparts, and the presence of only one polymerase complex, archaea has emerged as an excellent model system for structural studies of the transcriptional machinery in all nonprokaryotic organisms (2, 3).
The prototypical RNAPII is derived from Saccharomyces cerevisiae, with composite subunits named RPB1 to RPB12 in accordance with their order of increasing electrophoretic mobility (Table 1) (4, 5). Components of the archaeal RNAP are homologous to these subunits, and a parallel nomenclature from subunit A to P has developed to complement the numbering system used for RNAPII (3, 6). Four of the larger subunits in both eukarya and archaea (RPB1, RPB2, RPB3, and RPB11) have sequence and functional homology to the core α2ββ′ structure of eubacterial RNAP and thus likely represent the catalytic site and central scaffold of RNAPII. Several of the smaller subunits without eubacterial homologs (RPB5, RPB6, RPB8, RPB10, and RPB12) also show a high level of conservation among archaea and eukarya, are common to all three yeast RNAPs, and are essential for cell viability (7–9). Although the three-dimensional structures of RPB5, RPB6, and RPB8 have been reported (PDB ID codes 1HMJ/1EIK/1DZF, 1QKL, and 1A1D), the exact roles played by each of these subunits within RNAPII remain to be defined.
Table 1.
Relationship between homologous RNAP subunits
| Eukarya (Saccharomyces) | Archaea (Methanobacterium) | Eubacteria (E. coli) | ||
|---|---|---|---|---|
| RNAPI* | RNAPII | RNAPIII† | RNAP | RNAP |
| RPA190 | RPB1 | RPC160 | A′, A" | β′ |
| RPA135 | RPB2 | RPC128 | B′, B" | β |
| RPAC40 | RPB3 | RPAC40 | D | α‡ |
| — | RPB4 | — | F | — |
| RPB5 | RPB5 | RPB5 | H | — |
| RPB6 | RPB6 | RPB6 | K | — |
| — | RPB7 | — | E′, E" | — |
| RPB8 | RPB8 | RPB8 | — | — |
| — | RPB9 | — | — | — |
| RPB10 | RPB10 | RPB10 | N | — |
| RPAC19 | RPB11 | RPAC19 | L | α‡ |
| RPB12 | RPB12 | RPB12 | P | — |
RNAPI-specific subunits also include RPA49, RPA43, RPA34, RPA14, and RPA12.
RNAPIII-specific subunits also include RPC82, RPC53, RPC37, RPC34, RPC31, RPC25, and RPC11.
RPB3 and RPB11 are present in single copy within yeast RNAPII and appear to be homologous to the α2 homodimer in E. coli.
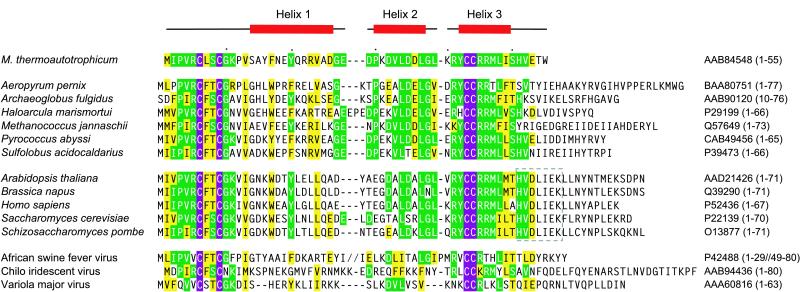
The RPB10 subunit (also referred to as ABC10β, hRPB7.6, and subunit N) is a zinc-binding protein (8) with an atypical CX2CXnCC metal binding motif (Fig. 1). Mutation of any of these four completely conserved Cys residues results in a lethal phenotype in yeast (10). However, apart from the observation that RPB10 is absolutely required for cell growth and is shared by all three yeast RNAPs, little is known about the role of this subunit in transcription. Its small size (55–80 amino acids) reasonably precludes a catalytic role. Therefore, it is likely that RPB10 functions in a structural capacity, either within the polymerase complex or with transcriptionally associated proteins and nucleic acids. Indeed, several studies have mapped part of the interaction network from RPB10 to other RNAP subunits (11–15), and mutational analysis has led to the identification of RNAPI-specific assembly defects in yeast (10). To gain better insight into the functional role of RPB10, we have used NMR spectroscopy to characterize structurally this protein isolated from Methanobacterium thermoautotrophicum (16).
Figure 1.
Alignment of RPB10 sequences from archaeal (top), eukaryal (middle), and viral (bottom) genomes. The four absolutely conserved Cys are highlighted in purple whereas green and yellow indicate residues that are identical or similar, respectively, between mtRPB10 and >40% of the homologous sequences shown. The NMR-derived secondary structure of mtRPB10 is illustrated above the alignment. GenBank accession numbers are listed on the right of each sequence, with the aligned residues indicated in parentheses. An HVDLIEK motif, conserved in eukarya, is boxed in light gray. The figure was made by using pileup (Genetics Computer Group, Madison, WI) and by visual inspection.
Materials and Methods
Protein Expression and Purification.
The DNA encoding the full-length MTH40 (mtRPB10) ORF from M. thermoautotrophicum (16) was cloned into pET15b (Novagen) as a fusion protein with an N-terminal His6 sequence and thrombin-cleavage site. Overexpression was carried out in an Escherichia coli BL21(λDE3) strain, harboring a plasmid coding for three rare tRNAs (agg, aga, and ata) and grown in Luria broth (LB) or in minimal M9 media supplemented appropriately with 15N-ammonium chloride (1 g⋅liter−1) or 13C6-glucose (3 g⋅liter−1). Purification of the protein from the cell pellet used Ni2+-affinity chromatography and a buffer of 50 mM Hepes (pH 7.5), 150 mM NaCl, and 5% (vol/vol) glycerol with 10, 30, and 500 mM imidazole for loading, washing, and elution, respectively. After dialysis in NMR buffer consisting of 20 mM Tris (pH 7.5), 150 mM NaCl, and 2 mM β-mercaptoethanol, the His6-tag was removed by incubation with thrombin followed by a second passage over the Ni2+-column. Electrospray ionization mass spectroscopy yielded a mass of 6,716 Da for the unlabeled protein, which is consistent with the expected value for the full length mtRPB10 with three residues (Gly-Ser-His) remaining at its N terminus after proteolytic cleavage. Circular dichroism spectrometry was measured by using a Jasco (Easton, MD) J-720 spectropolarimeter.
NMR Spectroscopy.
Protein samples typically contained 0.5 mM mtRPB10 in NMR buffer with 10 or 99% D2O added for the lock. Deuterated d9-Tris was used for homonuclear experiments. All spectra were recorded at 30°C by using a 500 MHz Varian Unity NMR spectrometer equipped with a triple resonance probe and a pulsed field gradient accessory, and were analyzed by using felix 95.0 (Biosym Technologies, San Diego). Assignments were obtained as described (17) from 13C- and 15N-HSQC, HNCACB, CBCACONH, HNCO, H(CCO)NH, C(CO)NH, HCCH-TOCSY, HNHB, 15N-TOCSY-HSQC, and long-range C′-Cγ and N-Cγ spin echo spectra. 15N T1, T2 and heteronuclear NOE experiments were carried out and analyzed as described (18). Under the experimental conditions, the three N-terminal residues Gly-Ser-His, as well as Ala15, Lys40, and Tyr42, display rapid hydrogen exchange and thus do not have detectable 1HN resonances.
Structure Calculations.
Distance restraints were acquired from three-dimensional (3D) 15N-HSQC-NOESY, 3D simultaneous 13C- and 15N-HSQC-NOESY, 2D 1H-NOESY, and constant time methyl-methyl NOESY spectra (17, 19), all with 150-ms mixing times. φ angles were constrained to −57° ± 30° for backbone amides with 3JHNHα < 6 Hz from 3D HNHA and 2D CT-HMQC-J experiments. ψ angles were constrained to −47° ± 30° for those amides in α-helical conformation, as judged from appropriate φ angles and CSI analysis. Two restraints per hydrogen bond were included for amides showing protection from hydrogen-deuterium exchange as measured by 1H-15N-HSQC spectroscopy 30 min after transfer in D2O buffer. Zinc coordination included six Sγ-Sγ restraints (3.6 ± 0.4 Å) and four Zn-Sγ restraints (2.35 ± 1 Å). One hundred and fifty unambiguous NOE and sixty-seven dihedral angle restraints were used to determine an initial family of structures by using distance geometry and simulated annealing with xplor 3.851 (20). Low energy members of this family were then used as input for nine iterations of the aria protocol (21), producing a total of 690 unambiguous and 376 ambiguous restraints. Hydrogen bond and zinc coordination restraints were included in iteration five. A final set of 60 structures was calculated by using the aria-generated restraints, yielding 20 lowest energy structures, each of which had no NOE or dihedral restraint violation greater than 0.5 Å or 5°, respectively. The N-terminal Gly-Ser-His is disordered in solution and is not included in structural models. procheck_nmr (22) shows that 91.7% of the nonglycine and nonproline residues fall within the most favored or additionally allowed regions of the Ramachandran plot, with bad contacts limited to termini and the unassigned amides between helix 2 and 3. All structural figures were created by using molscript (23) and raster3d (24).
Results and Discussion
RPB10 Contains a Three α-Helix Bundle.
Recombinant M. thermoautotrophicum RPB10 (mtRPB10) was expressed in E. coli and was purified to homogeneity. The isolated protein adopts a stable, folded structure under NMR conditions with a midpoint denaturation temperature of ≈85°C as monitored by circular dichroism spectroscopy. mtRPB10 is a monomeric protein under the experimental parameters used in this study, as evident by a correlation time of 4.6 ns for global tumbling measured by 15N relaxation analysis (data not shown).
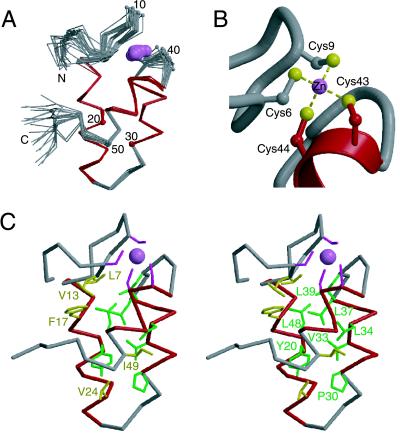
The ensemble of structures calculated for mtRPB10 is presented in Fig. 2, with statistical parameters summarized in Table 2. mtRPB10 is composed of three well defined α-helices (residues 14–26, 30–37, and 42–48), identified clearly by using a variety of NMR-derived parameters, including 3JHN-Hα coupling constants, sequential and short range NOE interactions, and secondary chemical shifts (not shown). These three amphipathic helices form a bundle around a common hydrophobic core comprised of residues with significant conservation across both archaea and eukarya (Fig. 2C). This sequence preservation strongly suggests that these homologous proteins all adopt a tertiary structure similar to that defined for mtRPB10. Some differences are noted with the viral proteins, particularly in the pattern of hydrophobic residues in the region corresponding to helix 1, which may indicate a degree of structural divergence.
Figure 2.
NMR-derived zinc-bundle structure of mtRPB10. (A) The α-trace of 20 structures of mtRBP10, superimposed by using the main chain helical atoms (14–26, 30–37, and 42–48). The helices are highlighted in red, and the zinc ion is shown as a pink ball. Every tenth residue is numbered. The N- and C-terminal residues Met1, Ile2, Glu53, Thr54, and Trp55 are conformationally flexible in solution, as evident by heteronuclear 1H{15N} NOE values less than 0.5. (B) Zinc binding site in mtRPB10. (C) Stereo view of a representative mtRPB10 structure displaying residues within the hydrophobic core: Leu7 and Val13 in the N-terminal region, Phe17, Tyr20, and Val24 of helix 1, Pro30, Val33, Leu34, and Leu37 of helix 2, Leu39 in the loop between helix 2 and 3, Leu48, Ile49 of helix 3, and Val52 in the C-terminal region. These conserved residues are shown according to the color scheme of Fig. 1.
Table 2.
NMR restraints and statistics for the ensemble of 20 structures calculated for mtRPB10
| Summary of restraints | |
| NOEs* | |
| Intraresidue | 487 (374) |
| Sequential | 211 (146) |
| Medium-range (1 < |i − j| < 5) | 168 (81) |
| Long-range (|i − j| ≥ 5) | 200 (89) |
| All | 1,066 (690) |
| Dihedral angles | |
| φ | 26 |
| ψ | 26 |
| χ1 | 15 |
| Hydrogen bonds | 11 × 2 |
| Zinc coordination | 10 |
| Deviation from restraints | |
| NOE restraints, Å | 0.019 ± 0.003 |
| Dihedral restraints, ° | 0.3 ± 0.2 |
| Deviation from idealized geometry | |
| Van der Waals, Å | 11 ± 3 |
| Bonds, Å | 0.0020 ± 0.0001 |
| Angles, ° | 0.38 ± 0.02 |
| Improper angles, ° | 13 ± 2 |
| Mean energies, kcal⋅mol−1 | |
| Evdw | 11 ± 3 |
| Ebonds | 3.9 ± 0.7 |
| Eangle | 37 ± 4 |
| ENOE | 16 ± 4 |
| Edih | 0.6 ± 0.6 |
| ELennard-Jones† | −141 ± 10 |
| Eelec† | −191 ± 9 |
| rmsd from average structure, Å |
| All
residues
|
Helical regions
|
|
|---|---|---|
| All heavy atoms | 1.33 | 0.71 |
| Backbone (C′, Cα, N) | 0.93 | 0.19 |
Number of aria total restraints (unambiguous and ambiguous), with number of unambiguous restraints in parenthesis.
Calculated by using the CHARMM energy function.
Zinc Binding Stabilizes the Tertiary Fold of mtRPB10.
A predominant feature of the mtRPB10 structure is the presence of a zinc ion chelated by four Cys residues. Examination of the M. thermoautotrophicum sequence reveals that these residues form a noncanonical zinc-binding motif CX2CXnCC that is absolutely conserved in every known archaeal and eukaryal RPB10, including three RPB10-like proteins found in viral genomes (Fig. 1). Mutation of any of these Cys residues to Ser or Thr leads to a lethal phenotype in yeast (10), providing further proof of their functional significance. Previous studies using radioactive zinc blot assays demonstrated that yeast RPB10 binds this metal ion in vitro (8), and the presence of zinc in our recombinant mtRPB10 was confirmed by using graphite furnace atomic absorption spectroscopy (data not shown). Initial structural calculations without inclusion of this metal ion revealed that NOE interactions unambiguously clustered Cys6, Cys9, Cys43, and Cys44 in close proximity and with a constant geometric arrangement. Thus, a zinc ion, tetrahedrally coordinated to these Cys residues, was included in the determination of the final ensemble of mtRPB10 structures. As evident in Fig. 2A, the zinc atom bridges residues at the N terminus of mtRPB10 with those near helix 3 and serves to stabilize the overall structure of this protein. In support of this conclusion, removal of the zinc by EDTA leads to a loss in circular dichroism intensity (222 nm), indicative of the unfolding of the metal-free protein (data not shown).
A close-up view of the mtRPB10 zinc binding site is illustrated in Fig. 2B. Tetrahedral coordination of zinc ions by four thiolates is commonly seen with protein modules involved in protein-protein (e.g., LIM domains and RING fingers) and protein-DNA interactions (e.g., steroid receptors). However, such metal binding involving two sequential Cys residues is relatively rare, with examples in the Protein Data Bank provided only by the potassium channel protein Shaw (PDB ID code 3KVT) and by Bruton's tyrosine kinase (PDB ID codes 1BTK and 1BWN). Distinct from these latter cases, mtRBP10 utilizes adjacent Cys that are both located within an α-helix. Although this appears to be the first structural description of such an atypical zinc-binding motif, the φ, ψ, and χ1 dihedral angles of (−51 ± 10, −29 ± 7, 58 ± 7 °) and (−87 ± 1, −55 ± 4, −33 ± 4 °) measured for Cys43 and Cys44, respectively, reveal that these amino acids are readily accommodated near the N terminus of helix 3 in mtRPB10. Note that Cys43 is the second residue of this helix and thus does not exhibit (i, i − 3) steric clashes that would otherwise disfavor its gauche− χ1 conformation within this element of secondary structure (25). In light of the integral role played by zinc in stabilizing the RPB10 three-helix bundle, we suggest that this fold be called a “zinc-bundle.” This name is in keeping with terms such as “zinc-finger” and “zinc-ribbon” used previously to classify families of small zinc-binding proteins.
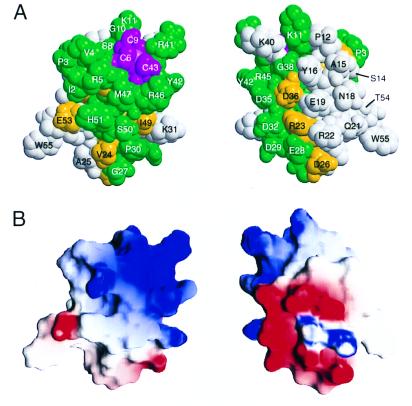
When the results of the sequence alignment in Fig. 1 are mapped onto the surface of mtRPB10 (Fig. 3A), a striking cluster of conserved exposed side chains are observed near the zinc binding site. Clearly, such conservation would be expected to maintain the chemical environment necessary for metal binding. Furthermore, there is a marked retention of basic residues in these two regions. Along with the macrodipole of helix 3, this clustering of positive charges may aid in lowering the pKa values of the four Cys to favor their thiolate form and, equivalently, to stabilize the net negatively charged sulfur-metal cluster that results on zinc binding (26). Similar basic regions are observed near other Cys4 zinc-binding sites, including those of the DNA-binding domains of GATA-1 and the steroid receptors, as well as LIM domains, alcohol dehydrogenase, and adenylate kinase.
Figure 3.
Conserved residues and charged regions on the surface of mtRPB10. (A) Space filling model of mtRPB10 with conserved surface residues highlighted according to the color scheme of Fig. 1. Note the pronounced clustering of such residues around the zinc-binding site. (B) Surface charge distribution calculated and drawn by using grasp (34), with red indicating regions of negative potential and blue of positive potential. The basic region, formed by Arg5, Lys11, Lys31, Lys40, Arg41, Arg45, and Arg46, is centered near the N terminus and along helix 3. The acidic region is located predominantly in helix 2 and the preceding loop because of Glu19, Asp26, Glu28, Asp29, Asp32, Asp35, and Asp36. The extended neutral region includes polar and hydrophobic residues near the N terminus (Met1, Ile2, Pro3, Val4, Pro12), along helix 1 (Ser14, Ala15, Tyr16, Asn18, Tyr20, Gln21), flanking the loop between helices 1 and 2 (Val24, Ala25, Pro30), and within or immediately after helix 3 (Tyr42, Met47, Ser50, His51). In both A and B, the two views differ by a rotation of 180° around the vertical axis.
Surface Features and Protein-Protein Interactions.
As indicated by chemical cross-linking, glutathione S-transferase fusion pulldown, far-Western, two-hybrid, and extragenic suppression studies (11–15), RPB10 participates in protein-protein interactions with additional components of the polymerase holoenzyme. Experimentally, the binding of RPB10 to an RPB3-RPB11 (or RPAC40-RPAC19 in RNAPI/III) heterodimer is well characterized and persists in both eukaryal and archaeal RNAPs (11, 13, 27). Binding of RPB10 to this α2-like heterodimer suggests an early role in holoenzyme assembly because the formation of the α2-complex is the first step in the assembly of the prokaryotic RNAPs (28). Additional contact to the β homolog RPB2 (12, 14, 15) further implicates RPB10 in a critical scaffold role within the archaeal and eukaryal polymerases.
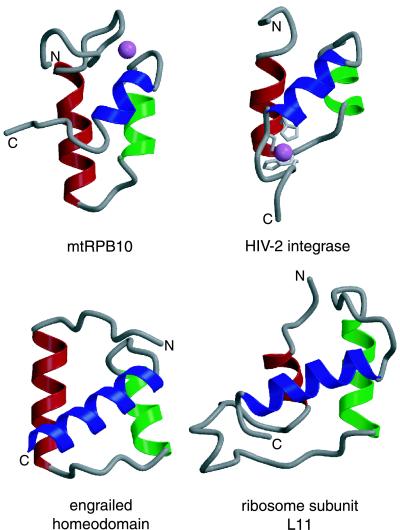
Insights into the possible nature of these scaffolding interactions may be gained through two avenues. First, inspection of the surface features of this RNAP subunit demonstrates that mtRPB10 has a distinctly dipolar charge distribution (Fig. 3B). Complementing the aforementioned patch of basic residues, there are also well defined acidic and neutral regions present on the surface of the protein. The acidic patch is centered along helix 2 and the loop after helix 1, with sequence conservation of Asp and Glu residues in this region (Fig. 1). The extended yet somewhat less conserved neutral area is comprised of polar residues located within or flanking helices 1 and 3, along with a strip of hydrophobic residues, primarily from the N-terminal region, that lie adjacent to the zinc-binding site. Second, structural comparisons reveal that RPB10 has an architecture similar to the N-terminal zinc-binding domains of HIV-1 and -2 integrase (IN1–55) (29, 30). Strikingly, both of these domains also use zinc to stabilize the same general three-helical zinc-bundle fold, but with the metal ion-binding motif HX3HXnCX2C located at the C terminus of helix 1 and the C-terminal region of the peptide (Fig. 4). In the case of HIV-1 IN1–55, helix 3 and the N-terminal end of helix 1 are involved in homodimerization (29). By analogy, it is reasonable that the corresponding neutral surface of RPB10 (Fig. 3) may also serve as a protein-binding interface. In support of this hypothesis, a preliminary inspection of the structure of the yeast RNAPII holoenzyme reveals that this neutral region contacts directly the RPB2 and RPB3 subunits whereas the acidic patch remains solvent exposed (R. Kornberg, personal communication).
Figure 4.
Structural similarity to protein and nucleic acid binding domains. mtRPB10 is shown with helices 1, 2, and 3 colored red, green, and blue, respectively. The zinc atom is shown as a pink ball. This color scheme is used to identify similar helices in the following structurally related proteins found in the Protein Data Bank: the protein-binding N-terminal domain of HIV-2 integrase (PDB ID code 1AUB), with the zinc-chelating Cys and His residues drawn in light gray, and the zinc atom shown as a pink ball; the DNA-binding homeodomain of engrailed (PDB ID code 1ENH); and the RNA-binding domain of ribosome subunit L11 (PDB ID code 1QA6).
The existence of a common association interface for archaeal and eukaryal RPB10 homologs is supported by two lines of evidence. First, the same RPB10 subunit is used by all three yeast RNAPs (Table 1), and, second, the archaeal RPB10 from Sulfolobus acidocaldarius exhibits largely equivalent behavior in substitution experiments with its yeast homolog in RNAPII and RNAPIII (10). In contrast, the lack of the eukaryotic-conserved HVDLIEK sequence in this archaeal RPB10 (Fig. 1) leads to RNAPI-specific assembly defects in yeast (10). Structurally, the presence of this motif would alter the surface properties of RPB10 in the region following the postulated protein-association interface involving helix 3. Because specificity is tied to variability in sequence, it is reasonable to ascribe different binding properties of RPB10 homologs toward various RNAPs to such changes in their C-terminal residues.
Structural Similarity to DNA- and RNA-Binding Domains.
A search of mtRPB10 against the structural database by using the vast (31) server yielded an interesting similarity to the helix-turn-helix (HTH) family of proteins. Several homeodomain representatives were found with a vast score ≥ 4 and an rmsd < 2 Å, including Oct3, Matα2, Dp2, and engrailed (Fig. 4). Within the homeodomain sequences, residues primarily in the N-terminal region and within helix 3 serve to make specific contacts within the minor and major grooves of DNA, respectively (32). A second class of nucleic acid binding proteins is represented by the RNA-binding ribosomal subunit L11 (Fig. 4), which aligns with mtRPB10 with an rmsd of 1.2 Å (13 residues) and a vast score of 4.2. Although it is tempting to speculate that RPB10 may contact directly DNA or possibly RNA during some stage in the transcription process, this would not be consistent with the proposed subunit-association role of residues within its N terminus and helices 1 and 3. Alternatively, it is possible that these structurally similar proteins, each of which is involved in the overall process of gene expression, may share a common evolutionary ancestry.
In summary, we have used NMR spectroscopy to determine that mtRPB10 adopts a three helix zinc-bundle fold with an atypical metal binding motif. This structure provides a framework for understanding the specific interactions of RPB10 with additional components of the transcriptional machinery and should aid in the determination of high-resolution models of entire RNAP holoenzyme complexes (33).
Acknowledgments
We are grateful to Akil Dharamsi for providing the MTH40 clone, Lewis Kay for providing NMR pulse sequences, Emmanuel Brun for help with felix and aria, and Bert Mueller for performing the graphite furnace atomic absorption spectroscopy. This work was funded by grants from the National Cancer Institute of Canada with funds from the Canadian Cancer Society (C.H.A., A.M.E., L.P.M.), the Medical Research Council of Canada (C.H.A.), and the Natural Sciences and Engineering Research Council of Canada (C.D.M.). Instrument support was provided by the Protein Engineering Network of Centres of Excellence (L.P.M.). L.P.M. acknowledges the Alexander von Humbolt-Stiftung for a Research Fellowship. C.H.A. and A.M.E. are Medical Research Council of Canada Scientists.
Abbreviations
- RNAP
RNA polymerase
- 3D
three-dimensional
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The coordinates and restraint tables have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1EF4). The NMR chemical shifts have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession no. 4571).
References
- 1.Baumann P, Qureshi S A, Jackson S P. Trends Genet. 1995;11:279–282. doi: 10.1016/s0168-9525(00)89075-7. [DOI] [PubMed] [Google Scholar]
- 2.Gaasterland T. Curr Opin Microbiol. 1999;2:542–547. doi: 10.1016/s1369-5274(99)00014-4. [DOI] [PubMed] [Google Scholar]
- 3.Langer D, Hain J, Thuriaux P, Zillig W. Proc Natl Acad Sci USA. 1995;92:5768–5772. doi: 10.1073/pnas.92.13.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young R A. Annu Rev Biochem. 1991;60:689–715. doi: 10.1146/annurev.bi.60.070191.003353. [DOI] [PubMed] [Google Scholar]
- 5.Sakurai H, Ishihama A. Gene. 1997;196:165–174. doi: 10.1016/s0378-1119(97)00222-9. [DOI] [PubMed] [Google Scholar]
- 6.Darcy T J, Hausner W, Awery D E, Edwards A M, Thomm M, Reeve J N. J Bacteriol. 1999;181:4424–4429. doi: 10.1128/jb.181.14.4424-4429.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woychik N A, Young R A. J Biol Chem. 1990;265:17816–17819. [PubMed] [Google Scholar]
- 8.Woychik N A, Liao S M, Kolodziej P A, Young R A. Genes Dev. 1990;4:313–323. doi: 10.1101/gad.4.3.313. [DOI] [PubMed] [Google Scholar]
- 9.Carles C, Treich I, Bouet F, Riva M, Sentenac A. J Biol Chem. 1991;266:24092–24096. [PubMed] [Google Scholar]
- 10.Gadal O, Shpakovski G V, Thuriaux P. J Biol Chem. 1999;274:8421–8427. doi: 10.1074/jbc.274.13.8421. [DOI] [PubMed] [Google Scholar]
- 11.Lalo D, Carles C, Sentenac A, Thuriaux P. Proc Natl Acad Sci USA. 1993;90:5524–5528. doi: 10.1073/pnas.90.12.5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Acker J, de Graaff M, Cheynel I, Khazak V, Kedinger C, Vigneron M. J Biol Chem. 1997;272:16815–16821. doi: 10.1074/jbc.272.27.16815. [DOI] [PubMed] [Google Scholar]
- 13.Kimura M, Ishiguro A, Ishihama A. J Biol Chem. 1997;272:25851–25855. doi: 10.1074/jbc.272.41.25851. [DOI] [PubMed] [Google Scholar]
- 14.Ishiguro A, Kimura M, Yasui K, Iwata A, Ueda S, Ishihama A. J Mol Biol. 1998;279:703–712. doi: 10.1006/jmbi.1998.1823. [DOI] [PubMed] [Google Scholar]
- 15.Flores A, Briand J-F, Gadal O, Andrau J-C, Rubbi L, Van Mullem V, Boschiero C, Goussot M, Marck C, Carles C, et al. Proc Natl Acad Sci USA. 1999;96:7815–7820. doi: 10.1073/pnas.96.14.7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, et al. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson P E, Joshi M D, Tomme P, Kilburn D G, McIntosh L P. Biochemistry. 1996;35:14381–14394. doi: 10.1021/bi961612s. [DOI] [PubMed] [Google Scholar]
- 18.Farrow N A, Muhandiram R, Singer A U, Pascal S M, Kay C M, Gish G, Shoelson S E, Pawson T, Forman-Kay J D, Kay L E. Biochemistry. 1994;19:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 19.Zwahlen C, Gardner K H, Sarma S P, Horita D A, Byrd R A, Kay L E. J Am Chem Soc. 1998;120:7617–7625. [Google Scholar]
- 20.Brünger A T. xplorVersion 3.1: A System for X-Ray Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1992. [Google Scholar]
- 21.Nilges M, Macias M J, O'Donoghue S I, Oschkinat H. J Mol Biol. 1997;269:408–422. doi: 10.1006/jmbi.1997.1044. [DOI] [PubMed] [Google Scholar]
- 22.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 23.Kraulis P J. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 24.Meritt E A, Murphy M E P. Acta Crystallogr D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 25.McGregor M J, Islam S A, Sternberg M J E. J Mol Biol. 1987;198:295–310. doi: 10.1016/0022-2836(87)90314-7. [DOI] [PubMed] [Google Scholar]
- 26.Christianson D W. Adv Protein Chem. 1991;42:281–355. doi: 10.1016/s0065-3233(08)60538-0. [DOI] [PubMed] [Google Scholar]
- 27.Eloranta J J, Kato A, Teng M S, Weinzierl R O J. Nucleic Acids Res. 1998;26:5562–5567. doi: 10.1093/nar/26.24.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishihama A. Adv Biophys. 1981;14:1–35. [PubMed] [Google Scholar]
- 29.Cai M, Zheng R, Caffrey M, Craigie R, Clore G M, Gronenborn A M. Nat Struct Biol. 1997;4:567–577. doi: 10.1038/nsb0797-567. [DOI] [PubMed] [Google Scholar]
- 30.Eijkelenboom A P A M, van den Ent F M I, Vos A, Doreleijers J F, Tullius T D, Plasterk R H A, Kaptein R, Boelens R. Curr Biol. 1997;7:739–746. doi: 10.1016/s0960-9822(06)00332-0. [DOI] [PubMed] [Google Scholar]
- 31.Madej T, Gibrat J-F, Bryant S H. Proteins. 1995;23:356–369. doi: 10.1002/prot.340230309. [DOI] [PubMed] [Google Scholar]
- 32.Wintjens R, Rooman M. J Mol Biol. 1996;262:294–313. doi: 10.1006/jmbi.1996.0514. [DOI] [PubMed] [Google Scholar]
- 33.Fu J, Gnatt A L, Bushnell D A, Jensen G J, Thompson N E, Burgess R R, David P R, Kornberg R D. Cell. 1999;98:799–810. doi: 10.1016/s0092-8674(00)81514-7. [DOI] [PubMed] [Google Scholar]
- 34.Nicholls A, Bharadwaj R, Honig B. Biophys J. 1993;64:A166. [Google Scholar]