Abstract
Background
Severe acute respiratory syndrome (SARS) is an infectious disease which was caused by a novel coronavirus (SARS‐CoV). SARS has caused an outbreak in the world during 2003 and 2004, with 8098 individuals being infected and a death toll of 774 in 28 regions around the world. Specific humoral responses to viral infection remain unclear.
Objective
To analyse the antigenicity of the SARS‐CoV genome and identify potential antigenic epitopes in the structural proteins.
Methods
Potential antigenic epitopes were identified in the structural proteins (nucleocapsid, membrane, spike, and small envelope proteins) and hypothetical proteins (SARS3a, 3b, 6, 7a, and 9b) that are specific for SARS‐CoV. A peptide chip platform was created and the profiles of antibodies to these epitopes were investigated in 59 different SARS patients' sera obtained 6–103 days after the onset of the illness. Serial sera from five additional patients were also studied.
Results
Epitopes at the N‐terminus of the membrane protein and the C‐terminus of nucleocapsid protein elicited strong antibody responses. Epitopes on the spike protein were only moderately immunogenic but the effects were persistent. Antibodies were also detected for some putative proteins, noticeably the C‐termini of SARS3a and SARS6.
Conclusions
Important epitopes of the SARS‐CoV genome that may serve as potential markers for the viral infection are identified. These specific antigenic sites may also be important for vaccine development against this new fatal infectious disease.
Keywords: SARS, coronavirus, antibody, peptide chip
An epidemic outbreak of severe acute respiratory syndrome (SARS) haunted 28 regions in the world from November to July 2003. In this epidemic, 8098 individuals were being infected, with a cumulative mortality of 9.2%.1 The clinical and pathological features of SARS have been studied extensively.2,3,4 The specific humoral responses against each of the proteins of the causative agent of SARS—a new coronavirus (SARS‐CoV)—are, however, poorly understood.5
The SARS‐CoV is an enveloped positive stranded RNA virus.6,7 The viral genome is ∼29.7 kb long and contains 14 open reading frames (ORFs). At the 5′ end of the genome, two large ORFs encoding replication proteins are present. These two proteins have relatively higher homology to the other members of the coronavirus. At the 3′ end, there are genes for structural proteins (envelope, membrane, nucleocapsid, and spike) as well as the other hypothetical proteins. This latter group of hypothetical proteins has no similarity to any known mammalian or viral proteins and may be specific target for treatment or vaccine developments.8
In this study, we analysed the 3′ end of the SARS‐CoV genome and identified some specific, unique, and potentially antigenic sites of sizes ∼20 amino acids on each of the structural and hypothetical proteins. Using a peptide chip platform, we determined the antibody profiles against these peptides in the sera of 59 SARS patients at different time points.
Materials
Chemicals
All chemicals were purchased from Sigma (St Louis, Missouri, USA) unless specified. Peptide synthesis was carried out by Abgent Inc. The keyhole limpet haemocyanin (KLH) conjugation kit was purchased from Pierce Biotechnology (Rockford, Illinois, USA). Goat Fc anti‐human IgG F(ab)2 (obtained from pepsin‐digested goat IgG which reacts specifically to the Fc fragment of human IgG) and goat anti‐human IgM Fμc (IgG that reacts with Fc5μ portion of the human IgM heavy chain) were purchased from the Jackson Laboratory (Bar Harbor, Maine, USA). Alexa 546 and Alexa 647 protein labelling kits were purchased from Molecular Probes (Eugene, Oregon, USA).
Patient selection
The study of the antibody in SARS patients was approved by local ethics committee. The clinical diagnosis of SARS followed the WHO case definition criteria.1 Fifty nine serum specimens from different patients collected 6–103 days after the onset of the illness were obtained from a single hospital. Only one serum was used if multiple specimens were obtained from the same patient on one single day. All these patients were treated with the steroid regime developed at the time of the epidemic.4 Samples with incomplete clinical data were discarded. Convalescence serum of 40 patients who had recovered from SARS (>180 days after the onset of the illness) were also studied. Five additional SARS patients with serial serum specimens (patient A with specimens from days 6, 10, 19, 31, and 68; patient B: days 4, 12, 18, 21, and 63; patient C: days 5, 11, 15, 22, and 57; patient D: days 5, 9, 15, 21, 31, and 81; and patient E: days 4, 10, 17, 22, and 71) were studied for heterogeneity of humoral responses. Controls for the experiments were obtained from 18 non‐SARS patients admitted to the two hospitals during the SARS epidemics.
Antigenic peptides in the SARS‐CoV genome
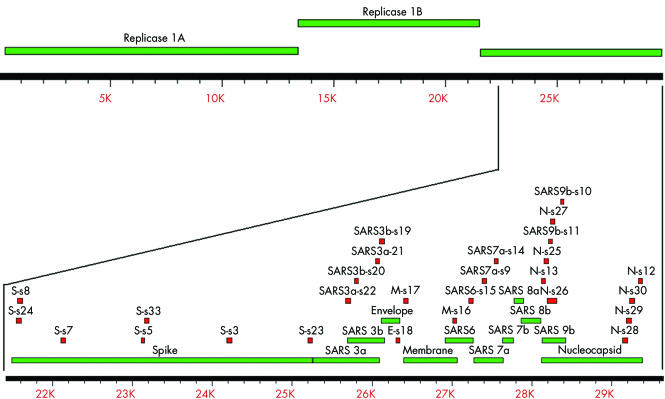
Antigenicities of epitopes on putative ORF of the SARS‐CoV (Genbank Accession No: NC_004718) were evaluated in silico using Clone Manager 5 (Scientific & Educational Software, Cary, North Carolina, USA), Peptide Companion (CSPS Pharmaceuticals Inc, San Diego, California, USA), and DS Gene (Accelrys, San Diego, California, USA), with the following criteria: protein homology and redundant sequences (mostly BLASTing against human and mouse sequences from RefSeq in GenBank); protein hydrophilicity; protein flexibility; protein structure—B‐turn, β‐sheet, and α‐helix; solvent accessibility; charge balance; protein stability; instability index; protein modification; amino acid composition; and peptide synthesis rules for coupling efficiency. These peptides were then compared with other published data of coronavirus (CoV) such as human group 1 and group 2 CoV, bovine‐CoV, porcine‐CoV, feline‐CoV, and canine‐CoV, and only SARS‐CoV specific sequences were selected. The genomic locations and sequences of the peptides selected are shown in fig 1 and table 1, respectively.
Figure 1 Location of the selected peptide relative to the SARS‐CoV genome. The upper section shows the whole SARS‐CoV genome. The portion of the structural proteins and hypothetical proteins is enlarged and shown in the lower section. The genomic locations of each peptide used in this study were shown as small red boxes and the predicted open reading frames are shown as green boxes. The scales of the upper and lower panels are shown in the rulers underneath each corresponding panels (unit: base pairs).
Table 1 Summary of peptides and their reactivity.
| SARS virus protein* | Peptide code | Position† | Sequence | Reactivity‡ | Conc§ |
|---|---|---|---|---|---|
| Envelope | E‐s18 | 60–76 | SRVKNLNSSEGVPDLLV | + | 1.0 |
| M‐s16 | 204–221 | KLNTDHAGSNDNIALLVQ | + | 1.0 | |
| Membrane | M‐s17 | 1–18 | MADNGTITVEELKQLLEQ | + | 0.5 |
| Spike | S‐s3 | 899–912 | YENQKQIANQFNKA | − | 1.0 |
| S‐s5 | 540–554 | PSSKRFQPFQQFGRD | + | 0.8 | |
| S‐s7 | 204–219 | DVVRDLPSGFNTLKPI | − | 1.0 | |
| S‐s8 | 24–39 | DVQAPNYTQHTSSMRG | − | 1.0 | |
| S‐s23 | 1236–1255 | CKFDEDDSEPVLKGVKLHYT | + | 1.0 | |
| S‐s24 | 19–36 | CTTFDDVQAPNYTQHTSS | − | 1.0 | |
| S‐s33 | 553–570 | RDVSDFTDSVRDPKTSEI | + | 1.0 | |
| Nucleocapsid | N‐s12 | 406–422 | RQLQNSMSGASADSTQA | − | 1.0 |
| N‐s13 | 1–17 | MSDNGPQSNQRSAPRIT | + | 1.0 | |
| N‐s25 | 13–30 | APRITFGGPTDSTDNNQN | + | 1.0 | |
| N‐s26 | 25–43 | TDNNQNGGRNGARPKQRRP | + | 1.0 | |
| N‐s27 | 38–55 | PKQRRPQGLPNNTASWFT | + | 1.0 | |
| N‐s28 | 341–360 | DDKDPQFKDNVILLNKHIDA | + | 1.0 | |
| N‐s29 | 356–375 | KHIDAYKTFPPTEPKKDKKK | + | 1.0 | |
| N‐s30 | 371–390 | KDKKKKTDEAQPLPQRQKKQ | + | 0.5 | |
| SARS3a | SARS3a‐s21 | 258–274 | AMDPIYDEPTTTTSVPL | + | 0.5 |
| SARS3a‐s22 | 134–148 | KSKNPLLYDANYFVC | − | 1.0 | |
| SARS3b | SARS3b‐s19 | 135–154 | KHKKVSTNLCTHSFRKKQVR | + | 1.0 |
| SARS3b‐s20 | 31–45 | KVTAFQHQNSKKTTK | + | 1.0 | |
| SARS6 | SARS6‐s15 | 45–63 | TKKNYSELDDEEPMELDYP | + | 1.0 |
| SARS7a | SARS7a‐s9 | 35–52 | CPSGTYEGNSPFHPLADN | − | 1.0 |
| SARS7a‐s14 | 85–100 | KLFIRQEEVQQELYSP | + | 1.0 | |
| SARS9b | SARS9b‐s10 | 78–92 | QMTKLATTEELPDEF | − | 1.0 |
| SARS9b‐s11 | 28–44 | EDAMGQGQNSADPKVYP | − | 1.0 |
*Nomenclatures of proteins of SARS‐CoV follow those listed in the reference sequence Genbank Accession No.: NC_004718).
†Positions of the epitopes are shown relative to the individual ORFs of each protein.
‡Positive reactive indicates detection of antibodies in the present study.
§Concentrations of peptides on the peptide chips are expressed in mg/ml.
Peptide chip platform
One milligram of peptide was conjugated with 1 mg of KLH carrier protein, purified by gel filtration chromatography. Conjugated peptides were spotted onto an in‐house developed multichambered Fast Slide (S&S) in an array of two, by a Cartesian Microsys 5100 fitted with a Telechem SMP3B pin. Negative controls were 1 mg/ml bovine serum albumin and 1 mg/ml KLH. The optimum concentration for each peptide (table 1) was determined empirically using serial diluted conjugated peptide from 1 mg/ml to 0.2 mg/ml. Diluted test serum (30 μl, 1/50 in blocking buffer) was incubated for 90 minutes at 37°C on peptide chip blocked with 5% non‐fat milk, 0.1% Tween‐20 in 20 mM Tris‐buffer saline, pH 7.6 for one hour at 37°C. After washing three times with 100 μl TBS buffer (20 mM Tris, pH 7.6), the reactions were then labelled simultaneously with Alexa 546 goat anti‐human IgG (1/500 dilution in blocking buffer) and Alexa 647 goat anti‐human IgM (1/5000 dilution in blocking buffer) for 30 minutes at 37°C. Fluorescence images were acquired using laser scanner SA5000 coupled with ScanArray software (Perkin Elmer, Norwalk, Connecticut, USA) and QuantArray software (Perkin Elmer). Data between experiments were analysed after signal normalisation with those of normal control samples and negative controls. Data analysis, modelling, and visualisation were undertaken using “Avadis” software (Strand Genomics, Bangalore, India).
Results
Specific antigenic sites on the SARS‐CoV genome
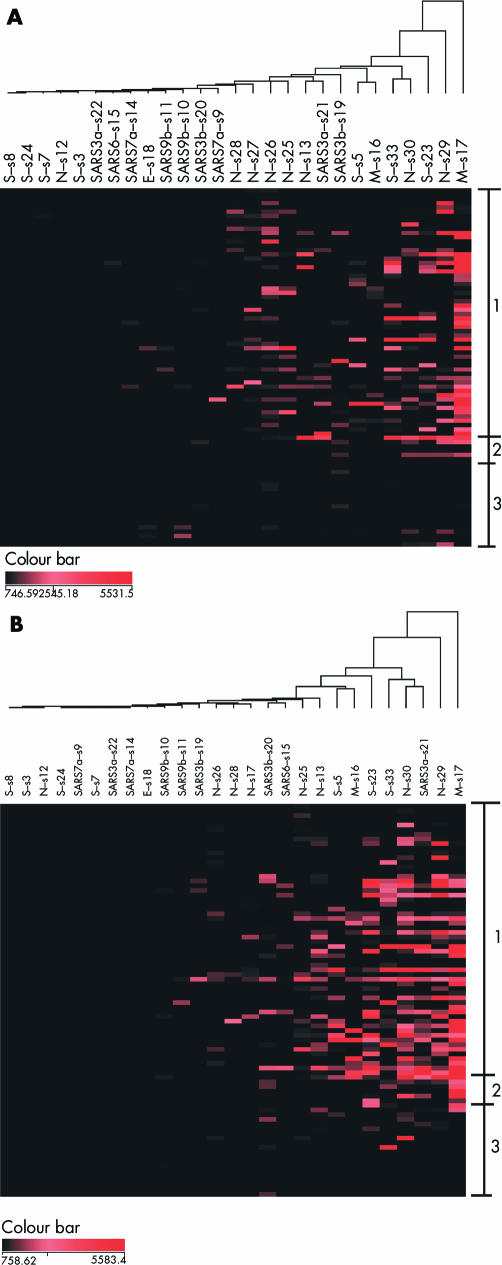
The reference genomic sequences of the SARS‐CoV were examined for antigenicity in silico. Among all potential epitopes, only those unique to the SARS‐CoV were selected for further analyses. At least one potential antigenic site was identified for each of the structural proteins and hypothetical proteins (fig 1; table 1). Corresponding peptides were synthesised and immobilised on membrane coated slides to create a peptide chip platform. Sera from 59 SARS patients, obtained from different patients at 6 to 103 days after the onset of the illness, were studied using this peptide chip platform to simultaneously record the levels of antibody against each of these epitopes. The IgM and IgG against specific epitopes were measured and displayed in array format as a function of time from disease onset (fig 2, panels A and B, respectively). The relative genomic location of each antigenic site and the average antibody responses over time intervals are shown in three dimensional histograms (fig 3, upper and lower panels for IgM and IgG, respectively). Antibodies against some of the specific epitopes were detected in the patient sera and showed different temporal sequences. In this 3′ end of the SARS‐CoV genome, there was no specific DNA segment in which antigenic epitopes were particularly concentrated.
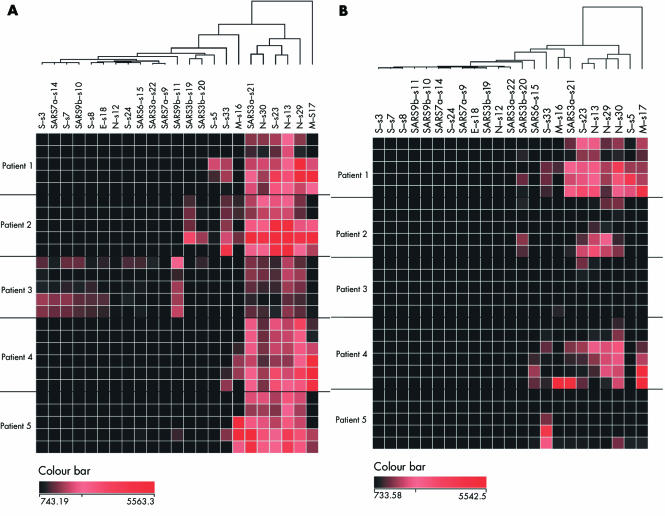
Figure 2 Antibody responses to specific epitopes expression pattern in SARS patients. The rows represent patient serum samples, arranged in ascending order from the day of onset of the disease. The columns represent antibody to each of the epitopes studied. The fluorescence signals for IgM (A) and IgG (B) were measured, and the dendrogram was created by hierarchical clustering analysis. Group 1 is the clinically affected SARS patients from day 6 to day 103 (top to bottom). Group 2 is the convalescence patient recovered and the serum collected >180 days after illness. Group 3 are non‐SARS controls. The colour bar represents the intensity of the fluorescence signals.
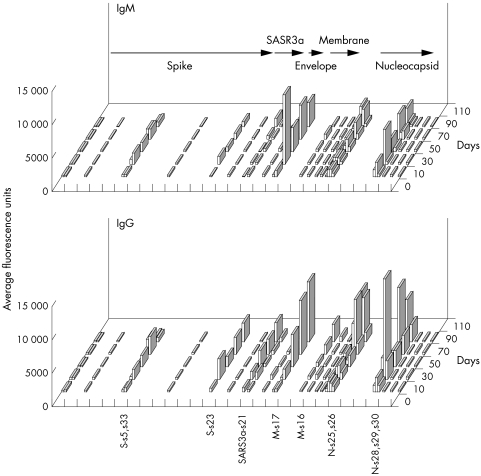
Figure 3 Genome view of the total antibody response profile against different structural proteins. Data collected from the peptide chip analyses were grouped into consecutive time series of 20 days for each epitope (y axis). The average fluorescence signals of IgM (upper panel) and IgG (lower panel) are shown (z axis). Each epitope was arranged to correspond to the individual genomic location (x axis). The arrows represent some of the open reading frames and those epitopes with significant signals are labelled along the x axis.
Antigenicity of the viral structural and putative proteins
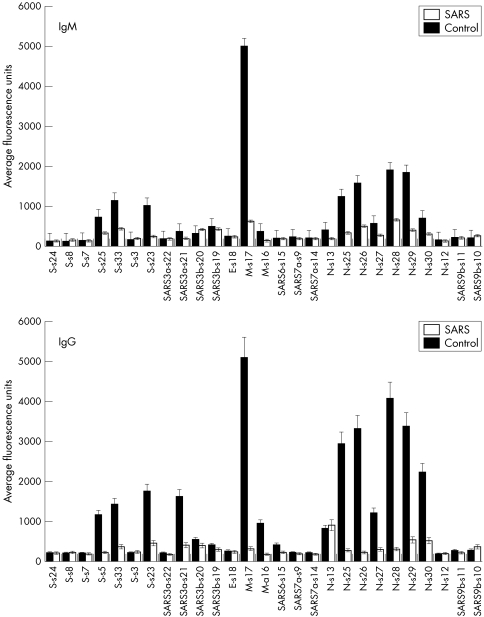
The average IgM and IgG responses over 6–103 days to each epitope were examined (fig 4, upper and lower panels, respectively). The control population consists of non‐SARS patients during the time of the epidemic. The N‐terminus epitope s17 of the membrane protein (M) and the C‐terminus epitope s29 of the nucleocapsid protein (N) elicited marked IgM and IgG responses. Multiple epitopes of the spike protein (S) evoked humoral responses but were relatively weaker. Antibodies against hypothetical proteins SARS3a, SARS3b, and SARS6 were also detected. All patients had low to non‐specific antibody responses to epitopes s3, s7, s8, and s24 of the S protein, s12 of the N protein, s18 of the envelope (E) protein, s22 of the hypothetical SARS3a, s9 and s14 of the hypothetical SARS7a, as well as s10 and s11 of SARS9b.
Figure 4 Average IgM and IgG responses for each of the epitopes. For each epitope, the average IgM (upper panel) and IgG (lower panel) over the observed period of time were calculated. The filled bars represent the SARS patient group and the empty bars represent the control group. Error bars = SEM.
Humoral responses to reactive epitopes of the SARS‐CoV
Hierarchical clustering analyses of the data showed single dendrogram organisations for the IgM and IgG responses (fig 2, panels A and B, respectively). Signals for each epitope are represented in descending order from the right side of the dendrogram. No segregation into major subgroups was observed. This result suggested homogeneity in the antibody reactions to all these epitopes. Minor subgroupings were, however, detected. The epitopes s30 of N, s33 of S, and s21 of SARS3a protein form a subcluster in the IgG responses (fig 2B). Epitopes s3, s7, s8, and s24 of S protein, s12 of the N protein, s18 of E protein, s22 of hypothetical SARS3a protein, s9 and s14 of hypothetical SARS7a protein, and s10 and s11 of SARS9b were grouped together at the left end of the cluster, owing to the common low mean antibody responses of these epitopes (see previous section and fig 3).
Antibody responses to immunogenic sites appeared to be biphasic. This phenomenon was better visualised by plotting the conventional time courses of the antibody responses for each of the epitopes (fig 5). The trough of antibody responses appeared in ∼50 days after the onset of the illness. Odds ratios of positivities of IgM and IgG responses to individual peptide at each time interval confirmed impressions of the graphic presentations (positives values for IgM and IgG are listed in tables 2 and 3, respectively).
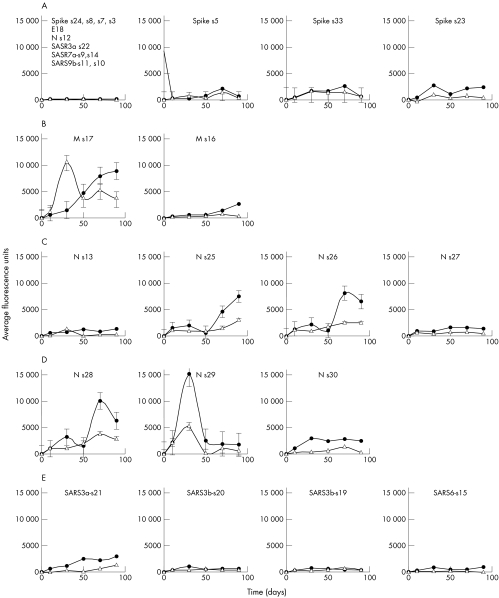
Figure 5 Profiles of antibody responses to individual epitopes. The temporal changes of IgM (empty triangles) and IgG (filled circles) for epitopes of spike (S) protein (first panel), membrane (M) protein (second panel), N‐terminus of nucleocapsid (N) protein (third panel), C‐terminus of nucleocapsid (N) protein (fourth panel), and hypothetical proteins (fifth panel). The x axes represent the day of onset of illness and the y axes represent the average fluorescence signals obtained from all specimens. SEMs are shown for each time point.
Table 2 Odds ratios IgM responses to selected epitopes of SARS‐CoV.
| Epitope | Time intervals (days) | ||||
|---|---|---|---|---|---|
| 1–20 | 21–40 | 41–60 | 61–80 | 81–103 | |
| E‐s18 | 0* | 0 | 0 | 2.00 | 0 |
| M‐s16 | 0 | 1.89 | 3.78 | 2.27 | 2.83 |
| M‐s17 | 6.80 | 68.00 | 170.00 | 40.80 | 102.00 |
| S‐s5 | 0 | 11.33 | 0 | 5.23 | 6.80 |
| S‐s23 | 1.31 | 17.00 | 6.38 | 9.27 | 12.75 |
| S‐s33 | 0 | 17.00 | 9.71 | 11.90 | 12.75 |
| N‐s13 | 1.31 | 7.29 | 0 | 2.27 | 2.83 |
| N‐s25 | 1.31 | 4.25 | 1.70 | 7.08 | 6.80 |
| N‐s26 | 45.00 | 19.29 | 25.71 | 24.55 | 60.00 |
| N‐s27 | 4.64 | 1.89 | 3.78 | 11.90 | 2.83 |
| N‐s28 | 6.80 | 1.89 | 0 | 3.64 | 0 |
| N‐s29 | 12.80 | 32.00 | 0 | 17.45 | 12.80 |
| N‐s30 | 8.73 | 8.00 | 3.20 | 17.45 | 5.33 |
| SARS3a‐s21 | 0 | 1.89 | 0 | 5.23 | 6.80 |
| SARS3b‐s19 | 2.46 | 0 | 3.20 | 4.27 | 12.80 |
| SARS3b‐s20 | 0 | 1.89 | 1.70 | 0.00 | 0 |
| SARS6‐s15 | 0 | 1.89 | 0 | 0.00 | 0 |
| SARS7a‐s9 | 0 | 0 | 0 | 1.06 | 0 |
| SARS7a‐s14† | 0 | 0 | 1.70 | 1.06 | 0 |
| Total No‡ | 14 | 10 | 11 | 17 | 7 |
*The odds ratio for IgM responses for individual epitope at different time intervals are shown. An arbitrary cut off of 800 average fluorescence units was used in the calculations.
†The epitopes listed are those showing positivity with IgG or IgM responses.
‡Total No of subjects per time interval is shown.
Table 3 Odds ratios of IgG responses to selected epitopes of SARS‐CoV.
| Epitope | Time intervals (days) | ||||
|---|---|---|---|---|---|
| 1–20 | 21–40 | 41–60 | 61–80 | 81–103 | |
| E‐s18 | 0* | 0 | 0 | 0 | 0 |
| M‐s16 | 0 | 1.89 | 6.38 | 5.23 | 42.50 |
| M‐s17 | 2.83 | 17.00 | 45.33 | 127.50 | 102.00 |
| S‐s5 | 0 | 1.89 | 6.38 | 11.90 | 12.75 |
| S‐s23 | 2.83 | 17.00 | 14.17 | 24.29 | 102.00 |
| S‐s33 | 1.31 | 25.50 | 9.71 | 15.11 | 2.83 |
| N‐s13 | 2.83 | 4.25 | 28.75 | 15.11 | 42.50 |
| N‐s25 | 4.64 | 1.89 | 1.70 | 11.90 | 6.80 |
| N‐s26 | 4.64 | 1.89 | 1.70 | 2.27 | 6.80 |
| N‐s27 | 0 | 0 | 1.70 | 5.23 | 0 |
| N‐s28 | 0 | 0 | 0 | 0 | 0 |
| N‐s29 | 17.78 | 48.00 | 56.00 | 35.00 | 192.00 |
| N‐s30 | 33.75 | 105.00 | 78.75 | 146.25 | 270.00 |
| SARS3a‐s21 | 4.64 | 7.29 | 14.17 | 19.13 | 102.00 |
| SARS3b‐s19 | 0 | 0 | 0 | 0 | 0 |
| SARS3b‐s20 | 0 | 11.25 | 10.00 | 9.64 | 7.50 |
| SARS6‐s15 | 0 | 4.25 | 1.70 | 2.27 | 2.83 |
| SARS7a‐s9 | 0 | 0 | 0 | 0 | 0 |
| SARS7a‐s14† | 0 | 0 | 0 | 0 | 0 |
| Total No‡ | 14 | 10 | 11 | 17 | 7 |
*The odds ratio for IgG responses, respectively, for individual epitope at different time intervals are shown. An arbitrary cut off of 800 average fluorescence units was used in the calculations.
†The epitopes listed are those showing positivity with IgG or IgM responses.
‡Total No of subjects per time interval is shown.
Heterogeneity of humoral responses in different individuals
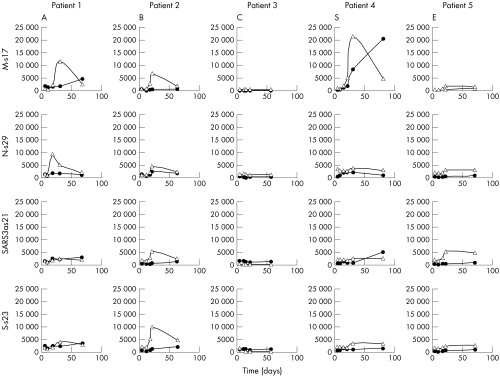
Heterogeneity of the humoral responses in different individuals can be seen in the longitudinal studies of IgM and IgG in five additional SARS patients (A to E) for whom serial samples of serum were available (figs 6 and 7). The major organisation of the dendrograms of individual patient is similar to the main dataset (fig 2). The presence of antibody responses to one epitope did not predict the occurrence of the other. The magnitude of responses to one epitope did not have any bearing on another reactive epitope. The underlying mechanisms of these differences remain unclear.
Figure 6 Profiles of antibody responses in five SARS patients. Serial samples were obtained from five patients (A–E) and the time points of each patient are arranged in rows of ascending order, 4 to 81 days after onset of the disease. Dendrograms for IgM (A) and IgG (B) were created by hierarchical clustering analysis. The colour bar represents the intensity of the fluorescence signals.
Figure 7 Temporal changes of antibodies against selected epitopes in 5 SARS patients. IgM (empty triangles) and IgG (filled circles) for epitopes of membrane protein (M‐s17) (upper panel), C‐terminus of nucleocapsid protein (N‐s29) (second panel), hypothetical protein SARS3a (SARS‐3a s21) (third panel), and spike protein (S‐s23) (fourth panel). A–E were five different SARS patients. The x axes represent the day of onset of illness and the y axes represent the average fluorescence signals.
Discussion
In this study, we have demonstrated humoral responses to some of the epitopes defined by oligopeptide specific to the SARS‐CoV genome. Antibodies against conformation or complex structures other than the primary amino acid sequences may not be captured by our peptide chip. For considerations of cost, only SARS‐CoV specific peptides were selected for this study. The epitopes identified are, therefore, only a portion of all the antigenic sites of SARS‐CoV. We reported strong humoral responses to the membrane, M, and nucleocapsid, N, proteins. Some of the putative hypothetical proteins, noticeably SARS3a and SARS6, were also shown to be important in the humoral responses to SARS‐CoV infection.
N protein has the strongest immunoreactivity among all other structural proteins.9,10,11,12,13,14 It is the most abundant protein in SARS‐CoV and this may explain the high immunoreactivity.14 To determine some of the immunodominant epitopes on the N protein that are specific to SARS‐CoV, eight epitopes from both the C‐ and N‐ termini, were studied. At the C‐terminus, epitopes s28–30 showed similar signal profiles, with a significant increase in intensity in the first 10 to 20 days and a second peak at >80 days (fig 5). Consistent with other studies, antibodies against the whole N protein were produced in all SARS patients and appeared in the early stages of the disease.13,15 Epitopes from the C‐terminus (N‐s28, s29 and s30) of N produced greater humoral responses than those from the N‐terminus (N‐s25 and s26). The epitopes N‐s30 and N‐s29 overlapped for only five amino acids but eight‐ to 10‐fold excess signal was detected on N‐s29. The N‐terminus of N protein is less well investigated. Our data suggested that a small region—defined by N‐s25 and N‐s26, excluding N‐s13 and N‐s27—was antigenic. The humoral responses appeared late in the course of the infection.
The spike (S) protein of SARS‐CoV is a type I transmembrane glycoprotein responsible for virus binding and fusion.16,17,18 Residue amino acids 318 to 510 of the S protein bound specifically to the functional receptor, angiotensin converting enzyme 2 (ACE2).19,20,21 In our study, seven epitopes were designed along the N‐ to C‐termini of the S protein. Only moderate signals were observed in the middle (S‐s5 and S‐s33, amino acids 540 to 570) and C‐terminus (S‐s23, amino acids 1236 to 1255). Unlike the N protein and consistent with earlier findings of reactivity to the whole S protein,14 very low signal was detected at the early stage. However, the signals increased exponentially after 80 days. Among the three epitopes, S‐s33 appeared to be the strongest immunodominant antigen in the S protein, supporting earlier reports with a finer resolution.16,17 The antigenicity detected in the current study for the C‐terminus (s23) is novel and, in view of the importance of S proteins in SARS‐CoV infection, further exploration of this area will be important.
The membrane (M) protein is a relatively small viral structural protein. Two potential epitopes were identified on this protein. Strong epitope was found in the N‐terminus with 100‐fold increased responses compared with the C‐terminus. Theoretically, sequence analysis predicts a single weak epitopic site for the M protein.13 Our study showed that antibody against M was in fact common among all the subjects and elicited strong, persistent responses. The humoral responses to M protein may be useful in diagnostic tests and in surveillance. Whether antibody against the M protein conveys protection against future SARS‐CoV infection awaits further clarification.
The biological basis of variable antibody responses to different epitopes of the SARS‐CoV in individual patient is not clear. Several lines of evidence suggest the influence of host genetic factors in SARS. Human leucocyte antigen (HLA) class I (B*0703) and class II (DRB1*0301) is associated with susceptibility of SARS in our locality.22 HLA class I (B*4601) is associated with severity of SARS in a cohort of Taiwanese.23 The difference in HLA phenotypes and, perhaps, in immune responses might be the key to the pathogenesis of SARS. It is therefore important to correlate HLA phenotypes of the patients and to explore the genetic basis of the observed variable antibody responses.
Take home points
Important epitopes of the SARS‐CoV genome have been identified. These may serve as potential markers for the viral infection.
These specific antigenic sites may also be important for vaccine development against this new fatal infectious disease.
Conclusions
We have identified some of the epitopes of SARS‐CoV in different structural proteins and putative proteins that elicited natural humoral responses. Owing to the relative simplicity of production of oligopeptides compared with whole protein, these SARS‐CoV specific antigenic sites are likely to be valuable for the design of bedside diagnostic tests. The epitopes may also be important in the development of vaccine for this new fatal human infectious disease.
Acknowledgements
The laboratory of AWIL was partially supported by the Research Grant Council of the Hong Kong Special Administrative Region (Project No CUHK4411/03M). The SARS project at Century Biotech Ltd is supported by Small Entrepreneur Research Assistance Programme of the Innovation and Technology Commission, the Government of Hong Kong Special Administrative Region (Project No S/P675/03). SCSC, CW, TC, DMYA, and AWIL are enlisted inventors of the patent applications (HK: 04104279.0; USA: 60/579,333).
Abbreviations
KLH - keyhole limpet haemocyanin
ORF - open reading frame
SARS - severe acute respiratory syndrome
References
- 1.WHO Severe acute respiratory syndrome (SARS). http://www.who.int/csr/sars/en/; last updated 2 July 2004
- 2.Tse G M, To K F, Chan P K.et al Pulmonary pathological features in coronavirus associated severe acute respiratory syndrome (SARS). J Clin Pathol 200457260–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsang K W, Ho P L, Ooi G C.et al A cluster of cases of severe acute respiratory syndrome in Hong Kong. N Engl J Med 20033481977–1985. [DOI] [PubMed] [Google Scholar]
- 4.Lee N, Hui D, Wu A.et al A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med 20033481986–1994. [DOI] [PubMed] [Google Scholar]
- 5.To K F, Lo A W. Exploring the pathogenesis of severe acute respiratory syndrome (SARS): the tissue distribution of the coronavirus (SARS‐CoV) and its putative receptor, angiotensin‐converting enzyme 2 (ACE2). J Pathol 2004203740–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marra M A, Jones S J, Astell C R.et al The Genome sequence of the SARS‐associated coronavirus. Science 20033001399–1404. [DOI] [PubMed] [Google Scholar]
- 7.Rota P A, Oberste M S, Monroe S S.et al Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 20033001394–1399. [DOI] [PubMed] [Google Scholar]
- 8.Stadler K, Masignani V, Eickmann M.et al SARS – beginning to understand a new virus. Nat Rev Microbiol 20031209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leung D T, Tam F C, Ma C H.et al Antibody response of patients with severe acute respiratory syndrome (SARS) targets the viral nucleocapsid. J Infect Dis 2004190379–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang L R, Chiu C M, Yeh S H.et al Evaluation of antibody responses against SARS coronaviral nucleocapsid or spike proteins by immunoblotting or ELISA. J Med Virol 200473338–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan P K, Liu E Y, Leung D T.et al Evaluation of a recombinant nucleocapsid protein‐based assay for anti‐SARS‐CoV IgG detection. J Med Virol 200575181–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Z, Pei D, Jiang L.et al Antigenicity analysis of different regions of the severe acute respiratory syndrome coronavirus nucleocapsid protein. Clin Chem 200450988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Wen J, Li J.et al Assessment of immunoreactive synthetic peptides from the structural proteins of severe acute respiratory syndrome coronavirus. Clin Chem 2003491989–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan Y J, Goh P Y, Fielding B C.et al Profiles of antibody responses against severe acute respiratory syndrome coronavirus recombinant proteins and their potential use as diagnostic markers. Clin Diagn Lab Immunol 200411362–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi Y, Yi Y, Li P.et al Diagnosis of severe acute respiratory syndrome (SARS) by detection of SARS coronavirus nucleocapsid antibodies in an antigen‐capturing enzyme‐linked immunosorbent assay. J Clin Microbiol 2003415781–5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He Y, Zhou Y, Wu H.et al Identification of immunodominant sites on the spike protein of severe acute respiratory syndrome (SARS) coronavirus: implication for developing SARS diagnostics and vaccines. J Immunol 20041734050–4057. [DOI] [PubMed] [Google Scholar]
- 17.Lu L, Manopo I, Leung B P.et al Immunological characterization of the spike protein of the severe acute respiratory syndrome coronavirus. J Clin Microbiol 2004421570–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Babcock G J, Esshaki D J, Thomas W D.et al Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. J Virol 2004784552–4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong S K, Li W, Moore M J.et al A 193‐amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin‐converting enzyme 2. J Biol Chem 20042793197–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li W, Moore M J, Vasilieva N.et al Angiotensin‐converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003426450–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prabakaran P, Xiao X, Dimitrov D S. A model of the ACE2 structure and function as a SARS‐CoV receptor. Biochem Biophys Res Commun 2004314235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng M H, Lau K M, Li L.et al Association of human‐leukocyte‐antigen class I (B*0703) and class II (DRB1*0301) genotypes with susceptibility and resistance to the development of severe acute respiratory syndrome. J Infect Dis 2004190515–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin M, Tseng H K, Trejaut J A.et al Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Med Genet 200349. [DOI] [PMC free article] [PubMed] [Google Scholar]