Abstract
Background
Integration of human papillomavirus (HPV) DNA has been considered a late event in cervical carcinogenesis. However, integrated forms of HPV were recently detected in cancer precursor lesions using a new real time polymerase chain reaction (PCR) to detect the deletions at the 3362–3443 region of HPV16 E2
Objective
To study the frequency of HPV16 DNA integration in cervical lesions and compare the sensitivity of an additional upstream region of the E2 ORF (2962–3138) in detecting HPV integration.
Methods
Using the TaqMan based PCR, HPV16 positive DNA samples were analysed in 164 cervical scrapings from women participating in a multicentre screening trial. Biopsy confirmation was available in 62 cases.
Results
Primers targeting the 3362–3443 region detected the majority of E2 deletions. In only 23% of the samples was the E2 upstream region equal or better target than the 3362–3443 region. Mixed (episomal/integrated) pattern was the most prevalent physical state of HPV16, also present in PAP smears with normal morphology. Pure integrated form was most prevalent in HSIL and cancer lesions, but also detectable in low grade abnormalities (NSIL, ASC‐US, LSIL). Women with only integrated HPV16 were almost 10 years older than those with episomal HPV16. Viral load of integrated HPV16 was related to cytological abnormality (p = 0.003) but not to histology.
Conclusions
Integrated HPV16 is present in low grade cervical lesions, mostly mixed with the episomal form. Women with the pure integrated form of HPV16 are older than those with the other forms.
Keywords: HPV16, CIN, cervical cancer, real time PCR
High risk human papillomaviruses (HR‐HPV) are the single most important risk factor of cervical cancer.1,2 The most common oncogenic HPV type in squamous cervical cancer is HPV16, found in over 50% of the cases.3,4 Integration of viral DNA into the host cell DNA is considered to provide a selective growth advantage to the infected cells, because integration usually disrupts the E1 and E2 open reading frames (ORF), while the E6 and E7 ORFs and long control region (LCR) remain intact.5,6,7,8 The elimination of E2 protein expression results in upregulation of the transcription of E6 and E7 oncogenes.9 Also, high viral loads in cervical intraepithelial neoplasia (CIN) lesions are associated with a 60‐fold increase in the risk of malignant progression.10
Early studies on virus integration suggested that the HPV genome in most of the benign lesions and CIN1 lesions was in episomal form,5,6,11,12 in contrast to cervical cancers and their derivate cell lines, where viral DNA was integrated into the host genome in almost all cases.7,11,13,14 Until now, integration of HPV16 has been regarded as a late event in cervical carcinogenesis.1 This view is challenged, however, by the recent cell culture studies suggesting that integration might be an early event in this process.5,6,15
We have found convincing evidence of early integration in the HPV 33 positive cell line (UT‐DEC‐1), which was followed by a selection of the cells containing integrated virus.16 This change from episomal to integrated virus took place between passages 21 and 22. In another HPV positive cell line, W12—established from an HPV16 positive CIN1 lesion17—the HPV genome was first in episomal form but was later integrated and accompanied by an increase in the grade of the lesion.18
We recently developed a sensitive method for detecting small amounts of integrated HPV16 DNA using real time polymerase chain reaction (PCR).15 In this system, E2 and E6 ORFs of HPV16 are amplified simultaneously. The primers are targeted to nn 3362–3443 of E2, which seems to be a common HPV integration site in cervical cancers.19
In the present study, we analysed 164 HPV16 positive DNA samples derived from cervical scrapings of women with different cervical pathology. We also compared the sensitivity of an additional upstream site of HPV16 E2 (2962–3138) in detecting HPV integration. This site has also been shown to be interrupted during HPV integration.20
Methods
Subjects and study design
The HPV16 positive cervical samples for this study were obtained from 164 women participating in a cross sectional cohort study, run in three new independent states (NIS) of the former Soviet Union, enrolled by six outpatient clinics in Moscow, Novgorod (Russia), Minsk (Belarus), and Riga (Latvia).21,22 The members of the NIS study group are given in the appendix. The study design has been detailed in two recent papers.21,22 All women had three tests: (1) PAP smear; (2) PCR; (3) hybrid capture II, the latter completed from the same sample. Women with cytological abnormality consistent with HPV‐CIN (atypical squamous cells (ASC) or above) and those testing positive with HCII were referred for colposcopy and biopsy confirmation. All women who presented with low grade lesions were subjected to prospective follow up, while high grade lesions (CIN2 or higher) were promptly treated.21,22
DNA samples
The cervical samples were taken with the Digene Cervical Sampler for HCII testing as described earlier.21,22 The same samples were then processed for DNA extraction with high salt method by Miller et al.23 The first 1500 DNA samples were further analysed using a novel real time PCR method for HPV typing, as described recently.24 This system detects several different HPV types and also HPV copy numbers can be calculated.24 Amplification and detection was performed using a 7700 Sequence Detection System (Applied Biosystems, Foster City, California, USA).
Integration assay
A novel real time PCR method developed by our group15 was used to analyse the physical status of HPV16 in the samples. The amplification conditions, primers, and probes were otherwise as described earlier, but for the test of another region within the E2, a new primer pair E2B was designed targeting the 2962–3138 region of HPV16 E2 as given in table 1. This was prompted by the previous detection of frequent deletions uniquely at this site in HPV16 positive carcinomas.20
Table 1 HPV16 upstream E2‐B primers and probe used in real time polymerase chain reaction assay for integration.
| Physical state | Name | Sequence (5′→3′) | Tm | Primer site | Amplimer length (bp) |
|---|---|---|---|---|---|
| (°C) | |||||
| EpisomalA15 | Probe:16E2‐APRO | (BODIPYR6G)‐CACCCCGCCGCGACCCATA‐(DQ) | 70 | ||
| Primer1:16E2‐AFP | AACGAAGTATCCTCTCCTGAAATTATTAG | 59 | 3362–3390 | ||
| Primer2:16E2‐ARP | CCAAGGCGACGGCTTTG | 60 | 3427–3443 | 82 | |
| EpisomalB | Probe:16E2‐BPRO | AAGACGTTAGCCTTGAAGTGTATTTAACTGCACC | 67 | ||
| Primer1:16E2‐BFP | ATTACAAGCAATTGAACTGCAACTAACGTT | 59 | 2962–2991 | ||
| Primer2:16E2‐BRP | GTATTGCATATGTCTCCATCAAACTG | 58 | 3113–3138 | 177 |
The E2‐B system is located upstream of the previous E2‐A system15 at HPV16 genomic primer positions 3362–3390 and 3427–3443.
Tm was determined using the “Primer Express™” program, version 2.0 (PE Applied Biosystems).
bp, base pairs; FP, forward primer; PRO, probe; RP, reverse primer; Tm, melting temperature.
The sensitivities of the E2A and E2B systems were comparable, as seen in a dilution series of SiHa cell DNA (not shown). The E2A and E2B probes were labelled with BODIPY R6G (the BODIPY dyes used are under license to Scandinavian Gene Synthesis from Molecular Probes Inc) at the 5′ end and Dark Quencher at the 3′ end. The E6 probe was labelled with 6‐carboxyfluorescein (6‐FAM) at the 5′ end and Dark Quencher (Scandinavian Gene Synthesis AB, Koping, Sweden) at the 3′ end. The primer and probe concentrations, in a total volume of 50 μl, were 0.3 and 0.1 μM, respectively. Ten to 50 nanograms (50 ng was used when possible) of target DNA from cervical swabs were added to the reaction mixture. Two standard curves were obtained by amplification of a dilution series of 50 million to 500 copies of a clone of HPV16 in pBR322. Three no‐template control reaction mixtures were included in each run.
The results were recorded as copy numbers in 50 ng of cellular DNA. The integrated E6 was calculated by subtracting the copy numbers of the E2 (episomal) from the total copy numbers of E6 (episomal and integrated). The ratio of E2 to integrated E6 represents the amount of the episomal form in relation to the integrated form. When episomal and integrated forms coexist, the ratio of E2 to E6 should be less than 1.0. When only the episomal form is present, equivalent copy numbers of E6 and E2 should be detected and the ratio of E2 to E6 should be 1.0. When only integrated form is present, no E2 PCR signal should be detected.
Statistical analyses
Statistical analyses were carried out using the SPSS® computer software (SPSS for Windows, version 11.5). Frequency tables were analysed using the χ2 test, with the likelihood ratio (LR) being used to assess the significance of the correlation between the categorical variables. Differences in the means of continuous variables (for example, the viral load between the three categories of HPV physical state) were analysed using non‐parametric tests (Kruskal–Wallis). Odds ratios (OR) and 95% confidence intervals (CI) were calculated using the exact method. In all tests, probability (p) values <0.05 were regarded as statistically significant.
Results
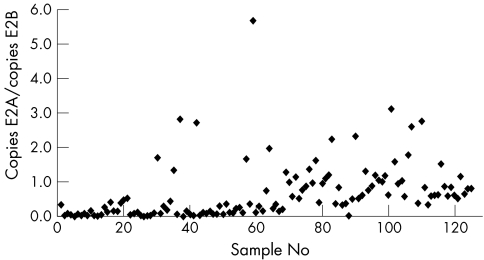
The ratio of copies E2A/E2B is shown in fig 1. The E2A/E2B ratio in 126 samples was below 1.0, indicating that the integration in the E2A region was more frequent than in the E2B region in these samples. In only 29 of 126 samples (23%) was E2B an equal or better marker of integration.
Figure 1 The individual ratios of copies of E2A and E2B in each of 124 clinical samples. The number of E2 copies was calculated for each fragment from the standard curves, obtained by a dilution series of a clone of the complete HPV16 genome.
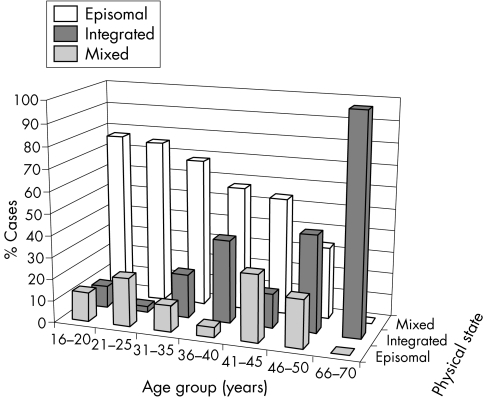
The physical state of HPV16 as related to PAP smear abnormality is summarised in table 2. A mixture of both episomal and integrated HPV16 was the most frequent pattern among these cases, with no statistical difference between the PAP smear categories (p = 0.056). The pure integrated form was mostly confined to high grade lesions, but noteworthy is its presence in samples with no signs of intraepithelial lesion and also in ASC‐US smears. Pure episomal forms were absent in all high grade smears.
Table 2 Physical state of HPV16 related to PAP smear abnormalities.
| PAP | Episomal | Integrated | Mixed |
|---|---|---|---|
| smear | (n/% total) | (n/% total) | (n/% total) |
| NSIL | 25/19.7 | 14/11.0 | 88/69.3 |
| ASC‐US | 0/0.0 | 2/18.2 | 9/81.8 |
| LSIL | 2/15.4 | 1/7.7 | 10/76.9 |
| HSIL | 0/0.0 | 0/0.0 | 6/100 |
| SCC | 0/0.0 | 3/42.9 | 4/57.1 |
| Total | 27/16.5 | 20/12.2 | 117/71.3 |
p = 0.056 between smear categories (χ2, LR statistics).
ASC‐US, atypical squamous cells of undetermined significance; HSIL, high grade squamous intraepithelial lesion; LSIL, low grade squamous intraepithelial lesion; NSIL, no sign of intraepithelial lesion; SCC, squamous cell carcinoma.
Table 3 relates the HPV16 physical state to the lesion grade in cervical biopsy. Integrated HPV16 is a good marker of high grade CIN and squamous cell cancer, never present in pure form in low grade lesions. Again, the mixed episomal/integrated pattern was the most frequent, and importantly, found in several cases of HPV‐NCIN and CIN1 lesions as well. Overall, the distribution of the three patterns among the biopsy categories was not statistically significant (p = 0.380).
Table 3 Physical state of HPV16 related to grade of cervical lesion.
| Lesion | Episomal | Integrated | Mixed |
|---|---|---|---|
| grade | (n/% total) | (n/% total) | (n/% total) |
| Normal | 1/14.3 | 0/0 | 6/85.7 |
| NCIN | 2/28.6 | 0/0 | 5/71.4 |
| CIN I | 4/28.6 | 0/0 | 10/71.4 |
| CIN II | 4/28.6 | 0/0 | 10/71.4 |
| CIN III | 1/7.7 | 1/7.7 | 11/84.6 |
| SCC | 0/0.0 | 2/33.3 | 4/66.7 |
| Total | 12/19.4 | 3/4.8 | 47/75.8 |
p = 0.380 between lesion grades (χ2, LR statistics).
CIN, cervical intraepithelial neoplasia; NCIN, HPV without CIN; SCC, squamous cell carcinoma.
The viral load of integrated HPV16 is related to PAP smear abnormality in table 4. The differences in viral loads between the five PAP smear categories were highly significant (p = 0.003; Kruskal–Wallis test), in that the values were lowest among NSIL cases, and highest among ASC‐US cases. Using biopsy as the reference, the difference in copy numbers was not significant (p = 0.274) (data not shown). The viral load of integrated HPV16 was higher in patients with mixed form of HPV16 than in those with pure integrated form of HPV16 (mean log_number = 16.48 (95% CI, 15.81 to 17.15) v 11.58 (10.17 to 13.00) in 50 ng of cellular DNA).
Table 4 Number of integrated copies of HPV16 related to epithelial abnormalities.
| PAP smear | Mean log_number* | No of cases |
|---|---|---|
| of integrated copies | ||
| NSIL | 14.93 | 60 |
| ASC‐US | 18.84 | 42 |
| LSIL | 17.73 | 11 |
| HSIL | 16.61 | 11 |
| SCC | 18.33 | 6 |
*In logarithmic scale; p = 0.003 (Kruskal–Wallis).
Figure 2 gives the age distribution of the women in the three categories of HPV16 physical state. The mean age of the women who had a purely integrated form of HPV16 was significantly greater than in the two other categories (p = 0.004, Kruskal–Wallis; p = 0.009, analysis of variance).
Figure 2 Age of the patients related to the physical state of HPV16.
Discussion
Because of its several advantages, quantitative real time PCR has been widely applied to study HPV in cervical carcinomas and other lesions.15,25,26,27,28,29,30 With new modifications in our real time PCR, we compared the previously described E2A method with a new upstream E2B system. Integration was more frequent in the E2A region than in the E2B region. Although a minor proportion of our samples showed deletions in the E2B region only, these data advocate the use of the previously described E2A system in the primary screening for E2 deletions in clinical samples. Our results are in agreement with a recent study by Kalantari et al, who showed that this specific region of the E2 was also deleted in the majority of cervical carcinomas.19
We have defined integration by the lack of E2 amplification with our novel real time PCR method. However, we must take into account that E2 cannot be amplified when there are mutations or deletions in the E2 region targeted by the selected primers. Another form of integration has also been described.31 In this integration event, multiple copies of HPV integrate in a head to tail tandem repeat and only the viral copy flanking the cellular DNA is interrupted in the E1 or E2 region, leaving the internal copies with intact E1 and E2 ORFs. This is the case in the cancer cell line CaSki.32
HPV integration into the host cell genome has been suggested to give a selective growth advantage to cells carrying mainly integrated virus as compared with those carrying episomal virus.16,25 In our series, purely episomal HPV16 was typically associated with negative or LSIL smears, and was never found in HSIL or cancer. However, negative and LSIL smears also contained mixed forms of episomal/integrated and pure integrated forms. These results are in contrast to many earlier reports which have mostly found only episomal HPV16 DNA in the normal or LSIL smears.33,34,35 Recently, Gallo et al36 and do Horto et al37 reported early integration of HPV16 DNA in 54% and 43% of LSIL cases, respectively. As in our study, Andersson and coworkers38 found mixed forms of HPV16 in CIN1 cases using our method. The results presented here are also consistent with recent data on HPV positive cell lines derived from low grade lesions.16,17 Detection of purely episomal form in the early passage cells and integrated in the late passage cells suggests a selection phase towards a more malignant phenotype in the late passages. This is consistent with the present observations, where HPV16 in pure episomal form was never found in HSIL or in cancer smears.
In our study, there were fewer HSIL/CIN3 and SCC samples with the pure integrated form of HPV16 than with the mixed form of HPV DNA compared with some earlier studies.14,34 One reason for this could be that we had only 15 samples with higher grade cytological and histological abnormalities. Also when collecting the cervical samples, the episomal signal could arise from cells that are not yet malignant. In cases of multiple infection, it is difficult to determine which virus is responsible for the cellular changes detected. To first identify the HPV16 positive samples derived from the 1500 women, we used an HPV testing method that detects 10 different HPV types.24 As multiple infections are known to be common, we also detected HPV types 18/45, 31, 33/52/58/67, 35, and 39 in some of the HPV16 positive samples selected for this study (data not shown).
Take home messages
Integration of human papillomavirus (HPV) DNA is present in low grade cervical lesions, mainly in older women.
The viral load of integrated HPV16 is related to cytological abnormality.
In a series of 62 histological biopsies, HPV16 integration was present in many of the early lesions (HPV‐NCIN and CIN1), and even in morphologically normal epithelium. However, HPV16 integration in these low grade cases was invariably as a mixed episomal/integrated form. This is substantiated by our observations on the cell lines,16 and this absence of purely integrated HPV16 in low grade lesions could implicate a similar “selection period” as found in the cell lines. Still present in one of the CIN3 lesions, the pure episomal HPV16 had disappeared when the lesions had progressed to invasive cancer. These data are in agreement with the report by Nagao et al,39 who showed an increase of integrated and mixed forms of HPV16 in parallel with the disease progression. In all studies, where HPV integration is studied in cellular samples and the results compared with the biopsy data, the possibility cannot be excluded that the biopsy does not represent the most severe lesion. In this respect, use of PAP smears as a reference might be more appropriate, and finding HSIL in the PAP smear is a highly specific indicator of a true high grade lesion in the cervix.
Viral load has been proposed as a sensitive indicator for progression to HSIL.10 Ylitalo et al30 found a consistent increase of HPV16 load present 13 years before the diagnosis of carcinoma in situ. Other investigators have not confirmed such an increased risk of developing CIN3,40,41 while yet others report a dramatic increase in the copy number of HPV16 with increasing abnormality.42,43 In the present study, the difference in copy numbers between the five diagnostic PAP smear categories was highly significant, but the trend was not linear. Interestingly, the largest number of integrated copies was present in ASC‐US smears. This controversial category in the Bethesda classification contains a substantial number of productive and evidently early non‐productive HPV infections which have not yet developed to LSIL. Viral loads decreased in parallel with the increasing grade of SIL, but increased again in cancer cases. These data support our concept on viral events in cervical carcinogenesis, where the initial high viral loads would increase the probability of integration. After selection of the integrated cell clones that progress towards cancer (LSIL, HSIL), viral load could be lower again. According to this hypothesis we found that the viral loads were higher in samples with the mixed form of HPV16 than with the integrated form.
Our novel real time PCR for the downstream E2A region of the SiHa deletion also detects the majority of E2 deletions in clinical samples, as compared with the upstream E2B system.
The present data show that integration of HPV16 is an early event in cervical carcinogenesis, present in cervical samples with no detectable abnormalities in the PAP smear or biopsy. The sequence of events from episomal to purely integrated HPV16 is a long process. Women with integrated HPV16 were 10 years older than those with episomal HPV. It has been suggested that the median incubation period from HPV infection to CIS in HPV16 positive women is 7–12 years.44 In the same study, a 30‐fold increased risk of CIS was ascribed to persistent HPV16 infections, compared with an occasional infection.44 During the incubation period, the virus is first assumed to integrate, after which a selection of integrated cell clones arises from a mixture of cells with both episomal and integrated HPV.
Acknowledgements
This study was supported by the INCO‐Copernicus Programme of the European Commission (Contract No ERB IC15‐CT98‐0321) and the Swedish Cancer Society (to UG). Special thanks to Digene Europe (London, UK), for providing the Hybrid Capture analyser, samplers, and test kits at our disposal, to Quantovir for providing the reagents for determining HPV titres, and to Niina Wahlroos, Tatjana Peskova, Inger Gustavsson, and Martin Moberg for their skilful technical assistance.
Abbreviations
ASC - atypical squamous cells
ASC‐US - atypical squamous cells of undetermined significance
CIN - cervical intraepithelial neoplasia
HPV - human papillomavirus
HSIL - high grade squamous intraepithelial lesion
LCR - long control region
LSIL - low grade squamous intraepithelial lesion
NSIL - no sign of intraepithelial lesion
ORF - open reading frame
Appendix
THE NIS COHORT STUDY GROUP
V Kozachenko, J Podistov, O Ivanchenko, S Zakharenko, R Nerovjna, L Kljukina, M Branovskaja, V Grunberga, A Juschenko, R Santopietro, T Zakharova, J Pajanidi, G Chemeris, L Sozaeva, E Lipova, I Tsidaeva, A Pshepurko, O Erokhina, M Nikitina, A Grunberg, M Cintorino.
References
- 1.zur Hausen H. Papillomaviruses causing cancer: evasion from host‐cell control in early events in carcinogenesis. J Natl Cancer Inst 200092690–698. [DOI] [PubMed] [Google Scholar]
- 2.Syrjänen K, Syrjänen S. Papillomavirus infections. In: Human pathology. Chichester: J Wiley & Sons, 20001–615.
- 3.Bosch F X, Manos M M, Munoz N.et al Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. J Natl Cancer Inst 199587796–802. [DOI] [PubMed] [Google Scholar]
- 4.Munoz N, Bosch F X, de Sanjose S.et al Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med 2003348518–527. [DOI] [PubMed] [Google Scholar]
- 5.Jeon S, Allen‐Hoffmann B L, Lambert P F. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J Virol 1995692989–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeon S, Lambert P F. Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: implications for cervical carcinogenesis. Proc Natl Acad Sci USA 1995921654–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El Awady M K, Kaplan J B, O'Brien S J.et al Molecular analysis of integrated human papillomavirus 16 sequences in the cervical cancer cell line SiHa. Virology 1987159389–398. [DOI] [PubMed] [Google Scholar]
- 8.Romanczuk H, Howley P M. Disruption of either the E1 or the E2 regulatory gene of human papillomavirus type 16 increases viral immortalization capacity. Proc Natl Acad Sci USA 1992893159–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durst M, Bosch F X, Glitz D.et al Inverse relationship between human papillomavirus (HPV) type 16 early gene expression and cell differentiation in nude mouse epithelial cysts and tumors induced by HPV‐positive human cell lines. J Virol 199165796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Josefsson A M, Magnusson P K, Ylitalo N.et al Viral load of human papilloma virus 16 as a determinant for development of cervical carcinoma in situ: a nested case‐control study. Lancet 20003552189–2193. [DOI] [PubMed] [Google Scholar]
- 11.Durst M, Kleinheinz A, Hotz M.et al The physical state of human papillomavirus type 16 DNA in benign and malignant genital tumours. J Gen Virol 1985661515–1522. [DOI] [PubMed] [Google Scholar]
- 12.Shirasawa H, Tomita Y, Kubota K.et al Detection of human papillomavirus type 16 DNA and evidence for integration into the cell DNA in cervical dysplasia. J Gen Virol 1986672011–2015. [DOI] [PubMed] [Google Scholar]
- 13.Boshart M, Gissmann L, Ikenberg H.et al A new type of papillomavirus DNA, its presence in genital cancer biopsies and in cell lines derived from cervical cancer. EMBO J 198431151–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cullen A P, Reid R, Campion M.et al Analysis of the physical state of different human papillomavirus DNAs in intraepithelial and invasive cervical neoplasm. J Virol 199165606–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peitsaro P, Johansson B, Syrjänen S. Integrated human papillomavirus type 16 is frequently found in cervical cancer precursors as demonstrated by a novel quantitative real‐time PCR technique. J Clin Microbiol 200240886–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peitsaro P, Hietanen S, Johansson B.et al Single copy heterozygote integration of HPV 33 in chromosomal band 5p14 is found in an epithelial cell clone with selective growth advantage. Carcinogenesis 2002231057–1064. [DOI] [PubMed] [Google Scholar]
- 17.Stanley M A, Browne H M, Appleby M.et al Properties of a nontumorigenic human cervical keratinocyte cell line. Int J Cancer 198943672–676. [DOI] [PubMed] [Google Scholar]
- 18.Alazawi W, Pett M, Arch B.et al Changes in cervical keratinocyte gene expression associated with integration of human papillomavirus 16. Cancer Res 2002626959–6965. [PubMed] [Google Scholar]
- 19.Kalantari M, Blennow E, Hagmar B.et al Physical state of HPV16 and chromosomal mapping of the integrated form in cervical carcinomas. Diagn Mol Pathol 20011046–54. [DOI] [PubMed] [Google Scholar]
- 20.Kalantari M, Karlsen F, Kristensen G.et al Disruption of the E1 and E2 reading frames of HPV16 in cervical carcinoma is associated with poor prognosis. Int J Gynecol Pathol 199817146–153. [DOI] [PubMed] [Google Scholar]
- 21.Syrjänen S, Shabalova I P, Petrovichev N.et al Human papillomavirus testing and conventional pap smear cytology as optional screening tools of women at different risks for cervical cancer in the countries of the former soviet union. J Lower Genital Tract Dis 2002697–110. [DOI] [PubMed] [Google Scholar]
- 22.Syrjänen S, Shabalova I P, Petrovichev N.et al Sexual habits and human papillomavirus (HPV) infections among women in three new independent states of the former Soviet Union. Sex Transm Dis 200330680–684. [DOI] [PubMed] [Google Scholar]
- 23.Miller S A, Dykes D D, Polesky H F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988161215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moberg M, Gustavsson I, Gyllensten U. Real‐time PCR‐based system for quantification of Human Papillomavirus types associated with high risk of cervical cancer. J Clin Microbiol 2003413221–3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ueda Y, Enomoto T, Miyatake T.et al Monoclonal expansion with integration of high‐risk type human papillomavirus is an initial step for cervical carcinogenesis: association of clonal status and human papillomavirus infection with clinical outcome in cervical intraepithelial neoplasia. Lab Invest 2003831517–1527. [DOI] [PubMed] [Google Scholar]
- 26.Mellin H, Dahlgren L, Munck‐Wikland E.et al Human papillomavirus type 16 is episomal and high viral load may be correlated to better prognosis in tonsillar cancer. Int J Cancer 2002102152–158. [DOI] [PubMed] [Google Scholar]
- 27.van Duin M, Snijders P J, Schrijnemakers H F.et al Human papillomavirus 16 load in normal and abnormal cervical scrapes: an indicator of CIN II/III and viral clearance. Int J Cancer 200298590–595. [DOI] [PubMed] [Google Scholar]
- 28.Si H X, Tsao S W, Poon C S.et al Viral load of HPV in esophageal squamous cell carcinoma. Int J Cancer 2003103496–500. [DOI] [PubMed] [Google Scholar]
- 29.Ha P K, Pai S I, Westra W H.et al Real‐time quantitative PCR demonstrates low prevalence of human papillomavirus type 16 in premalignant and malignant lesions of the oral cavity. Clin Cancer Res 200251203–1209. [PubMed] [Google Scholar]
- 30.Ylitalo N, Sorensen P, Josefsson A M.et al Consistent high viral load of human papillomavirus 16 and risk of cervical carcinoma in situ: a nested case‐control study. Lancet 20003552194–2198. [DOI] [PubMed] [Google Scholar]
- 31.Wagatsuma M, Hashimoto K, Matsukura T. Analysis of integrated human papillomavirus type 16 DNA in cervical cancers: amplification of viral sequences together with cellular flanking sequences. J Virol 199064813–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baker C, Phelps W, Lindgren V.et al Structural and transcriptional analysis of human papillomavirus type 16 sequences in cervical carcinoma cell lines. J Virol 198761962–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Das B, Sharma J, Gopalakrishnaet al Analysis by polymerase chain reaction of the physical state of human papillomavirus type 16 DNA in cervical preneoplastic and neoplastic lesions. J Gen Virol 1992732327–2336. [DOI] [PubMed] [Google Scholar]
- 34.Klaes R, Woerner S, Ridder R.et al Detection of high‐risk cervical intraepithelial neoplasia and cervical cancer by amplification of transcripts derived from integrated papillomavirus oncogenes. Cancer Res 1999596132–6136. [PubMed] [Google Scholar]
- 35.Tonon S, Picconi M, Bos P.et al Physical status of the E2 human papilloma virus 16 viral gene in cervical preneoplastic and neoplastic lesions. J Clin Virol 200121129–134. [DOI] [PubMed] [Google Scholar]
- 36.Gallo G, Bibbo M, Bagella L.et al Study of viral integration of HPV‐16 in young patients with LSIL. J Clin Pathol 200356532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.do Horto dos Santos Oliveira L, Rodrigues Ede V, de Salles Lopes A P.et al HVP 16 detection in cervical lesions, physical state of viral DNA and changes in p53 gene. Sao Paulo Med J 200312167–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andersson S, Safari H, Mints M.et al Type distribution, viral load and integration status of high‐risk human papillomaviruses in pre‐stages of cervical cancer (CIN). Br J Cancer 2005922195–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagao S, Yoshinouchi M, Miyagi Y.et al Rapid and sensitive detection of physical status of human papillomavirus type 16 DNA by quantitative real‐time PCR. J Clin Microbiol 200240863–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lorincz A T, Castle P E, Sherman M E.et al Viral load of human papillomavirus and risk of CIN3 or cervical cancer. Lancet 2002360228–229. [DOI] [PubMed] [Google Scholar]
- 41.Clavel C, Masure M, Levert M.et al Human papillomavirus detection by the hybrid capture II assay: a reliable test to select women with normal cervical smears at risk for developing cervical lesions. Diagn Mol Pathol 20009145–150. [DOI] [PubMed] [Google Scholar]
- 42.Swan D C, Tucker R A, Tortolero‐Luna G.et al Human papillomavirus (HPV) DNA copy number is dependent on grade of cervical disease and HPV type. J Clin Microbiol 1999371030–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hernandez‐Hernandez D M, Ornelas‐Bernal L, Guido‐Jimenez M.et al Association between high‐risk human papillomavirus DNA load and precursor lesions of cervical cancer in Mexican women. Gynecol Oncol 200290310–317. [DOI] [PubMed] [Google Scholar]
- 44.Ylitalo N, Josefsson A, Melbye M.et al A prospective study showing long‐term infection with human papillomavirus 16 before the development of cervical carcinoma in situ. Cancer Res 2000606027–6032. [PubMed] [Google Scholar]