Abstract
Background and aims
Faecal bile acid elimination greatly contributes to cholesterol homeostasis. Synthesised from cholesterol in the liver, bile acids are actively reclaimed in the ileum by the apical sodium dependent bile acid transporter (ASBT). Although the expression level of ASBT affects body cholesterol balance, the impact of cholesterol on ASBT gene expression remains unclear. In this study, the effect of cholesterol on ASBT expression and ileal bile acid uptake was explored in vivo and in vitro.
Methods
ASBT gene expression was assessed by real time quantitative polymerase chain reaction and northern or western blotting, or both, in mice subjected to a 2% cholesterol diet for two weeks, in mouse ileal explants, or in human enterocyte‐like Caco‐2 cells cultured in sterol enriched or depleted media. Bile acid uptake was determined by measuring [3H]‐taurocholic acid influx into in situ isolated ileal loops from mice or into differentiated Caco‐2 cells. Molecular analysis of mouse and human ASBT promoters was undertaken with reporter assays, site directed mutagenesis, and electrophoretic mobility shift assays.
Results
In mice, cholesterol enriched diet triggered a downregulation of ASBT expression (mRNA and protein), a fall in ileal bile acid uptake, and a rise in the faecal excretion of bile acids. This effect was direct as it was reproduced ex vivo using mouse ileal explants and in vitro in differentiated Caco‐2 cells.
Conclusions
This regulation, which involves an original partnership between SREBP‐2 and HNF‐1α transcription factors, affects ileal bile acid recycling and thus might participate in the maintenance of body cholesterol homeostasis.
Keywords: cholesterol homeostasis, bile acids, ASBT, HNF‐1α, SREBP‐2
Hepatic synthesis of bile acids and their subsequent elimination in faeces constitute the main physiological way for the removal of body cholesterol. Thus they contribute greatly to the maintenance of cholesterol homeostasis.1 The regulation of hepatic bile acid biosynthesis, which has been extensively studied, has highlighted the crucial role played by the cholesterol‐7α‐hydroxylase (CYP7A1) in cholesterol balance.2 In rodents, this rate limiting enzyme—which promotes the conversion of cholesterol into bile acids2—is upregulated by sterols through an LXRα dependent pathway.3,4,5 Conversely, the CYP7A1 gene is downregulated by bile acids through a feedback loop involving SHP dependent and independent pathways.6,7,8,9,10,11 In humans, only bile acid mediated repression is present.12,13,14 Paradoxically, the regulation of intestinal bile acid reclamation remains poorly known, though this step is crucial for maintaining a constant bile acid pool. Indeed, in healthy humans and rodents, more than 90% of bile acids are reabsorbed by the small intestine and return to the liver to be secreted again into the bile.15,16,17 This efficient bile acid recycling mainly takes place in the ileum through an active process involving a 48 kDa integral brush border membrane glycoprotein termed apical sodium dependent bile acid transporter (ASBT), or ileal‐bile acid transporter (I‐BAT).18,19,20
There is compelling evidence that ASBT is essential for efficient bile acid uptake by the ileum. First, the primary bile acid malabsorption syndrome—caused by inherited mutations in human ASBT gene—is associated with a defective enterohepatic bile acid circulation, congenital steatorrhoea, and a reduced blood cholesterol concentration.21,22 Second, the targeted deletion of the ASBT gene in mice leads to a dramatic rise in faecal bile acid excretion and a decrease both in bile acid pool size and hepatic cholesterol ester levels.23 Third, ASBT inhibitors strongly enhance faecal bile acid outputs, reduce plasma cholesterol level and atherosclerosis risk in various species.24,25,26,27,28 This important homeostatic gene appears be tightly downregulated by bile acids in both mice and human.29,30 By contrast, although the basal expression of the ASBT gene is highly dependent on the transcription factor HNF‐1α31,32—a major player in cholesterol homeostasis31—the direct impact of cholesterol on the ASBT gene remains controversial. Indeed, the mild rise in ileal ASBT protein levels found in rabbits subjected to cholesterol feeding was not reproduced in rats.33 Moreover, experiments done in mice are conflicting as cholesterol mediated upregulation and downregulation of the ASBT gene have both been reported.34,35 However, the fact that bile acid influx into Caco‐2 cells is depressed by oxysterols strongly suggests that ASBT is a sterol target gene, at least in vitro.36
By using a combination of physiological, cellular, and molecular approaches we show in this report that ASBT gene expression is repressed by cholesterol in both mice and human enterocyte‐like Caco‐2 cells. This effect is direct and leads to a decrease in the cellular uptake of bile acids. Analysis of both human and mice ASBT promoters strongly suggests that this cholesterol mediated downregulation takes place at a transcriptional level through an original regulatory pathway involving a partnership between the transcription factors SREBP‐2 and HNF‐1α.
Methods
Animals and experimental treatments
French guidelines for the use and care of laboratory animals were followed. Protocols were approved by the ethics committee of the University of Burgundy. Eight week old male C57BL6/J mice were purchased from Charles River (France). In a first set of experiments, the impact of body cholesterol status on ASBT gene expression was explored in mice. To induce a cholesterol overload, mice were fed ad libitum on a 2% cholesterol enriched diet for 14 days, while the control animals were subjected to a standard laboratory chow containing 0.02% cholesterol (UAR A04, Saint Genest, France). Body cholesterol depletion was induced by force feeding with the HMG‐CoA reductase inhibitor simvastatin (100 mg/kg/d of simvastatin) for four days. Controls received the vehicle alone (1% carboxymethyl cellulose) by the same route, as described previously.37
The second set of experiments was carried out to evaluate impact of cholesterol on intestinal bile acid reclamation. To assess faecal bile acid elimination accurately, mice were placed in individual metabolic cages and were fed either a low or a high cholesterol diet (0.02% or 2%, respectively) for 14 days. Bile acid content of the faeces was determined by gas chromatography.38 At the end of this period, an in situ isolated ileal loop was formed to evaluate the level of intestinal bile acid uptake.24,39 In brief, a rapid laparotomy was done on isofluorane anaesthetised animals and a 5 cm segment of the terminal ileum was isolated in situ between two catheters. The ileal segment was gently rinsed with a prewarmed phosphate buffered saline (PBS) (0.9% NaCl, 0.01% sodium phosphate, 37°C) before being infused with 200 μl of [3H]‐taurocholic acid (TCA) (3 mM, 1.19 mCi/mmol). At the end of the incubation period, the ileal loop was rapidly removed and rinsed with 10 ml of cold PBS (4°C) to terminate cellular bile acid uptake.
[3H]‐Taurocholic acid uptake by intestinal cells
Mucosa from in situ isolated intestinal loops was lysed in 0.2 N NaOH (for four hours at 37°) before the determination of bile acid uptake. For in vitro experiments, 21day post‐confluent human enterocyte‐like Caco‐2 cells were cultured in a sterol enriched (10 μg/ml cholesterol, 1 μg/ml 25‐hydroxycholesterol) Dulbecco modified Eagle's medium (DMEM) for 48 hours. The choice of the 10:1 ratio between cholesterol and 25‐hydroxycholesterol was based on previous work from Brown and Goldstein's laboratory.40 Control cultures received vehicle alone (11 μl ethanol). Cells were washed three times with prewarmed PBS (37°C), then incubated at 37°C for 20 minutes in the presence of 25 μM [3H]‐TCA (25 mCi/mmol). At the end of incubation period, cells were washed four times with cold PBS containing 1 mM of unlabelled TCA, then once with cold PBS alone. Cells were next lysed in 0.2 N NaOH (for four hours at 37°C). The radioactivity incorporated into ileal and Caco‐2 samples was counted with a liquid scintillation counter (Tri‐CARB, Packard Instruments), and the protein concentration was assayed using the BCA protein reagent (Interchim). Bile acid uptake was expressed as the ratio of the incorporated [3H]‐TCA to the protein content of the sample.
Plasmid constructions and site directed mutagenesis
The −1042/+118 mouse ASBT promoter, kindly provided by Dr BL Shneider (Mount Sinai Medical Center, New York, USA), was subcloned upstream of the chloramphenicol acetyltransferase (CAT) reporter gene into the pCAT3 basic vector (Promega). The mASBT−552/+118 and mASBT−212/+118 plasmids were generated by endonuclease restriction. The shorter constructs were PCR generated (forward primers: M‐108, M‐65; reverse primers: M+118, M+26; table 1). Human HNF‐1α−473/+4 promoter, −1688/+539, and +26/+539 versions of human ASBT promoter—kindly provided by Dr GA Kullak‐Ublick (University Hospital, Zurich, Switzerland)—were subcloned into the pCAT3 basic vector. hASBT+292/+539 and hASBT+324/+539 constructs were PCR generated (forward primers: H+292, H+324; reverse primer: H+539; table 1). The ASBT−50/−24‐SV40‐CAT plasmid was constructed by ligating a dimerised oligonucleotide (mASBT−50/−24; table1). Deletion of the proximal HNFE (position −40/−24) of mASBT−65/+26 plasmid was achieved by site directed mutagenesis (Quickchange™ site directed mutagenesis kit, Stratagene) using the MHNFEdel primer (table 1). For all experiments, activity of the CAT reporter gene was quantified by an enzyme linked immunosorbent assay (ELISA) kit (Roche) and normalised to β‐galactosidase activity. pCMV‐β‐galactosidase expression vector was introduced in equal amount in each transfection mix, as described below.
Table 1 Oligonucleotides used for plasmid contructions, EMSA, and real time PCR.
| Oligonucleotides for mouse and human ASBT promoter constructions | |
| M‐108 | 5′‐ttacgagcTCCCAGCAAACTTTCCTGGC |
| M‐65 | 5′‐ttacgagCTCTGTGGCATAGTTCATTATC |
| M+118 | 5′‐attaCTCGAGCAAAAGCCACGTTAGAGAAGC |
| M+26 | 5′‐attactcgagATGAATCATAAAACGTCCAGAG |
| MHNFEdel | 5′‐GGCATAGTTCATTATCATACCATTATAACTTTCTGTCTTGGCC |
| H+292 | 5′‐ttacgagctcGGCATGGTTCATTATCATGCC |
| H+324 | 5′‐aatcgagctCTCTGTCTTGACCAATAATTT |
| H+539 | 5′‐attactcgagGCTGCTGGTTGAGTTAAGCAA |
| Oligonucleotide for heterologous construction | |
| mASBT−50/−24 | 5′‐CATTATCATACCAATAAATGATTAAT |
| Oligonucleotides for EMSA | |
| mASBT−50/−24 | 5′‐ccgagctCATTATCATACCAATAAATGATTAATagatctcc |
| SRE LDLr | 5′‐GATCAAAATCACCCCACTGC |
| Oligonucleotides for real time PCR | |
| hASBT fw | 5′‐GCCCCAAAAAGCAAAGATCA |
| hASBT rv | 5′‐GCTATGAGCACAATGAGGATGG |
| mHNF‐1α fw | 5′‐GACTTGACCATCTTCGCCACG |
| mHNF‐1α rv | 5′‐CAGAAAGCCGTGGTGGAGTC |
ASBT, apical sodium dependent bile acid transporter; EMSA, electrophoretic mobility shift assay; PCR, polymerase chain reaction.
Electrophoretic mobility shift assay (EMSA)
SREBP‐2 and HNF‐1α were synthesised using the TNT rabbit reticulocyte lysate coupled to an in vitro transcription/translation system (Promega). The doubled strand oligonucleotide probes were obtained by hybridising single strand complementary oligonucleotides (Eurogentec). The sequences used are listed in table 1. The dimers were labelled with [γ‐32P]ATP using the T4 polynucleotide kinase (Invitrogen). For the gel mobility shift assays, HNF‐1α or SREBP‐2 proteins (1 and 2 μl, respectively), or both, were incubated on ice for 15 minutes with 10 ng of [γ‐32P] end labelled dimerised oligonucleotides, and 0.5 μg of sonicated salmon sperm DNA in 20 mM HEPES (pH 7.8), 120 mM KCl, 0.4% nonidet P‐40, 12% glycerol, and 2 mM dithiothreitol. Equivalent gel loading was ensured by addition of processed empty vector. For the supershift experiment, 1 μl of anti‐SREBP‐2 antibody (N‐19: sc‐8151, Santa Cruz Biotechnology), or 1 μl of anti‐HNF‐1 antibody (H205: sc‐8986, Santa Cruz Biotechnology) was added to the reaction mixes, then incubated on ice for 30 minutes. Reactions were analysed by electrophoresis through a 4% polyacrylamide gel in 0.5 M TBE (90 mM Tris, 90 mM boric acid, 2 mM EDTA). Gels were dried, then subjected to autoradiography at −70°C.
Cell culture and transfection assays
The human Caco‐2 cells were maintained in DMEM supplemented with 4 mM glutamine, 1% non‐essential amino acids, 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% fetal calf serum (FCS). For the transactivation assays, cells were plated in six‐well plates at 40–50% confluency, then transfected by the calcium phosphate precipitation method. The transfection mixes contained 4 μg of mouse or human ASBT promoter reporter plasmids, 500 ng of pCMV‐βgalactosidase expression vector, 50 to 250 ng SREBP‐2 expression vector (gift from Dr TF Osborne, University of California, Irvine, California, USA), and/or 100 ng HNF‐1α expression vector (gift from Dr W Wahli, University of Lausanne, Switzerland) according to the experiment. For SREBP‐2 experiments, 10 μg/ml cholesterol and 1 μg/ml 25‐hydroxycholesterol were added 12 hours after the transfection to inhibit endogenous SREBP maturation.40,41 For the cholesterol sensitivity assessment, the culture medium was changed 12 hours after transfection with an experimental medium containing 10% lipoprotein‐free FCS plus 10 μg/ml of simvastatin (sterol− condition) or 10 μg/ml cholesterol plus 1 μg/ml 25‐hydroxycholesterol (sterol+ condition) for 24 hours. Control cultures received the vehicle alone (11 µl ethanol). Cellular extracts were assayed for CAT and β‐galactosidase activities. For RNA isolation experiments, Caco‐2 cells were plated onto 60 mm diameter dishes at 60% confluency and transfected using JetPEI (PolyPlus tranfection) with 2 μg SREBP‐2 or 2 μg HNF‐1α expression vectors, or both; or they were differentiated for 21 days and then cultured in sterol enriched medium (cholesterol and 25‐hydroxycholesterol for 48 hours, 10:1 ratio) or in sterol depleted medium (simvastatin 5 μg/ml for 48 hours).
Organ culture of ileal explants
Ileal explants from mice were obtained and cultured according to a previously published procedure.42 Explants were cultured in a medium (DMEM, 10% NCTC 135, 2 mM L‐glutamine, 2.5 μg/ml fungizone, 100 μg/ml gentamicin) supplemented with 10% lipoprotein‐free FCS in the presence of 10 μg/ml cholesterol and 1 μg/ml 25‐hydroxycholesterol for 18 hours. Control cultures were treated with vehicle alone (11 μl ethanol).
Northern blotting
Total RNA from mouse ileal explants and Caco‐2 cells was extracted with the RNeasy kit (Qiagen) and Trizol reagent (Invitrogen), respectively; 20 μg of RNAs were electrophoresed on a 1% agarose gel, then transferred onto Genescreen membranes (PerkinElmer Life Sciences). ASBT cDNA probe, and 18S rRNA oligonucleotide were [α‐32P]dCTP and [γ‐32P]ATP labelled, respectively. Blot size was quantified using a GS‐800 calibrated densitometer (Biorad).
Real time quantitative PCR
cDNA was produced from 1 μg total RNA by reverse transcription (Superscript II reverse transcriptase, Invitrogen). Real time PCR was undertaken using the ICycler IQ detection system (Biorad) with 50 ng cDNA, and qPCR MasterMix Plus SYBR (Eurogentec). The primers used for mRNA quantification are listed in the table 1. Quantification of data was done by the comparative ΔΔCt method.43
Western blotting
Twenty micrograms of total proteins extracted from the ileal mucosa were prepared in ice cold buffer (0.154 M KCl, 0.01 M phosphate buffer, pH 7.4), separated on 10% SDS‐PAGE, then blotted onto a Polyscreen membrane (PerkinElmer Life Sciences). Polyclonal ASBT antibodies were generated by Eurogentec. Anti‐rabbit peroxidase conjugated secondary antibodies were purchased from Sigma. Antibodies were diluted at 1:5000, and 1:20 000, respectively. Anti‐β‐actin antibodies (Sigma) was used as controls (1:20 000 dilution). Detection was undertaken using the ECL blotting kit (PerkinElmer Life Sciences). Blot size was quantified using GS‐800 calibrated densitometer (Biorad).
Statistical analysis
The results are expressed as means (SEM). The significance of the differences between groups was determined by Student's t test.
Results
ASBT expression is downregulated by cholesterol
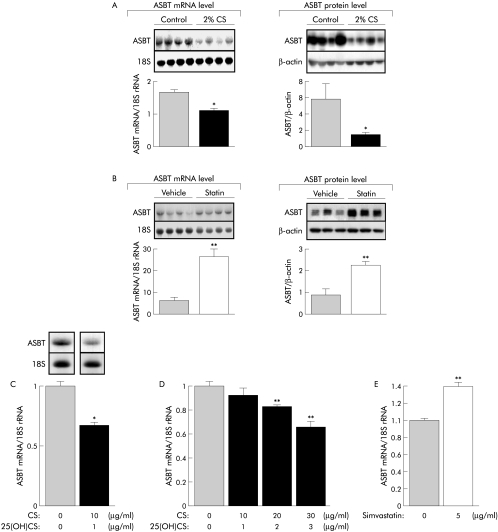
The impact of body cholesterol levels on ASBT gene expression was assessed in mice fed either a 2% cholesterol diet or force fed the hypocholesterolaemic drug simvastatin. Chronic dietary cholesterol overload significantly decreased ASBT mRNA and the protein levels in ileal mucosa (fig 1A). Conversely, simvastatin mediated cholesterol depletion led to a rise in ASBT mRNA and protein level (fig 1B). In the mouse, cholesterol mediated stimulation of bile acid synthesis3,4,5 raises the possibility that repression of ASBT gene in ileal mucosa is an indirect event secondary to its inhibition by bile acids.29,30 To address this question, mouse ileal explants were cultured in a sterol enriched medium. As shown in fig 1C, a 40% fall in ASBT mRNA levels occurred in the presence of sterols. A similar regulation was also found in human Caco‐2 cells in which sterols decreased ASBT gene expression in a dose dependent manner (fig 1D). As reported in the mouse, cholesterol depletion induced by simvastatin reduced the ASBT mRNA levels in Caco2 cells (fig 1E). Altogether these data show that mouse and human ASBT genes are sterol targets.
Figure 1 Body cholesterol status directly affects ASBT gene expression in the ileum. (A) ASBT mRNA and protein levels in mice subjected to a standard laboratory chow (control) or 2% cholesterol enriched diet (2% CS) for 14 days (n = 4). (B) Force fed with simvastatin (100 mg/d/kg) or vehicle alone (1% carboxymethylcellulose, 200 μl) for four days (n = 4). (C) Effect of a sterol enriched medium on ASBT mRNA levels in mouse ileal explants. Explants were incubated with 10 μg/ml cholesterol and 1 μg/ml 25 hydroxycholesterol (25(OH)CS) for 18 hours. Control cultures received vehicle alone (11 μl ethanol) (n = 4). (D) Effect of a sterol enriched (cholesterol + 25(OH)CS for 48 hours) or (E) sterol depleted (simvastatin for 48 hours) media on ASBT mRNA level in 21 day differentiated Caco‐2 cells. Control culture received vehicle alone (ethanol). The bar graphs represent ASBT mRNA levels normalised to 18S rRNA as determined by real time quantitative polymerase chain reaction (n = 3). Values are means with SEM; *p<0.05; **p<0.01. ASBT, apical sodium dependent bile acid transporter.
Cholesterol decreases bile acid uptake in intestinal cells
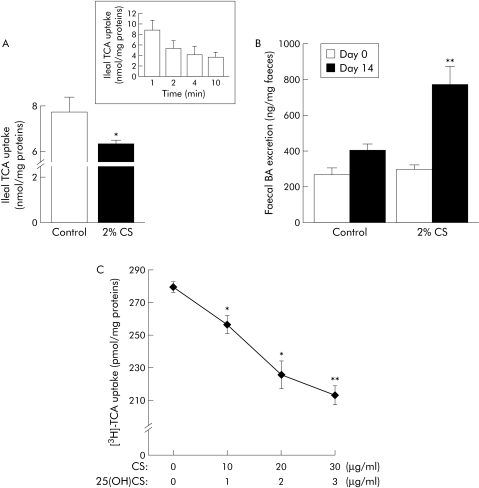
To determine whether the cholesterol mediated changes in ASBT gene expression affect the influx of bile acid into intestinal cells, an emulsion containing 3 mM [3H]‐TCA was infused into an in situ isolated ileal loop from mice fed a standard chow or a cholesterol enriched diet. As this method—which keeps the enteric blood circulation intact—is highly sensitive to the incubation time, a preliminary experiment was undertaken to determine the time needed for optimal [3H]‐TCA uptake. Because the highest uptake of TCA occurred at one minute (fig 2A, insert), this time was used for subsequent experiments. As expected, the cholesterol mediated decrease in ASBT gene expression was associated with a significant fall in ileal bile acid uptake (fig 2A). In line with this result, the faecal bile acid output was increased in mice subjected to chronic cholesterol feeding (fig 2B). In differentiated Caco‐2 cells, an inverse correlation between [3H]‐TCA uptake capacity and sterol concentration was found (fig 2C). Again, these functional changes paralleled the ASBT expression level quite well (fig 1A).
Figure 2 Cholesterol decreases the uptake of bile acid by the ileum. (A, insert) Time course of uptake of [3H]‐taurocholic acid ([3H]‐TCA, 3mM, 1.19 mCi/mmol) by the mucosa of in situ isolated ileal loops in mice fed a standard laboratory chow. (A, graph) Uptake of [3H]‐TCA within a one minute period by the ileal mucosa of mice fed a standard laboratory chow (control) or 2% cholesterol enriched diet (2% CS) for 14 days (n = 5). (B) Evolution of faecal bile acid outputs in controls and 2% cholesterol (CS) fed mice (n = 6). (C) Dose effect of sterols (48 hour exposure) on [3H]‐TCA influx into 21 day differentiated Caco‐2 cells. Plots represent the ratio of [3H]‐TCA uptake (25 μM, 25 mCi/mmol) for 20 minutes upon cellular protein content (n = 3). Values are means with SEM; *p<0.05, **p<0.01.
Mouse and human ASBT promoters are SREBP‐2 sensitive
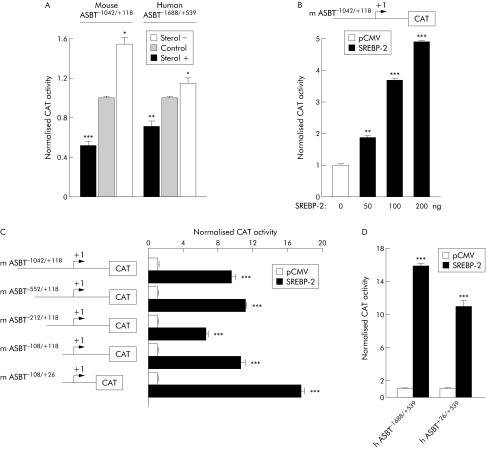
The transcriptional origin of this regulation of the ASBT gene was suggested by transfection assays. Indeed, sterols induced a significant fall in the activity of CAT reporter gene driven by the mouse or human ASBT promoters (fig 3A). Conversely, sterol depletion mediated by the addition of simvastatin (sterol−) increased the transactivation of the reporter gene (fig 3A).
Figure 3 SREBP‐2 is responsible for cholesterol sensitiveness of mouse and human ASBT promoters. (A) Sterol dependent effects on the CAT reporter gene activity in Caco‐2 cells transiently transfected with mouse or human ASBT promoter constructs. Cells were transfected in 10% FCS medium for 12 hours, and then cultured for an additional 24 hours in a medium containing 10% lipoprotein deprived serum in presence of 10 μg/ml simvastatin (sterol−), or 1 μg/ml 25‐hydroxycholesterol, and 10 μg/ml cholesterol (sterol+). Control cells received vehicle alone (11 μl ethanol). CAT activity was normalised to β‐galactosidase activity. (B) Effect of active SREBP‐2 peptide on the transactivation of CAT reporter gene driven by mouse ASBT promoter. Caco‐2 cells were co‐transfected with mouse ASBT promoter construct and increasing amounts of an expression vector encoding for SREBP‐2 active peptide. (C) Effect of a progressive 5′ deletion of mASBT promoter on the SREBP‐2 mediated transactivation of CAT reporter gene. Caco‐2 cells were co‐transfected with different constructs of mouse ASBT promoter together with 200 ng of the expression vector encoding for SREBP‐2 active peptide. (D) Co‐transfection of Caco‐2 cells with human ASBT−1688/+539 or ASBT+26/+539 promoters and 200 ng of mature SREBP‐2 expression vector. CAT activity was normalised to β‐galactosidase activity. Values are means with SEM; **p<0.01, ***p<0.001 (n = 3). ASBT, apical sodium dependent bile acid transporter; CAT, chloramphenicol acetyltransferase; SREBP, sterol regulatory element binding protein.
Cholesterol is known to regulate the expression of target genes involved in its own metabolism through the activation of the sterol responsive element binding protein‐2 (SREBP‐2). Cellular depletion of cholesterol triggers a proteolytic cleavage and the nuclear translocation of mature SREBP‐2, which transactivates target genes by binding to specific SREBP responsive elements (SRE).44,45 To determine whether sterol mediated control of ASBT gene expression takes place through this pathway, mouse ASBT−1042/+118 promoter was co‐transfected with an expression vector encoding for the active SREBP‐2 peptide. As expected, the activity of the reporter gene was increased in the presence of SREBP‐2. Moreover, this effect was dose dependent (fig 3B). The progressive 5′ deletions of mouse ASBT promoter showed that the proximal mASBT−108/+26 sequence was sufficient to confer SREBP‐2 responsiveness (fig 3C). Interestingly, human ASBT promoter showed a similar pattern of regulation (fig 3D).
ASBT responsiveness to SREBP‐2 is HNFE dependent
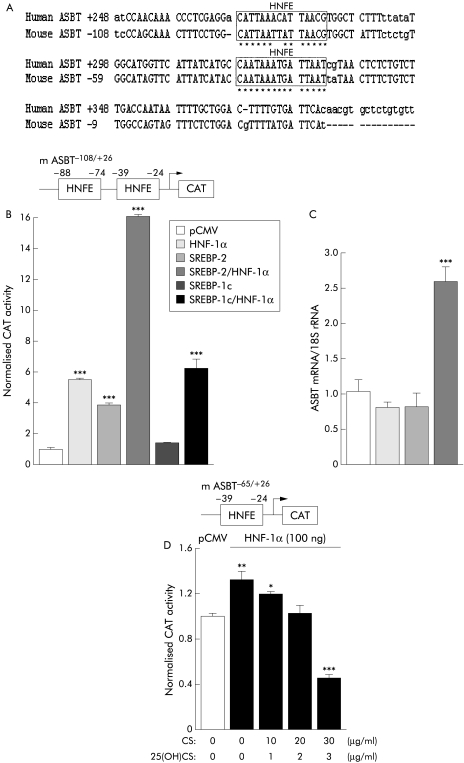
Paradoxically, bioinformatic analysis (MatInspector, www.genomatix.de) and EMSA failed to reveal the existence of an SRE sequence in the proximal sequence of ASBT promoter. By contrast, the nucleotide sequence alignments of mouse and human ASBT promoters showed that two responsive elements for HNF‐1α (HNFE) are highly conserved (fig 4A). As HNF‐1α is a major regulator for ASBT gene expression,31,32 and as interactions between HNFs and SREBPs are known,46,47 a putative involvement of these HNFE sequences in cholesterol mediated regulation of the ASBT gene was explored. Both SREBP‐2, and HNF‐1α transactivated the mASBT−108/+26 sequence (fig 4B). Surprisingly, co‐transfection with SREBP‐2 and HNF‐1α expression vectors together led to a large rise in CAT activity. This synergic action was SREBP‐2 specific as it was not reproduced with SREBP‐1c (fig 4B). In line with these data, only the concomitant transfection of SREBP‐2 and HNF‐1α was able to produce a significant increase in endogenous ASBT expression in human Caco‐2 cells (fig 4C). Conversely, the transactivation of CAT reporter gene in presence of HNF‐1α was decreased when SREBP activation was inhibited by the use of the sterol (+) condition (fig 4D). Taken together these data strongly suggest that a partnership between SREBP‐2 and HNF‐1α allows the responsiveness of ASBT promoter to cholesterol. In addition, HNFE sequences seem to play a crucial role in this regulation as SREBP‐2 did not directly interact with ASBT promoter. To explore this hypothesis, the involvement of HNFE sequences found in mouse and human ASBT promoters were analysed by 5′ sequence deletions or targeted mutagenesis. Deletion of the distal HNFE−89/−75 sequence of mouse ASBT promoter decreased the synergic response to SREBP‐2 and HNF‐1α, the double deletion of HNFE sequences fully abolishing the transactivation (fig 5A). Similar results were also obtained with human promoter (fig 5B).
Figure 4 SREBP‐2 and HNF‐1α synergistically transactivate the mouse and human ASBT promoters. (A) Sequence alignments, and analysis of the proximal human and mouse ASBT promoter regions. (B) Cooperative effect of SREBP‐2 and HNF‐1α on the transcriptional activity of mouse ASBT promoter. Caco‐2 cells were transiently co‐transfected with mASBT−108/+26 construct and 100 ng of SREBP‐2 or SREBP‐1c and/or HNF‐1α expression vectors. (C) Impact of SREBP‐2 and/or HNF‐1α overexpression on endogenous ASBT mRNA levels in Caco‐2 cells. Cells were transiently transfected with 2 μg SREBP‐2, and/or HNF‐1α expression vectors. ASBT mRNA was assessed by real time quantitative polymerase chain reaction. The bar graphs represent ASBT mRNA levels normalised to 18S rRNA. (D) HNF‐1α is involved in responsiveness of mASBT–65/+26 construct to sterols. Caco‐2 cells were transiently co‐transfected with mASBT−65/+26 construct and 100 ng of HNF‐1α expression vector, and cultured during 24 hours with the indicated amounts of cholesterol and 25‐hydroxycholesterol (25(OH)CS). CAT activity was normalised to β‐galactosidase activity (n = 3). Values are means with SEM; *p<0.05, **p<0.01, ***p<0.001 (n = 3). ASBT, apical sodium dependent bile acid transporter; CAT, chloramphenicol acetyltransferase; HNF, hepatocyte nuclear factor; SREBP, sterol regulatory element binding protein.
Figure 5 ASBT responsiveness to SREBP‐2 is HNFE dependent. Role of HNFE sequences in sterol sensitivity of mouse (A) and human (B) ASBT promoters. Caco‐2 cells were transiently co‐transfected with either native or HNFE deleted versions of ASBT promoters plus 100 ng of SREBP‐2 and/or HNF‐1α expression vectors. (C) Exploration of HNFE function in an heterologous promoter context. The minimum ASBT−50/−24 SV40‐CAT construct containing the proximal HNFE of mouse ASBT promoter was co‐transfected in presence of 100 ng of SREBP‐2, and/or HNF‐1α expression vectors. CAT activity was normalised to β‐galactosidase activity. (D) Binding capacities of mouse ASBT−50/−24 sequence to in vitro translated SREBP‐2 or HNF‐1α proteins. Supershift experiments were carried out by incubating for 15 minutes on ice with SREBP‐2 protein (2 μl) or HNF‐1α protein (1 μl), indicating [32P]‐end labelled probes with or without specific SREBP‐2 or HNF‐1α antibodies (1 μl). Values are means with SEM; **p<0.01, ***p<0.001 (n = 3). ASBT, apical sodium dependent bile acid transporter; CAT, chloramphenicol acetyltransferase; HNF, hepatocyte nuclear factor; HNFE, HNF‐1α responsive element; SREBP, sterol regulatory element binding protein.
To explore further the role of HNFE sequences in this regulation, the mASBT−50/−24 sequence containing the proximal HNFE−39/−24—sharing 100% homology with human ASBT proximal HNFE+308/+323 (fig 4A)—was cloned upstream of a reporter gene driven by the strong SV40 promoter, then co‐transfected together with SREBP‐2 and/or HNF‐1α expression vectors in Caco‐2 cells. As shown fig 5C, this short sequence was sufficient to reproduce the cooperative action of SREBP‐2 and HNF‐1α fully, demonstrating the involvement of this promoter region containing a canonical HNFE sequence in cholesterol mediated regulation of ASBT gene. The absence of a physical interaction between SREBP‐2 and the mASBT−50/−24 sequence was further confirmed by EMSA (fig 5D). While in vitro translated SREBP‐2 was bound to the canonical SRE sequence of the LDL receptor (lanes 2 and 3), no shift was detected with the mASBT−50/−24 probe (lane 5). This data show the inability of SREBP‐2 to bind the sterol sensitive sequence of mouse ASBT promoter, in contrast to HNF‐1α (lanes 6 and 7).
Cholesterol mediated regulation of ASBT gene is not due to a sterol dependent induction of HNF‐1α expression
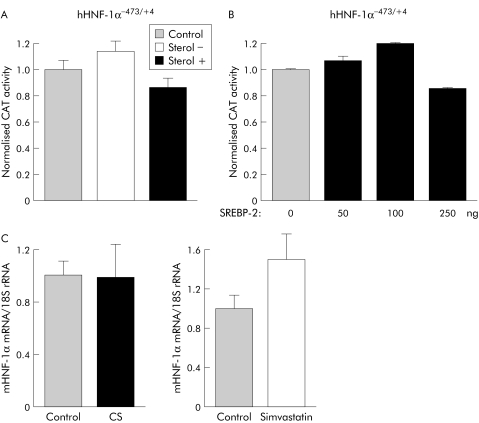
The present data clearly show the involvement of HNF‐1α in cholesterol mediated regulation of the ASBT gene. To explore whether the transactivation of ASBT promoter by SREBP‐2 resulted from an induction of the HNF‐1α gene by cholesterol, we subcloned human HNF‐1α−473/+4 promoter upstream of a CAT reporter gene and assessed its responsiveness to sterols or SREBP‐2 in Caco‐2 cells. In contrast to ASBT promoter, neither sterol+ nor sterol− conditions (fig 6A), nor SREBP‐2 were able to affect the reporter gene activity significantly when it was driven by HNF‐1α promoter (fig 6B). In agreement with these in vitro data, no change in HNF‐1α mRNA levels was found in mice subjected to a dietary cholesterol overload, or force fed with the hypocholesterolaemic drug simvastatin for four days (fig 6C).
Figure 6 HNF‐1α gene expression is not regulated by cholesterol. (A) Effect of sterols on human HNF‐1α−473/+4 promoter activity. Caco‐2 cells were transfected with human HNF‐1α−473/+4 promoter and cultured for 24 hours in a medium containing 10% lipoprotein‐free serum in the presence of 10 μg/ml simvastatin (sterol−) or 10 μg/ml cholesterol and 1 μg/ml 25 hydroxycholesterol (sterol+). Control cells received vehicle alone (11 μl ethanol). (B) Human HNF‐1α−473/+4 promoter is SREBP‐2 insensitive. Caco‐2 cells were co‐transfected with hHNF‐1α−473/+4and various amount of SREBP‐2 expression vector. CAT activity was normalised to β‐galactosidase activity (n = 3). (C) Ileal HNF‐1α mRNA levels assessed by real time quantitative polymerase chain reaction in mice fed a 2% cholesterol diet for 14 days or daily force fed simvastatin for four days (n = 4). Values are means with SEM. CAT, chloramphenicol acetyltransferase; HNF, hepatocyte nuclear factor; SREBP, sterol regulatory element binding protein.
Discussion
Adjustment of hepatic synthesis and ileal reclamation of bile acid contribute greatly to the maintenance of cholesterol homeostasis, and therefore to the protection against gallstone formation and cardiovascular diseases. While it is well known that cholesterol directly affects hepatic bile acid synthesis in rodents,2,3,5 its impact on intestinal uptake and subsequent faecal elimination of bile acids is still unclear. The present data provide the first demonstration that a dietary cholesterol overload leads to a strong downregulation of ASBT expression, associated with a 20% fall in ileal bile acid uptake and a twofold rise in faecal bile acid elimination in the mouse. These changes are in good agreement with previous data obtained in rats treated with pharmacological inhibitors of ASBT. Indeed, the use of the drug 264W94 produces to a twofold increase in faecal bile acid outputs despite a 24% inhibition of bile acid uptake.24 Thus a moderate decrease in ileal bile acid uptake markedly affects bile acid balance in rodents.
The origin of this regulation remains elusive in vivo, as a cholesterol mediated rise in hepatic bile acid synthesis by itself could account for the decreased ASBT expression.29 The use of mouse ileal explants shows that ASBT gene can also be directly affected by sterols. Indeed, this ex vivo approach allows the study of gene expression independently from exogenous (that is, luminal) and endogenous (blood and nervous) influences. Thus both bile acid and cholesterol downregulate ASBT gene expression in mice fed a cholesterol enriched diet. This concomitant repression might explain the high efficiency of faecal bile acid wasting and hence contribute to the well known resistance of the mouse to diet induced hypercholesterolaemia.1,13,48,49 As ASBT mRNA levels and bile acid uptake are also inversely correlated with sterol levels in human enterocyte‐like Caco‐2 cells, it is tempting to speculate that there is a common regulation pattern in the mouse and the human. This assumption, supported by the recent results from Alrefai et al,36 is reinforced by the fact that both mouse and human ASBT promoters are sterol sensitive (fig 3A).
The molecular mechanism whereby this regulation takes place is not fully elucidated. The transcription factor SREBP‐2 was a plausible candidate for such a function. Indeed, SREBP‐2 is known to regulate several key genes involved in the control of cholesterol homeostasis (for example, LDL receptor, HMG CoA reductase), secondary to a sterol dependent proteolytic activation.44,45 Moreover, this maturation of SREBP‐2 is functional in the ileum of rodents.50,51 Finally, initial transfection studies using mouse ASBT reporter gene constructs showed that the promoter activity was induced in a dose dependent manner by the active SREBP‐2 peptide (fig 3B). Paradoxically, the CAT activity remains significantly high when SREBP‐2 is co‐transfected with the mASBT−108/+26 sequence in which a SREBP responsive element was lacking. This finding strongly suggests that the impact of SREBP‐2 on ASBT gene expression is indirect. It is noteworthy the mASBT−108/+26 sequence contains two binding sites for HNF‐1α (ΗΝFE), a nuclear receptor recently highlighted as a major regulator of mouse and human ASBT genes.31,32 Co‐transfections with expression vectors encoding for HNF‐1α and active SREBP‐2 peptide synergistically transactivated the reporter gene driven by the mASBT−108/+26 sequence. The functional importance of this cooperation is supported by the fact that only a combination of HNF‐1α and SREBP‐2 was capable of inducing the endogenous ASBT mRNA levels in enterocyte‐like Caco‐2 cells (fig 4C). There is compelling evidence to support the crucial role exerted by HNFE sites in this regulation. First, in a heterologous promoter context, the proximal HNFE motif found in mouse and human ASBT promoters was sufficient to enhance the reporter gene activity of an SV40 promoter‐CAT construct in presence of SREBP‐2. This effect was dramatically strengthened when SREBP‐2 and HNF‐1α expression vectors were co‐transfected together. Second, the double loss of HNFE sequences in mouse and human promoters fully abolished the responsiveness of the ASBT gene to SREBP‐2 and SREBP‐2/HNF‐1α.
How SREBP‐2 cooperates with HNF‐1α is not yet established. However, an indirect regulation of ASBT gene through SREBP‐2 mediated control of HNF‐1α expression is unlikely as the HNF‐1α gene is not sterol sensitive (fig 6A). Likewise, the hypothesis of a bile acid mediated downregulation of the HNF‐1α gene52 was discarded as its mRNA level did not change in 2% cholesterol‐fed mice fed (fig 6B). A cooperative interaction between HNF‐1α and SREBP‐2 provides an alternative possibility. Indeed, recent studies have reported that physical protein–protein interactions between SREBP‐2 and HNF‐4α were responsible for synergic regulation of several of the key genes determining cholesterol metabolism, such as Δ8‐isomerase, HMG‐CoA synthase, and LDL receptor.46 Similarly, interaction between SREBP‐1 and HNF‐4α interferes with the recruitment of the transcriptional co‐activator PGC‐1 to suppress hepatic gluconeogenic genes.47 The existence of an interaction between SREBP‐2 and HNF‐1α is conceivable but requires further investigation to fully delineate the mechanism of this cooperation.
Conclusions
Our data provide the first evidence that mouse and human ASBT genes are regulated by cholesterol through an original pathway involving a partnership between SREBP‐2, and HNF‐1α. This regulation appears to be direct and leads to sustained faecal bile acid output. These new findings also raise the possibility of a coordinated regulation of intestinal and hepatic bile acid transfer following cholesterol feeding, as hepatic bile acid transporters such as NTCP or OATP1 are HNF‐1α target genes.16,17,31,53,54. Molecular dissection of such a regulatory pathway might provide new insights for the development of original hypocholesterolaemic treatments in the future.
Acknowledgements
This work was supported by funds from Conseil Régional de Bourgogne (to PB). CT is supported by a PhD fellowship from the Ministère de l'Education Nationale, de la Recherche et de la Technologie. We thank Dr Maâmar Souidi (Institut de Radioprotection, Fontenay‐aux‐Roses, France) for the generous gift of anti‐ASBT antibody and Dr Laurent Lagrost (INSERM U 498, Dijon, France) for critical reading of the manuscript.
Abbreviations
ASBT - apical sodium dependent bile acid transporter
CAT - chloramphenicol acetyltransferase
CYP7A1 - cholesterol 7α‐hydroxylase
EMSA - electrophoretic mobility shift assay
FCS - fetal calf serum
FXR - farnesoid‐X receptor
HMG‐CoA reductase - hydroxymethylglutaryl‐coenzyme A reductase
HNF - hepatocyte nuclear factor
HNFE - HNF‐1α responsive element
25(OH)CS - 25‐hydroxycholesterol
LDL - low density lipoprotein
LXR - liver X‐receptor
NCTC - National Collection of Type Cultures
NTCP - Na+‐taurocholate co‐transporting polypeptide
OATP - organic anion transport protein
PGC‐1 - PPAR γ co‐activator 1
PPAR - peroxisome proliferator activated receptor
SHP - small heterodimer partner
SRE - sterol responsive element
SREBP - sterol regulatory element binding protein
TCA - taurocholic acid
Footnotes
Conflict of interest: None declared.
References
- 1.Dietschy J M, Turley S D. Control of cholesterol turnover in the mouse. J Biol Chem 20022773801–3804. [DOI] [PubMed] [Google Scholar]
- 2.Russell D W. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem 200372137–174. [DOI] [PubMed] [Google Scholar]
- 3.Gupta S, Pandak W M, Hylemon P B. LXR alpha is the dominant regulator of CYP7A1 transcription. Biochem Biophys Res Commun 2002293338–343. [DOI] [PubMed] [Google Scholar]
- 4.Chiang J Y, Kimmel R, Stroup D. Regulation of cholesterol 7alpha‐hydroxylase gene (CYP7A1) transcription by the liver orphan receptor (LXRalpha). Gene 2001262257–265. [DOI] [PubMed] [Google Scholar]
- 5.Peet D J, Turley S D, Ma W.et al Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell 199893693–704. [DOI] [PubMed] [Google Scholar]
- 6.Kerr T A, Saeki S, Schneider M.et al Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev Cell 20022713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu T T, Makishima M, Repa J J.et al Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell 20006507–515. [DOI] [PubMed] [Google Scholar]
- 8.Goodwin B, Jones S A, Price R R.et al A regulatory cascade of the nuclear receptors FXR, SHP‐1, and LRH‐1 represses bile acid biosynthesis. Mol Cell 20006517–526. [DOI] [PubMed] [Google Scholar]
- 9.Wang L, Lee Y K, Bundman D.et al Redundant pathways for negative feedback regulation of bile acid production. Dev Cell 20022721–731. [DOI] [PubMed] [Google Scholar]
- 10.De Fabiani E, Mitro N, Anzulovich A C.et al The negative effects of bile acids and tumor necrosis factor‐alpha on the transcription of cholesterol 7alpha‐hydroxylase gene (CYP7A1) converge to hepatic nuclear factor‐4: a novel mechanism of feedback regulation of bile acid synthesis mediated by nuclear receptors. J Biol Chem 200127630708–30716. [DOI] [PubMed] [Google Scholar]
- 11.Abrahamsson A, Gustafsson U, Ellis E.et al Feedback regulation of bile acid synthesis in human liver: importance of HNF‐4alpha for regulation of CYP7A1. Biochem Biophys Res Commun 2005330395–399. [DOI] [PubMed] [Google Scholar]
- 12.Goodwin B, Watson M A, Kim H.et al Differential regulation of rat and human CYP7A1 by the nuclear oxysterol receptor liver X receptor‐alpha. Mol Endocrinol 200317386–394. [DOI] [PubMed] [Google Scholar]
- 13.Chen J Y, Levy‐Wilson B, Goodart S.et al Mice expressing the human CYP7A1 gene in the mouse CYP7A1 knockout background lack induction of CYP7A1 expression by cholesterol feeding and have increased hypercholesterolemia when fed a high fat diet. J Biol Chem 20022828. [DOI] [PubMed] [Google Scholar]
- 14.Chen W, Owsley E, Yang Y.et al Nuclear receptor‐mediated repression of human cholesterol 7alpha‐ hydroxylase gene transcription by bile acids. J Lipid Res 2001421402–1412. [PubMed] [Google Scholar]
- 15.Hofmann A F. Bile secretion in mice and men. Hepatology 200134848–850. [DOI] [PubMed] [Google Scholar]
- 16.Trauner M, Boyer J L. Bile salt transporters: molecular characterization, function, and regulation. Physiological reviews 200383633–671. [DOI] [PubMed] [Google Scholar]
- 17.Meier P J, Stieger B. Bile salt transporters. Annu Rev Physiol 200264635–661. [DOI] [PubMed] [Google Scholar]
- 18.Shneider B L, Dawson P A, Christie D M.et al Cloning and molecular characterization of the ontogeny of a rat ileal sodium‐dependent bile acid transporter. J Clin Invest 199595745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong M H, Oelkers P, Craddock A L.et al Expression cloning and characterization of the hamster ileal sodium‐ dependent bile acid transporter. J Biol Chem 19942691340–1347. [PubMed] [Google Scholar]
- 20.Craddock A L, Love M W, Daniel R W.et al Expression and transport properties of the human ileal and renal sodium‐ dependent bile acid transporter. Am J Physiol 1998274G157–G169. [DOI] [PubMed] [Google Scholar]
- 21.Oelkers P, Kirby L C, Heubi J E.et al Primary bile acid malabsorption caused by mutations in the ileal sodium‐ dependent bile acid transporter gene (SLC10A2). J Clin Invest 1997991880–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heubi J E, Balistreri W F, Fondacaro J D.et al Primary bile acid malabsorption: defective in vitro ileal active bile acid transport. Gastroenterology 198283804–811. [PubMed] [Google Scholar]
- 23.Dawson P A, Haywood J, Craddock A L.et al Targeted deletion of the ileal bile acid transporter eliminates enterohepatic cycling of bile acids in mice. J Biol Chem 200327833920–33927. [DOI] [PubMed] [Google Scholar]
- 24.Root C, Smith C D, Sundseth S S.et al Ileal bile acid transporter inhibition, CYP7A1 induction, and antilipemic action of 264W94. J Lipid Res 2002431320–1330. [PubMed] [Google Scholar]
- 25.Lewis M C, Brieaddy L E, Root C. Effects of 2164U90 on ileal bile acid absorption and serum cholesterol in rats and mice. J Lipid Res 1995361098–1105. [PubMed] [Google Scholar]
- 26.Huff M W, Telford D E, Edwards J Y.et al Inhibition of the apical sodium‐dependent bile acid transporter reduces LDL cholesterol and apoB by enhanced plasma clearance of LDL apoB. Arterioscler Thromb Vasc Biol 2002221884–1891. [DOI] [PubMed] [Google Scholar]
- 27.Telford D E, Edwards J Y, Lipson S M.et al Inhibition of both the apical sodium‐dependent bile acid transporter and HMG‐CoA reductase markedly enhances the clearance of LDL apoB. J Lipid Res 200344943–952. [DOI] [PubMed] [Google Scholar]
- 28.Bhat B G, Rapp S R, Beaudry J A.et al Inhibition of ileal bile acid transport and reduced atherosclerosis in apoE−/− mice by SC‐435. J Lipid Res 2003441614–1621. [DOI] [PubMed] [Google Scholar]
- 29.Chen F, Ma L, Dawson P A.et al Liver receptor homologue‐1 mediates species‐ and cell line‐specific bile acid‐dependent negative feedback regulation of the apical sodium‐dependent bile acid transporter. J Biol Chem 200327819909–19916. [DOI] [PubMed] [Google Scholar]
- 30.Neimark E, Chen F, Li X.et al Bile acid‐induced negative feedback regulation of the human ileal bile acid transporter. Hepatology 200440149–156. [DOI] [PubMed] [Google Scholar]
- 31.Shih D Q, Bussen M, Sehayek E.et al Hepatocyte nuclear factor‐1alpha is an essential regulator of bile acid and plasma cholesterol metabolism. Nat Genet 200127375–382. [DOI] [PubMed] [Google Scholar]
- 32.Jung D, Fried M, Kullak‐Ublick G A. Human apical sodium‐dependent bile salt transporter gene (SLC10A2) is regulated by the peroxisome proliferator‐activated receptor alpha. J Biol Chem 200227730559–30566. [DOI] [PubMed] [Google Scholar]
- 33.Xu G, Shneider B L, Shefer S.et al Ileal bile acid transport regulates bile acid pool, synthesis, and plasma cholesterol levels differently in cholesterol‐fed rats and rabbits. J Lipid Res 200041298–304. [PubMed] [Google Scholar]
- 34.Torchia E C, Cheema S K, Agellon L B. Coordinate regulation of bile acid biosynthetic and recovery pathways. Biochem Biophys Res Commun 1996225128–133. [DOI] [PubMed] [Google Scholar]
- 35.Wang L, Han Y, Kim C S.et al Resistance of SHP‐null mice to bile acid‐induced liver damage. J Biol Chem 200327844475–44481. [DOI] [PubMed] [Google Scholar]
- 36.Alrefai W A, Sarwar Z, Tyagi S.et al Cholesterol modulates human intestinal sodium‐dependent bile acid transporter. Am J Physiol Gastrointest Liver Physiol 2005288G978–G985. [DOI] [PubMed] [Google Scholar]
- 37.Landrier J F, Thomas C, Grober J.et al Statin induction of liver fatty acid‐binding protein (L‐FABP) gene expression is peroxisome proliferator‐activated receptor‐alpha‐dependent. J Biol Chem 200427945512–45518. [DOI] [PubMed] [Google Scholar]
- 38.Batta A K, Salen G, Rapole K R.et al Highly simplified method for gas‐liquid chromatographic quantitation of bile acids and sterols in human stool. J Lipid Res 1999401148–1154. [PubMed] [Google Scholar]
- 39.Root C, Smith C D, Winegar D A.et al Inhibition of ileal sodium‐dependent bile acid transport by 2164U90. J Lipid Res 1995361106–1115. [PubMed] [Google Scholar]
- 40.Adams C M, Reitz J, De Brabander J K.et al Cholesterol and 25‐hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and Insigs. J Biol Chem 200427952772–52780. [DOI] [PubMed] [Google Scholar]
- 41.Zaghini I, Landrier J F, Grober J.et al Sterol regulatory element‐binding protein‐1c is responsible for cholesterol regulation of ileal bile acid‐binding protein gene in vivo. Possible involvement of liver‐X‐receptor. J Biol Chem 20022771324–1331. [DOI] [PubMed] [Google Scholar]
- 42.Mallordy A, Besnard P, Carlier H. Research of an in vitro model to study the expression of fatty acid‐binding proteins in the small intestine. Mol Cell Biochem 199312385–92. [DOI] [PubMed] [Google Scholar]
- 43.Livak K J, Schmittgen T D. Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) method. Methods 200125402–408. [DOI] [PubMed] [Google Scholar]
- 44.Brown M S, Goldstein J L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane‐bound transcription factor. Cell 199789331–340. [DOI] [PubMed] [Google Scholar]
- 45.Shimano H. Sterol regulatory element‐binding proteins (SREBPs): transcriptional regulators of lipid synthetic genes. Prog Lipid Res 200140439–452. [DOI] [PubMed] [Google Scholar]
- 46.Misawa K, Horiba T, Arimura N.et al Sterol regulatory element‐binding protein‐2 interacts with hepatocyte nuclear factor‐4 to enhance sterol isomerase gene expression in hepatocytes. J Biol Chem 200327836176–36182. [DOI] [PubMed] [Google Scholar]
- 47.Yamamoto T, Shimano H, Nakagawa Y.et al SREBP‐1 interacts with hepatocyte nuclear factor‐4 alpha and interferes with PGC‐1 recruitment to suppress hepatic gluconeogenic genes. J Biol Chem 200427912027–12035. [DOI] [PubMed] [Google Scholar]
- 48.Schwarz M, Davis D L, Vick B R.et al Genetic analysis of cholesterol accumulation in inbred mice. J Lipid Res 2001421812–1819. [PubMed] [Google Scholar]
- 49.Phan J, Pesaran T, Davis R C.et al The Diet1 locus confers protection against hypercholesterolemia through enhanced bile acid metabolism. J Biol Chem 2002277469–477. [DOI] [PubMed] [Google Scholar]
- 50.Field F J, Born E, Murthy S.et al Regulation of sterol regulatory element‐binding proteins in hamster intestine by changes in cholesterol flux. J Biol Chem 200127617576–17583. [DOI] [PubMed] [Google Scholar]
- 51.Field F J, Born E, Murthy S.et al Gene expression of sterol regulatory element‐binding proteins in hamster small intestine. J Lipid Res 2001421–8. [PubMed] [Google Scholar]
- 52.Jung D, Kullak‐Ublick G A. Hepatocyte nuclear factor 1 alpha: a key mediator of the effect of bile acids on gene expression. Hepatology 200337622–631. [DOI] [PubMed] [Google Scholar]
- 53.Jung D, Hagenbuch B, Fried M.et al Role of liver‐enriched transcription factors and nuclear receptors in regulating the human, mouse, and rat NTCP gene. Am J Physiol Gastrointest Liver Physiol 2004286G752–G761. [DOI] [PubMed] [Google Scholar]
- 54.Kullak‐Ublick G A, Stieger B, Meier P J. Enterohepatic bile salt transporters in normal physiology and liver disease. Gastroenterology 2004126322–342. [DOI] [PubMed] [Google Scholar]