Abstract
Background
Despite intent to cure surgery with negative resection margins, locoregional recurrence is common in pancreatic cancer.
Aims
To determine whether detection of K‐ras gene mutation in the histologically negative surgical margins of pancreatic cancer reflects unrecognised disease.
Patients
Seventy patients who underwent curative resection for pancreatic ductal adenocarcinoma were evaluated.
Methods
All patients had surgical resection margins (pancreatic transection and retroperitoneal) that were histologically free of invasive cancer. DNA was extracted from these paraffin embedded surgical margins and assessed by quantitative real time polymerase chain reaction to detect the K‐ras gene mutation at codon 12. Detection of K‐ras mutation was correlated with standard clinicopathological factors.
Results
K‐ras mutation was detected in histologically negative surgical margins of 37 of 70 (53%) patients. A significant difference in overall survival was demonstrated between patients with margins that were K‐ras mutation positive compared with negative (median 15 v 55 months, respectively; p = 0.0008). By univariate and multivariate analyses, detection of K‐ras mutation in the margins was a significant prognostic factor for poor survival (hazard ratio (HR) 2.8 (95% confidence interval (CI) 1.5–5.3), p = 0.0009; and HR 2.8 (95% CI 1.4–5.5), p = 0.004, respectively).
Conclusions
Detection of cells harbouring K‐ras mutation in histologically negative surgical margins of pancreatic cancer may represent unrecognised disease and correlates with poor disease outcome. The study demonstrates that molecular‐genetic evaluation of surgical resection margins can improve pathological staging and prognostic evaluation of patients with pancreatic ductal adenocarcinoma.
Keywords: K‐ras, pancreatic cancer, surgical margin, quantitative polymerase chain reaction
Pancreatic ductal adenocarcinoma has one of the worst five year survival rates of any cancer.1 A contributing factor is frequent locoregional failure after curative resection.2,3,4,5 The high rate of such failures may reflect inadequate removal of all preneoplastic cells or residual tumour (for example, occult cancer cells) despite histologically negative surgical resection margins. During the past decade, increasingly sophisticated molecular techniques have identified genetic, epigenetic, and molecular aberrancies in benign appearing cells in tissues surrounding primary tumours or at the surgical margins.6,7 These aberrancies have been implicated in locoregional cancer recurrences.8,9,10,11,12
Genetic aberrancies may have an insidious role in cancer recurrence in pancreatic cancer because its genotypic features cannot be correctly assessed in surgical margins by conventional histological haematoxylin and eosin (H&E) and light microscopy techniques. Therefore, molecular‐genetic assessment of surgical resection margins in pancreatic cancer would have particular clinical relevance. Accordingly, more sophisticated analysis of histologically negative surgical margins might improve pathological staging and prognostic evaluation of patients undergoing surgery for pancreatic cancer.
For this report, we examined the incidence and potential prognostic significance of K‐ras gene mutation in the surgical margins of pancreatic cancer. Recently, Hingorani and colleagues directed endogenous expression of K‐ras in a mouse model and recapitulated the entire progression of pancreatic cancer, demonstrating the key role of K‐ras mutation in pancreatic cancer pathogenesis.13 Since the earliest reports of K‐ras mutation in pancreatic cancer, innovative methods to utilise the mutation for diagnostic evaluation of blood, bile, or stool have been developed but have had little impact on disease outcome.14,15,16,17,18,19,20 Here we hypothesised that detection of K‐ras mutation in histologically negative surgical margins of pancreatic cancer correlates with less favourable clinical outcomes. Our findings indicate that K‐ras mutation in surgical resection margins may be a clinically relevant surrogate for unrecognised disease.
Methods
Patients with pancreatic cancer
Twenty three patients who underwent curative resection for pancreatic ductal adenocarcinoma from 1996 to 2004 were initially evaluated as a pilot cohort for this research study. Patient specimens were obtained from the John Wayne Cancer Institute (JWCI, Santa Monica, California, USA) and the David Geffen School of Medicine (UCLA, Los Angeles, California, USA). After analysis of the pilot cohort to ensure the feasibility of detection of K‐ras gene mutation in paraffin embedded surgical margin tissues, 47 additional patients who underwent intent to cure surgery for pancreatic cancer were added to form a cohort of 70 patients for the study. Clinicopathological data were analysed for all 70 patients. Specimens from 30 patients, including the longest survivors, underwent independent pathologic re‐review (by SY) and were reaffirmed to have histology consistent with pancreatic ductal adenocarcinoma. Only patients with an adequate follow up interval (that is, ⩾36 months or until expiration if follow up was ⩽36 months) were selected. Patients were excluded if the final permanent section of the pancreatic transection or retroperitoneal surgical resection margin was histologically positive by H&E (R1 resection; that is, microscopic evidence of invasive ductal adenocarcinoma), if a surgical margin was unavailable for analysis, or if the patient had expired within 30 postoperative days. Therefore, only R0 resections (that is, absence of microscopic invasive cancer cells in the margins) were included. None of the additional 47 patients had these exclusion criteria.
All patients, regardless of stage or lymph node status, were offered adjuvant radiotherapy and chemotherapy with various combinations of 5‐fluorouracil, leucovorin, mitomycin C, dipyridamole, and gemcitabine at the discretion of their individual physicians. Institutional Review Board approval was obtained at the respective institutions (JWCI and UCLA) for the purposes of this study. Patient records, including radiographic films (when available), were reviewed. Patient demographics and clinicopathological factors are shown in table 1.
Table 1 Patient demographics and clinicopathological factors.
| Clinicopathological factor | Patients (n (%) |
|---|---|
| Total No of patients | 70 |
| Males | 35 (50%) |
| Females | 35 (50%) |
| Age (y) | |
| Median (range) | 67 (42–90) |
| Surgical procedures | |
| Pancreaticoduodenectomy | 68 (97%) |
| Distal pancreatectomy | 2 (3%) |
| UICC/TNM stage* | |
| pI† | 26 (37%) |
| pII† | 44 (63%) |
| Primary tumour | |
| T1 | 16 (23%) |
| T2 | 46 (66%) |
| T3 | 8 (11%) |
| Lymph node metastasis | |
| N0 | 30 (43%) |
| N1 | 40 (57%) |
| Pathological grade | |
| Well | 11 (16%) |
| Moderate | 33 (47%) |
| Poor | 26 (37%) |
| Tumour size (cm) | |
| 0–2 | 20 (28%) |
| >2 | 50 (72%) |
| Perineural invasion | |
| Absent | 24 (34%) |
| Present | 46 (66%) |
| Lymphovascular invasion | |
| Absent | 52 (74%) |
| Present | 18 (26%) |
*UICC/TNM 6th edition.
†pI, pII, pathological stage I and pathological stage II.
Pancreatic cancer specimens
Uniform methods to assess and process surgical margins (pancreatic transection and retroperitoneal) were not implemented as standard practice. In the current study, the surgical margins were evaluated and processed by different methods at the two institutions (JWCI and UCLA). At JWCI, pancreatic transection and retroperitoneal margins were submitted as separate tissue specimens by the surgeons and analysed by frozen section analysis. Once analysis was completed, the frozen sections were paraffin embedded and stored. At UCLA, only pancreatic transection margins were routinely evaluated by frozen section; retroperitoneal margins were evaluated only on final permanent section. Once the retroperitoneal margin was identified (by ink or suture or in relation to the location of the primary tumour), a perpendicular section to include the pancreatic tumour and margin was procured and paraffin embedded as a single retroperitoneal margin block. Each surgical margin was submitted in a separate block. In rare cases, more than one margin was submitted (for example, patients whose original transection margin was positive for invasive carcinoma). In these cases all blocks were clearly submitted and labelled, and only the final negative margin was analysed.
At both institutions, the tissue immediately adjacent to the superior mesenteric artery was generally considered the retroperitoneal margin. For the two patients with distal pancreatectomy, we defined the retroperitoneal margin as the pancreatic tissue radial from the primary tumour extending to the posterior peripancreatic soft tissues. Inclusion criteria for this study required the use of only paraffin embedded surgical margin tissues that were clearly marked and stored as the margins (except for the two patients who underwent distal pancreatectomy).
The archived H&E margin slide sections of the study cohort were reviewed under microscopy (by SMD) to confirm the absence of invasive carcinoma cells in the surgical margins (pancreatic transection and retroperitoneal). These margins were designated as “negative”. However, many surgical margins had evidence of low grade pancreatic intraepithelial neoplasia (PanIN; 1A and 1B) and inflammatory changes; it was generally not reported as a pathological finding in the surgical pathology reports. From available data, high grade PanIN (2 and 3) was identified in 10 patients (table 2). Detection of PanIN‐3 was, by definition, not considered “positive” for invasive cancer in the margins; none of the patients underwent additional resection for its clearance. However, two patients did require additional resection for the presence of invasive cancer cells in the pancreatic transection margin. Additional pathological abnormalities were not characterised in the margin specimens. Immunohistochemistry (IHC) was not utilised to re‐evaluate the surgical resection margins.
Table 2 Comparison of clinicopathological factors for patients with K‐ras mutation negative and positive margins.
| Clinicopathological factor | K‐ras mutation in margin | *p Value | |
|---|---|---|---|
| Negative (n = 33) | Positive (n = 37) | ||
| Age (y) | 0.075 | ||
| Mean (SD) | 65 (9) | 69 (11) | |
| Sex | 1.0 | ||
| Female | 17 (52%) | 18 (49%) | |
| Male | 16 (48%) | 19 (51%) | |
| UICC/TNM stage† | 0.33 | ||
| pI‡ | 10 (30%) | 16 (43%) | |
| pII‡ | 23 (70%) | 21 (57%) | |
| Primary tumour | 0.17 | ||
| T1 | 11 (33%) | 5 (14%) | |
| T2 | 19 (58%) | 27 (72%) | |
| T3 | 3 (9%) | 5 (14%) | |
| Lymph node metastasis | 0.15 | ||
| N0 | 11 (33%) | 19 (51%) | |
| N1 | 22 (67%) | 18 (49%) | |
| Tumour size (cm) | 0.20 | ||
| 0–2 | 12 (36%) | 8 (22%) | |
| >2 | 21 (64%) | 29 (78%) | |
| Pathological grade | 0.037 | ||
| Well | 3 (9%) | 8 (22%) | |
| Moderate | 21 (64%) | 12 (32%) | |
| Poor | 9 (27%) | 17 (46%) | |
| Perineural invasion | 0.024 | ||
| Absent | 16 (48%) | 8 (22%) | |
| Present | 17 (52%) | 29 (78%) | |
| Lymphovascular invasion | 0.027 | ||
| Absent | 29 (88%) | 23 (62%) | |
| Present | 4 (12%) | 14 (38%) | |
| PanIN in margin¶ | 1.0 | ||
| Absent | 1 (7%) | 2 (12.5%) | |
| Present | 13 (93%) | 14 (87.5% | |
| 1 | 9 | 8 | |
| 2–3 | 4 | 6 | |
| K‐ras mutation in tumour | 0.36 | ||
| Absent | 8 | 25 | |
| Present | 5 | 32 | |
*Comparison for age was performed by the Student's t test; the remaining clinical factors were compared by Fisher's exact test.
†UICC/TNM 6th edition.
‡pI and pII, pathological stage I and pathological stage II.
¶For patients with available pancreatic intraepithelial neoplasia data.
From the paraffin embedded primary tumour and margins, 30 μm sections (10 μm×3) were cut from each block and collected in sterile microcentrifuge tubes (Eppendorf Biopur, Westbury, New York, USA). After procurement of paraffin sections for polymerase chain reaction (PCR) analysis, an additional section (5 μm) from the block was cut and stained by H&E and examined by a surgical pathologist to further confirm the absence of invasive carcinoma cells in the resection margins. The entire 30 μm of sections cut from the paraffin embedded tissue blocks were first deparaffinised with xylene and then washed with 100% ethanol. DNA was extracted and purified from these paraffin sections using a modified assay (QIAamp DNA Mini Kit, Qiagen Inc., Valencia, California, USA), as previously described.21,22 DNA was quantified by the PicoGreen assay. Due to the protocol for sampling retroperitoneal margins at UCLA, in some cases carcinoma was present within the pancreas but was not present at the inked retroperitoneal margin. In these cases, paraffin embedded sections (4×5 μm) were cut, placed on microscope slides, and histologically benign appearing tissue was microdissected with laser capture microdissection (Arcturus, Mountain View, California, USA), as previously described.23 Due to the logistics of the study, IHC and PCR could not be performed on the same paraffin embedded sections. PCR analysis of all primary tumour and surgical resection margins was performed at JWCI.
K‐ras mutation in pancreatic cancer cell lines
Established pancreatic cancer cell lines (MIA PaCa2, PANC‐1, Hs 766T, and B×PC‐3) were obtained from the American Type Culture Collection (Manassas, Virginia, USA) and cultured as recommended. MIA PaCa2 and PANC‐1 have K‐ras gene mutations (GGT→TGT and GGT→GAT, respectively) at codon 12 whereas Hs 766T and B×PC‐3 have wild‐type (wt) K‐ras. These cell lines were used as positive and negative cancer controls for the PCR assay. Mutations in codons 13 and 61 were not assessed because of the overwhelming predominance of codon 12 mutations in pancreatic cancer.24,25 All cells were incubated at 37°C with 5% CO2. DNA from cell lines was extracted, isolated, and purified using DNAZol (Molecular Research Center Inc., Cincinnati, Ohio, USA) and then quantified as previously described.26
Primers and probes and PCR assay
Because of the difficulties in detecting a small number of K‐ras mutant copies among thousands of copies of wtK‐ras, detection of the K‐ras gene mutation was performed using a peptide nucleic acid (PNA) quantitative real time PCR assay which was previously established to specifically detect K‐ras mutation at codon 12 in paraffin embedded tissue sections.27 The PNA clamp, which has higher binding affinity for DNA than PCR primers, was designed for complementary hybridisation to the wtK‐ras allele.28 By hybridising to the wtDNA template, it inhibits annealing of the partially overlapping reverse primer and thus inhibits the amplification of the wtK‐ras. Because the PNA/DNA hybrid is unstable due to base‐pair mismatch, it does not anneal to and inhibit the amplification of mutant K‐ras. The high sensitivity of the PCR assay to detect micrometastases with K‐ras mutation among normal cells bearing the wtK‐ras allele has been previously demonstrated.27 The PCR assay was analysed and expressed as binary (+/−) values.
Quantitative real time PCR was performed using the following primers: K‐ras, 5′‐CGC TCA CTG CGC TCA ACA C‐3′ (forward) and 5′‐TCA GGC GGC CGC ACA CCT‐3′ (reverse); FRET probe, 5′‐FAM‐CAT TCT GTG CCG CTG AGC CG‐BHQ‐1‐3′; PNA, 5′‐TAC GCC ACC AGC TCC‐3′. The PCR assay was performed with the iCycler iQ RealTime PCR Detection System (Bio‐Rad Laboratories, Hercules, California, USA). Genomic DNA (20 ng) was amplified in a 20 μl reaction containing 1 μM of each primer, 1.75 μM PNA, 200 μM of each deoxynucleotide triphosphate, 4.0 mM MgCl2, 10× AmpliTaq Buffer, and 1 unit AmpliTaq Gold Polymerase (Applied Biosystems, Branchburg, New Jersey, USA). Each PCR reaction was subjected to 40 cycles at 94°C for 60 seconds, 70°C for 50 seconds, and 58°C for 50 seconds and 72°C for 60 seconds. PCR was also performed without PNA to amplify wtK‐ras and verify DNA integrity. Each sample was assayed in triplicate with positive and negative PCR controls.
K‐ras sequencing
K‐ras mutation was assessed initially on specimens using the PNA PCR assay. Representative K‐ras mutation positive and negative specimens (n = 16) were directly sequenced on both strands to confirm the accuracy of the PNA clamp method. A nested PCR assay was then performed. It was used to amplify K‐ras mutation so that sequencing could be performed from the paraffin embedded tissue sections. This assay approach was specifically designed to detect occult cancer cells with K‐ras mutation with minimal enrichment of mutant DNA. Then the following K‐ras primers were used: 5′‐GGT ACT GGT GGA GTA TTT GAT AGT G‐3′ (forward) and 5′‐TGG ATC ATA TTC GTC CAC AAA A‐3′ (reverse). Each PCR reaction mixture was initially heated to 94°C for 10 minutes and was then subjected to 32–40 cycles at 94°C for 30 seconds, 64°C for 30 seconds, and 72°C for 7 minutes. PCR products were purified with QIAquick PCR Product Purification Kit (Qiagen) and direct sequenced using DTCS Quick Start kit (Beckman Coulter; Fullerton, California, USA) with an annealing temperature of 58°C. Dye terminated products were precipitated by ethanol and separated by capillary array electrophoresis on a CEQ8000XL automated sequencer (Beckman Coulter).
Statistical analysis
Patient characteristics and detection of K‐ras mutation were summarised using mean, median, and frequency. Clinicopathological factors of patients with positive or negative K‐ras mutation were compared by Fisher's exact test and the Student's t test. Survival curves with respect to K‐ras mutation were constructed using Kaplan‐Meier's method. The log rank test was used to compare the equality of the two curves. Univariate analysis of prognostic factors included age, sex, stage, tumour extent, lymph node status, grade, tumour size, perineural invasion, and lymphovascular invasion. The presence of PanIN in the surgical margins was also assessed. The Cox proportional hazard regression model was used to evaluate the prognostic significance of K‐ras mutation when clinical prognostic factors were adjusted. A stepwise method was chosen for covariate selection. All analyses were performed using SAS (SAS/STAT User's Guide, version 8; SAS Institute Inc, Cary, North Carolina, USA) and tests were two sided and were considered significant when p values were ⩽0.05.
Results
Validation of PCR assay
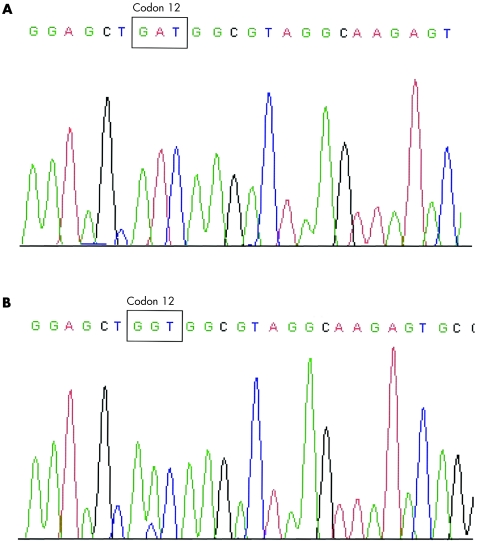
The accuracy and sensitivity of the PNA clamp method for detection of K‐ras mutation have been previously established.27 To validate the accuracy of the PNA quantitative real time PCR assay in our current study, we analysed 16 representative K‐ras mutation positive (n = 8) and negative (n = 8) paraffin embedded pancreatic cancer specimens. Representative sequences of K‐ras mutation are presented in fig 1. The wild‐type K‐ras DNA sequence at codon 12 is GGT.
Figure 1 Representative sequencing of pancreatic cancer tissues to validate polymerase chain reaction results. (A) Pancreatic cancer primary tumour with mutant K‐ras sequence (GGT→GAT). (B) Pancreatic cancer surgical resection margin specimen with mutant K‐ras sequence (GGT→GTT).
K‐ras mutation in patients with pancreatic cancer
Twenty three patients with pancreatic cancer were initially analysed as the pilot cohort. K‐ras mutation was detected in 19 of 23 (83%) primary tumours and in 11 of 23 (48%) surgical resection margins (pancreatic transection and/or retroperitoneal). The pancreatic transection and retroperitoneal margins were positive for K‐ras mutation in four and eight patients, respectively; both margins were positive in one patient. After analysis of this pilot cohort, an additional 47 patients were accrued and assessed. In assessment of all 70 patients, median survival was 21 months at a median follow up of 17 months. The five year overall survival rate was 19%. Patients were treated with pancreaticoduodenectomy (n = 68) or distal pancreatectomy (n = 2); no patient underwent total resection of the pancreas. One patient had segmental resection of the superior mesenteric‐portal vein confluence. At the time of analysis, 45 of 70 (64%) patients had succumbed to disease.
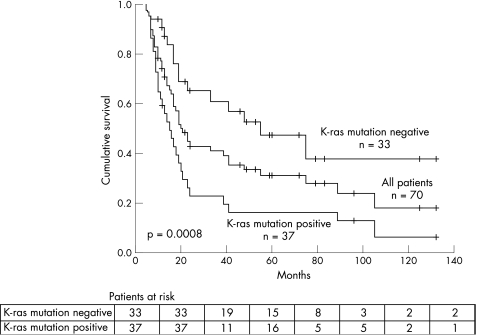
Overall, 57 of 70 (81%) patients had K‐ras gene mutation in the primary tumour. The mutation was detected in either margin (pancreatic transection and/or retroperitoneal) in 37 of 70 (53%) patients. K‐ras mutation was detected in the pancreatic transection (n = 17), retroperitoneal (n = 27), or both (n = 7) margins. Comparison of patients based on K‐ras mutation status of the surgical margins revealed a higher rate of perineural invasion, lymphovascular invasion, and poorly differentiated tumours, when K‐ras mutation was detected in the margin. There was no significant difference in the presence of K‐ras mutation in the margin by TNM classification, tumour size, age, sex, or presence of PanIN (table 2). Kaplan‐Meier curves showed a significant difference in overall survival for patients with K‐ras mutation positive versus negative surgical margins (median 15 v 55 months, respectively; log rank, p = 0.0008) (fig 2).
Figure 2 Kaplan‐Meier curves comparing overall survival between patients with K‐ras mutation positive and negative surgical margins.
By univariate analysis, K‐ras mutation in the surgical margins, grade, and perineural invasion were significant factors for disease outcome (table 3). When clinicopathological factors were adjusted, multivariate analysis identified K‐ras mutation in the surgical margins as a significant prognostic factor for poor survival (hazard ratio (HR) 2.8 (95% confidence interval (CI) 1.4–5.5); p = 0.004) (table 3). Tumour grade and perineural invasion were also significant for poor survival (HR 6.7 (95% CI 2.4–18.5), p = 0.0001; and HR 2.1 (95% CI 1.0–4.2), p = 0.04, respectively).
Table 3 Univariate and multivariate analyses.
| Death/n | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| HR (95% CI) | p Value | HR (95% CI) | p Value | ||
| Age (y) | 0.41 | NS | |||
| ⩽70 | 27/41 (66%) | 1.0 | |||
| >70 | 18/29 (62%) | 1.3 (0.7–2.4) | |||
| Sex | 0.63 | NS | |||
| Female | 22/35 (63%) | 1.0 | |||
| Male | 23/35 (66%) | 1.2 (0.6–2.1) | |||
| UICC/TNM stage* | 0.59 | NS | |||
| pI† | 17/26 (65%) | 1.0 | |||
| pII† | 28/44 (64%) | 1.2 (0.6–2.2) | |||
| Tumour extent | 0.28 | NS | |||
| T1 | 9/16 (56%) | 1.0 | |||
| T2 | 32/46 (70%) | 1.5 (0.7–3.2) | |||
| T3 | 4/8 (50%) | 0.8 (0.2–2.6) | |||
| Lymph node disease | 0.23 | NS | |||
| N0 | 18/30 (60%) | 1.0 | |||
| N1 | 27/40 (68%) | 1.5 (0.8–2.7) | |||
| Tumour size (cm) | 0.47 | NS | |||
| 0–2 | 12/20 (60%) | 1.0 | |||
| >2 | 33/50 (66%) | 1.3 (0.7–2.5) | |||
| Tumour grade | |||||
| Well | 6/11 (55%) | 1.0 | 0.0001 | 1.0 | 0.0001 |
| Moderate | 16/33 (48%) | 1.4 (0.5–3.5) | 2.6 (0.9–7.3) | ||
| Poor | 23/26 (88%) | 5.0 (1.9–13.0) | 6.7 (2.4–18.5) | ||
| Perineural invasion | |||||
| No | 13/24 (54%) | 1.00 | 0.03 | 1.0 | 0.04 |
| Yes | 32/46 (70%) | 2.2 (1.1–4.2) | 2.1 (1.0–4.2) | ||
| Lymphovascular invasion | |||||
| No | 33/52 (63%) | 1.0 | NS | NS | |
| Yes | 12/18 (67%) | 1.5 (0.8–2.9) | |||
| K‐ras in margins | |||||
| No | 15/33 (45%) | 1.00 | 0.0013 | 1.0 | 0.004 |
| Yes | 30/37 (81%) | 2.8 (1.5–5.3) | 2.8 (1.4–5.5) | ||
| PanIN in margins‡ | |||||
| No | 1/3 | 1.0 | 0.67 | NS | |
| Yes | 11/27 | 0.72 (0.16–3.2) | |||
| K‐ras in tumour | |||||
| No | 11/13 (85%) | 1.0 | 0.15 | NS | |
| Yes | 34/57 (60%) | .60 (.30–1.2) | |||
*UICC/TNM 6th edition.
†pI and pII, pathological stage I and pathological stage II.
‡For patients with available pancreatic intraepithelial neoplasia data.
HR (95% CI), hazard ratio (95% confidence interval).
Discussion
The value of obtaining histologically negative surgical resection margins by H&E has been established for pancreatic cancer.29,30 Neoptolemos et al identified a six month survival advantage for patients with negative compared with positive surgical margins.29 Our study delineates further this subgroup of patients with H&E negative surgical margins into those with PCR negative and positive surgical margins. Using a quantitative PNA PCR approach, we detected K‐ras mutation in the surgical resection margins of 37 of 70 (53%) patients, all of whom had negative margins by H&E. Ohigashi et al have also detected K‐ras mutation in retroperitoneal tissues of patients with pancreatic cancer.31 Here we correlated K‐ras margin status with disease outcome and demonstrated a significant difference in overall survival (fig 2). These findings therefore suggest that the current techniques to determine the adequacy of surgical margins may not be sensitive to identify relevant genetic aberrancies which may be the surrogates of unrecognised disease. Molecular‐genetic aberrancies in the surgical margins may be indicative of field cancerisation or occult cancer cells, both of which appear benign under the microscope.6,7,8,9,10,11,12,32 Further investigation will be necessary to identify the specific occult cells harbouring these aberrancies.
To correlate PCR detection of K‐ras mutation with clinical cancer recurrence, we reviewed radiographic reports and interventional studies for patients with positive K‐ras mutation in the margins and poor survival. Because most patients were referred from outside institutions, radiographic data was, for the most part, unavailable and we were unable to determine disease free intervals for the entire cohort. However, in nine patients with positive K‐ras mutations in the surgical margins and poor survival, radiographic imaging studies and peritoneal cytology revealed local recurrence (n = 6) or malignant ascites (n = 3), respectively. In studies of patients with head and neck cancers, Brennan et al have provided evidence to support the implications of our study.8 They found that patients whose histologically negative surgical margins harboured p53 mutation had recurrence at those margins and had worse disease outcome than patients without such genetic aberrancies.
K‐ras mutation was chosen for this study because of its frequent occurrence in pancreatic cancer and its potential pathogenetic role.13,14,15,16 Although recent evidence demonstrates that K‐ras gene mutation may be an essential precursor of pancreatic malignancy, there are reports of K‐ras mutation in benign pancreatic disorders.13,33,34 Moreover, K‐ras mutation has been detected in PanIN lesions, which were mostly present in the surgical margins of this study cohort.35 It is therefore feasible that detection of K‐ras mutation may be a surrogate feature of PanIN. However, there are no current reports that propose or identify a clinical significance for PanIN in the surgical margins. At the participating institutions of this study, detection of any grade PanIN in a surgical margin was designated as a histopathologically “negative” margin; however, detection of PanIN‐3 generally prompted consideration for further surgical resection.
K‐ras mutation in the surgical margins could be a reflection of tumours with more aggressive biology. The prognostic significance of grade and perineural invasion by multivariate analysis correlates with the high number of PCR positive retroperitoneal margins (table 3). Differences in tumour grade, perineural invasion, and K‐ras mutation in the margins were manifest as a 40 month survival advantage in the K‐ras mutation negative group. Median survival of 55 months for the K‐ras mutation negative patients may seem indiscriminately high; however, this survival figure is derived from the entire cohort which had an overall median survival of 21 months and a five year overall survival rate of 19%. These outcomes are consistent with survival data from large prospective and retrospective studies.29,30,36 Furthermore, prolonged survival is not uncommon for a small percentage of patients with pancreatic cancer. This has been reported from both large national cancer registries and smaller retrospective reviews.1,37,38,39 Our findings specifically relate to PCR positivity and negativity in R0 margins. We acknowledge that the power of our study appears to be limited inasmuch as univariate and multivariate analyses did not find lymph node metastasis and tumour size to be prognostic factors for survival, despite the fact that 40 of 70 (57%) patients had lymph node disease and mean tumour size was >2 cm. These factors have been found to be significant predictors of prognosis in other studies.29,30,36
Although unrecognised disease in the surgical margins may occur in many cancers, the anatomical limitations of the pancreas make it particularly problematic to determine whether wider PCR negative surgical margins could affect outcome.6,7 Even when total pancreatectomy has been performed for pancreatic cancer, survival data are no better, an outcome which, perhaps, can be explained by our high number of K‐ras mutation positive retroperitoneal margins.40 Adjuvant therapy may appear attractive when surgical margins are PCR positive but a recent large randomised controlled trial demonstrated no survival advantage in patients receiving radiation therapy.41 However, a meta‐analysis of chemotherapy has demonstrated a survival advantage.42 Furthermore, the development of promising new therapeutic agents provides a potential avenue of treatment for high risk patients, as defined by the detection of genetic mutation in surgical resection margins.
Expansion of genotypically altered benign or malignant cells has important clinical consequences. We have demonstrated in patients with pancreatic cancer that detection of K‐ras mutation in the surgical margins may represent unrecognised disease and correlates strongly with clinical outcomes. We are currently investigating whether additional genetic or epigenetic aberrancies are present in the margin tissues. We have relied on PCR assays to detect such defects because sensitive and efficient diagnostic antibodies are still lacking. A recent study by Bogoevski et al further demonstrates the importance of molecular techniques in identifying occult spread of cancer cells in pancreatic cancer.43 Here we present an argument to implement measures to assess surgical resection margins beyond the standard H&E techniques. Our study findings have direct and immediate clinical implications for pathological staging and prognostic evaluation of patients undergoing surgery for pancreatic cancer. We suggest that molecular‐genetic evaluation of surgical margins should be considered to better define “negative” surgical margins. Moreover, such characterisation of surgical resection margins can provide valuable data that may potentially lead to treatment strategies to alter outcomes in patients undergoing surgery for pancreatic cancer.
Abbreviations
JWCI - John Wayne Cancer Institute
H&E - haematoxylin and eosin
PanIN - pancreatic intraepithelial neoplasia
IHC - immunohistochemistry
PNA - peptide nucleic acid
PCR - polymerase chain reaction
wt - wild‐type
HR - hazard ratio
Footnotes
Supported by Harold J McAlister Charitable Foundation, Los Angeles, California, USA; Martin H Weil Research Laboratories, John Wayne Cancer Institute, Santa Monica, California, USA; and the Hirshberg Foundation for Pancreatic Cancer Research, UCLA School of Medicine, Los Angeles, California, USA. These study sponsors had no role in: study design; collection, analysis, and interpretation of data; writing of the report; or the decision to submit the paper for publication.
Conflict of interest: None declared.
References
- 1.Jemal A, Murray T, Ward E.et al Cancer statistics, 2005. CA Cancer J Clin 20055510–30. [DOI] [PubMed] [Google Scholar]
- 2.Evans D B, Lee J E, Pisters P W T. Periampullary cancer. In: Cameron JL, ed. Current surgical therapy. St Louis: Mosby, 2001558
- 3.Westerdahl J, Andren‐Sandberg A, Ihse I. Recurrence of exocrine pancreatic cancer—local or hepatic? Hepatogastroenterology 199340383–387. [PubMed] [Google Scholar]
- 4.Griffin J F, Smalley S R, Jewell W.et al Patterns of failure after curative resection of pancreatic carcinoma. Cancer 19906656–61. [DOI] [PubMed] [Google Scholar]
- 5.Willett C G, Lewandrowski K, Warshaw A L.et al Resection margins in carcinoma of the head of the pancreas. Implications for radiation therapy. Ann Surg 1993217144–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tabor M P, Brakenhoff R H, Ruijter‐Schippers H J.et al Genetically altered fields as origin of locally recurrent head and neck cancer: a retrospective study. Clin Cancer Res 2004103607–3613. [DOI] [PubMed] [Google Scholar]
- 7.Braakhuis B J, Tabor M P, Kummer J A.et al A genetic explanation of Slaughter's concept of field cancerization: evidence and clinical implications. Cancer Res 2003631727–1730. [PubMed] [Google Scholar]
- 8.Brennan J A, Mao L, Hruban R H.et al Molecular assessment of histopathological staging in squamous‐cell carcinoma of the head and neck. N Engl J Med 1995332429–435. [DOI] [PubMed] [Google Scholar]
- 9.Deng G, Lu Y, Zlotnikov G.et al Loss of heterozygosity in normal tissue adjacent to breast carcinomas. Science 19962742057–2059. [DOI] [PubMed] [Google Scholar]
- 10.Masasyesva B G, Tong B C, Brock M V.et al Molecular margin analysis predicts local recurrence after sublobar resection of lung cancer. Int J Cancer 20051131022–1025. [DOI] [PubMed] [Google Scholar]
- 11.Guo M, House M G, Hooker C.et al Promoter hypermethylation of resected bronchial margins: a field defect of changes? Clin Cancer Res 2004105131–5136. [DOI] [PubMed] [Google Scholar]
- 12.Nathan C A, Franklin S, Abreo F W.et al Analysis of surgical margins with the molecular marker eIF4E: a prognostic factor in patients with head and neck cancer. J Clin Oncol 1999172909–2914. [DOI] [PubMed] [Google Scholar]
- 13.Hingorani S R, Petricoin E F, Maitra A.et al Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 20034437–450. [DOI] [PubMed] [Google Scholar]
- 14.Hirai H, Okabe T, Anraku Y.et al Activation of the c‐K‐ras oncogene in a human pancreas carcinoma. Biochem Biophys Res Commun 1985127168–174. [DOI] [PubMed] [Google Scholar]
- 15.Almoguera C, Shibata D, Forrester K.et al Most human carcinomas of the exocrine pancreas contain mutant c‐K‐ras genes. Cell 198853549–554. [DOI] [PubMed] [Google Scholar]
- 16.Smit V T, Boot A J, Smits A M.et al KRAS codon 12 mutations occur very frequently in pancreatic adenocarcinomas. Nucleic Acids Res 1988167773–7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abbruzzese J L, Evans D B, Raijman I.et al Detection of mutated c‐Ki‐ras in the bile of patients with pancreatic cancer. Anticancer Res 199717795–801. [PubMed] [Google Scholar]
- 18.Caldas C, Hahn S A, Hruban R H.et al Detection of K‐ras mutations in the stool of patients with pancreatic adenocarcinoma and pancreatic ductal hyperplasia. Cancer Res 1994543568–3573. [PubMed] [Google Scholar]
- 19.Tada M, Omata M, Ohto M. Clinical application of ras gene mutation for diagnosis of pancreatic adenocarcinoma. Gastroenterology 1991100233–238. [DOI] [PubMed] [Google Scholar]
- 20.Sorenson G D. Detection of mutated KRAS2 sequences as tumor markers in plasma/serum of patients with gastrointestinal cancer. Clin Cancer Res 200062129–2137. [PubMed] [Google Scholar]
- 21.Hoon D S, Spugnardi M, Kuo C.et al Profiling epigenetic inactivation of tumor suppressor genes in tumors and plasma from cutaneous melanoma patients. Oncogene 2004234014–4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujimoto A, Takeuchi H, Taback B.et al Allelic imbalance of 12q22–23 associated with APAF‐1 locus correlates with poor disease outcome in cutaneous melanoma. Cancer Res 2004642245–2250. [DOI] [PubMed] [Google Scholar]
- 23.Hoon D S, Fujimoto A, Shu S.et al Assessment of genetic heterogeneity in tumors using laser capture microdissection. Methods Enzymol 2002356302–309. [DOI] [PubMed] [Google Scholar]
- 24.Hruban R H, Wilentz R E, Kern S E. Genetic progression in the pancreatic ducts. Am J Pathol 20001561821–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kitago M, Ueda M, Aiura K.et al Comparison of K‐ras point mutation distributions in intraductal papillary‐mucinous tumors and ductal adenocarcinoma of the pancreas. Int J Cancer 2004110177–182. [DOI] [PubMed] [Google Scholar]
- 26.Spugnardi M, Tommasi S, Dammann R.et al Epigenetic inactivation of RAS association domain family protein 1 (RASSF1A) in malignant cutaneous melanoma. Cancer Res 2003631639–1643. [PubMed] [Google Scholar]
- 27.Taback B, Bilchik A J, Saha S.et al Peptide nucleic acid clamp PCR: a novel K‐ras mutation detection assay for colorectal cancer micrometastases in lymph nodes. Int J Cancer 2004111409–414. [DOI] [PubMed] [Google Scholar]
- 28.Faruqi A F, Egholm M, Glazer P M. Peptide nucleic acid‐targeted mutagenesis of a chromosomal gene in mouse cells. Proc Natl Acad Sci U S A 1998951398–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neoptolemos J P, Stocken D D, Dunn J A.et al Influence of resection margins on survival for patients with pancreatic cancer treated by adjuvant chemoradiation and/or chemotherapy in the ESPAC‐1 randomized controlled trial. Ann Surg 2001234758–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeo C J, Cameron J L, Sohn T A.et al Six hundred fifty consecutive pancreatico‐duodenectomies in the 1990s: pathology, complications, and outcomes. Ann Surg 1997226248–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohigashi H, Ishikawa O, Sasaki Y.et al K‐ras point mutation in the nerve plexuses around the superior mesenteric artery in resectable adenocarcinoma of the pancreatic head. Arch Surg 20001351450–1455. [DOI] [PubMed] [Google Scholar]
- 32.Slaughter D P, Southwick H W, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer 19536963–968. [DOI] [PubMed] [Google Scholar]
- 33.Luttges J, Schlehe B, Menke M A.et al The K‐ras mutation pattern in pancreatic ductal adenocarcinoma usually is identical to that in associated normal, hyperplastic, and metaplastic ductal epithelium. Cancer 1999851703–1710. [PubMed] [Google Scholar]
- 34.Rivera J A, Rall C J, Graeme‐Cook F.et al Analysis of K‐ras oncogene mutations in chronic pancreatitis with ductal hyperplasia. Surgery 199712142–49. [DOI] [PubMed] [Google Scholar]
- 35.Hruban R H, Takaori K, Klimstra D S.et al An illustrated consensus on the classification of pancreatic intraepithelial neoplasia and intraductal papillary mucinous neoplasms. Am J Surg Pathol 200428977–987. [DOI] [PubMed] [Google Scholar]
- 36.Balcom JH I V, Rattner D W, Warshaw A L.et al Ten‐year experience with 733 pancreatic resections: changing indications, older patients, and decreasing length of hospitalization. Arch Surg 2001136391–398. [DOI] [PubMed] [Google Scholar]
- 37.Cleary S P, Gryfe R, Guindi M.et al Prognostic factors in resected pancreatic adenocarcinoma: Analysis of actual 5‐year survivors. J Am Coll Surg 2005198722–731. [DOI] [PubMed] [Google Scholar]
- 38.Shimizu Y, Yasui K, Matsueda K.et al Small carcinoma of the pancreas is curable: new computed tomography finding, pathological study and postoperative results from a single institute. J Gastroenterol Hepatol 2005201591–1594. [DOI] [PubMed] [Google Scholar]
- 39.Allison D C, Piantadosi S, Hruban R H.et al DNA content and other factors associated with ten‐year survival after resection of pancreatic carcinoma. J Surg Oncol 199867151–159. [DOI] [PubMed] [Google Scholar]
- 40.Karpoff H M, Klimstra D S, Brennan M F.et al Results of total pancreatectomy for adenocarcinoma of the pancreas. Arch Surg 200113644–47. [DOI] [PubMed] [Google Scholar]
- 41.Neoptolemos J P, Stocken D D, Friess H.et al A randomized trial of chemoradiotherapy and chemotherapy after resection of pancreatic cancer. N Engl J Med 20043501200–1210. [DOI] [PubMed] [Google Scholar]
- 42.Stocken D D, Buchler M W, Dervenis C.et al Meta‐analysis of randomized adjuvant therapy trials for pancreatic cancer. Br J Cancer 2005921372–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bogoevski D, Yekebas E F, Schurr P.et al Mode of spread in the early phase of lymphatic metastasis in pancreatic ductal adenocarcinoma: prognostic significance of nodal microinvolvement. Ann Surg 2004240993–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]