Abstract
HIV-1 reverse transcriptase (RT) catalyzes the conversion of genomic RNA into cDNA. The enzyme is a heterodimer of p66 and p51 subunits, and the dimerization of these subunits is required for optimal enzyme activity. To analyze this process at the genetic level, we developed constructs that permit the detection of the interaction between these subunits in the yeast two-hybrid system. Genetic analysis of RT subdomains required for heterodimerization revealed that the fingers and palm of p66 were dispensable for p51 interaction. However, as little as a 26-amino acid deletion at the C terminus of p51 prevented dimerization with p66. A primer grip mutation, L234A, previously shown to inhibit RT dimerization by biochemical assays, also prevented RT dimerization in the yeast two-hybrid system. Second-site mutations that restored RT dimerization in yeast to the L234A parent were recovered in the tryptophan repeat region at the dimer interface and at the polymerase active site, suggesting the involvement of these sites in RT dimerization. In vitro binding experiments confirmed the effects of the L234A mutation and the suppressor mutations on the interaction of the two subunits. The RT two-hybrid assay should facilitate the extensive genetic analysis of RT dimerization and should make possible the rapid screening of potential inhibitors of this essential process.
The HIV type 1 (HIV-1) reverse transcriptase (RT) is required for the conversion of genomic RNA into double-stranded proviral DNA, catalyzed by the RNA- and DNA-dependent polymerase and ribonuclease H activities of the enzyme. HIV-1 RT is an asymmetric dimer formed by the association of p66 and p51 polypeptides, which are cleaved from a large Pr160GagPol precursor by the viral protease during virion assembly. p51 contains identical N-terminal sequences as p66, but lacks the C-terminal ribonuclease H (RNase H) domain (1). The structure of HIV-1 RT has been elucidated by x-ray crystallography in a variety of configurations, including unliganded (2), complexed to nonnucleoside RT inhibitors (3), or complexed with double-stranded DNA either with (4) or without deoxynucleotide triphosphate (5, 6). Such analyses have shown that p66 can be divided structurally into the polymerase and RNase H domains, with the polymerase domain further divided into the fingers, palm, thumb, and connections subdomains (6). Although p51 has the same polymerase domains as p66, the relative orientations of these individual domains differ markedly, resulting in p51 assuming a closed structure.
The RT heterodimer represents the biologically relevant form of the enzyme; the monomeric subunits have only low catalytic activity (7). Structural analysis reveals three major contacts between p66 and p51, with most of the interaction surfaces being largely hydrophobic (8, 9). The three contacts comprise an extensive dimer interface that includes the fingers subdomain of p51 with the palm of p66, the connection subdomains of both subunits, and the thumb subdomain of p51 with the RNase H domain of p66 (9).
Several single amino acid substitutions in HIV-1 RT have been shown to inhibit heterodimer association (10–12). These include the mutations L234A (10, 11), G231A (11), and W229A (11), all located in the primer grip region of the p66 subunit, and L289K (12) in the thumb subdomain. Remarkably, these mutations are not located at the dimer interface and probably mediate their effects indirectly through conformational changes in the p66 subunit.
Several biochemical assays have been used previously to specifically measure RT dimerization. Some are based on the physical separation of monomers and dimers as determined by analytical ultracentrifugation (8) and gel filtration (7). Other assays include intrinsic tryptophan fluorescence (13), chemical crosslinking (14), the use of affinity tags (15), and polymerase activity itself (7). Although these methods detect dimerization, they either lack specificity or are not easy to perform. Moreover, these assays do not facilitate the rapid genetic analysis of protein-protein interactions under physiological conditions nor are they suitable for high throughput screening for RT dimerization inhibitors.
The yeast two-hybrid (Y2H) system (16) has been exploited to study the homomeric interactions of several retroviral proteins (see, e.g., ref. 17) and heteromeric interactions between viral proteins and various cellular partners (see, e.g., ref. 18). We have used this system to perform a genetic analysis of the determinants of RT dimerization. In addition, we have identified second-site mutations that restore heterodimerization to a noninteracting mutant p66.
Materials and Methods
Bacterial and Yeast Strains.
Saccharomyces cerevisiae strain CTY10-5d (MATa ade2 trp1-901 leu2-3,112 his3-200 gal4-gal80-URA3∷lexA-lacZ) contains an integrated GAL1-lacZ gene with the lexA operator (a gift from Stanley Fields, State University of New York, Stony Brook). The yeast strain HF7c contains CYC1-lacZ gene with three copies of the GAL4 responsive UASG 17-mer operator (CLONTECH). Escherichia coli mutator strain XL1-Red (Stratagene) was used for random mutagenesis whereas XL1-Blue (Stratagene) was used to amplify the mutated library. KC8 (CLONTECH), an auxotrophic leuB, trpC, and hisB E. coli strain, was used to isolate plasmids from yeast. E. coli strains M15 and BL21 were used to express p66-His and glutathione S-transferase-tagged p51 (GST-p51), respectively (see below).
Yeast Methods.
Transformation of yeast and the qualitative β-galactosidase (β-gal) colony lift assay were as published (19). Quantification of protein-protein interactions was determined by using the β-gal liquid assay performed on permeabilized yeast grown from three independent transformants by using orthonitrophenyl-β-d-galactopyranoside as substrate (19).
Protein Expression and RT Activity.
Fusion protein expression in yeast was determined by Western blot analysis of lysates with Gal4AD polyclonal antibodies (Upstate Biotechnology, Lake Placid, NY), anti-lexA polyclonal antibodies (Invitrogen), and HIV-1 RT polyclonal (Intracel, Cambridge, MA) or 5B2 monoclonal antibody (20). Immunodetection was with ECL-Plus (Amersham). To measure RT activity, yeast lysates were prepared by glass bead disruption (19), and enzyme activity was determined in exogenous assays (21) and was quantified by phosphoimager analysis.
Yeast Shuttle Vectors.
pSH2-1 (22) and pLex202-PL (23) express the lexA DNA binding domain (lexA87) and the full-length lexA protein (lexA202), respectively. pGBT9 and pAS2-1, both containing the GAL4 DNA binding domain (GAL4 BD), were purchased from CLONTECH. pNLexA allows expression of proteins fused to the N terminus of full-length lexA202 (OriGene Technologies, Rockville, MD). pGADNOT (18) and pACTII (24) allow expression of proteins fused to the Gal4 activation domain (GAL4 AD). pACTII also contains the influenza hemagglutinin (HA) epitope tag located between GAL4AD and the polylinker.
Construction of HIV-1 RT Fusions in Yeast Vectors.
Constructs and expressed fusion proteins are as described in Fig. 1. The RT sequence for constructing the following expression vectors was amplified from HIV-1 molecular clone pNLenv-1 (containing the HIVNL43 sequence) (25). The p66 amplimers were cloned into the BamHI-SalI sites of pGBT9, pSH2-1, pLex202-PL, pACTII, and pGADNOT; the BamHI-XhoI sites of pACTII; and the EcoRI-BamHI sites of pNLexA. p51 amplimers were cloned into the BamHI-SalI sites of these vectors except for cloning into pACTII, where the BamHI-XhoI sites were used. The HXB2 RT sequence from pHXB2gpt (26) was used to construct p66HXAlaLex202 and p51HXGADNOT.
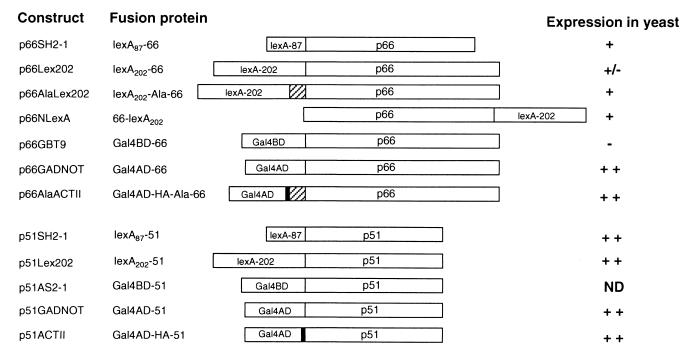
Figure 1.
RT fusion constructs, encoded fusion proteins, and expression of fusions in yeast reporter strains. The six-alanine linker is denoted by the hatched box, and the HA epitope by black shaded regions. p66 and p51 indicate the 66- and 51-kDa subunits of HIV-1 RT, respectively. Expression of fusion proteins was determined by introducing the indicated plasmids into CTY10-5d, except for p66GBT9 and p51AS2-1, which were introduced into HF7c. Fusion protein expression was detected by probing yeast protein lysates with anti-RT antibodies, as described in Materials and Methods. ++, high; +, moderate; +/−, low; −, undetectable protein expression. ND, not done.
Construction of HIV-1 RT Deletion Mutants.
All p66 deletion mutants were prepared by cloning PCR amplimers into the BamHI-SalI sites of pSH2-1. Fingers, palm, connection, thumb, and RNase H domains of HIV-1 RT are denoted F, P, C, T, and R, respectively. pT+C+RSH2-1 (encoding lexA87-T+C+R) contains RT (from HIVNL43) codons 236–560. pC+RSH2-1 (encoding lexA87-C+R) contains codons 322–560 whereas pRSH2-1 (encoding lexA87-R) comprises codons 425–560. All p51 deletion mutants were prepared by cloning of PCR amplimers into the BamHI-XhoI sites of pACTII. pF+P+T-ACTII (encoding Gal4AD-HA-F+P+T) includes RT codons 1–325, and pF+P-ACTII (encoding Gal4AD-HA-F+P) has codons 1–244. p51Δ13ACTII (encoding Gal4AD-HA-51Δ13) contains RT codons 1–426. p51Δ26GADNOT (encoding Gal4AD-51Δ26) was obtained by random mutagenesis of p51GADNOT in XL1-Red.
Construction of RT Fusions with the L234A Mutation and Random Mutagenesis of p66Ala234Lex202 and Selection of Revertants.
p66Ala234Lex202 (encoding lexA202-Ala-66L234A) was made by inserting p66 from p6HprotL234A (a gift from Vinayaka Prasad, Albert Einstein College of Medicine, New York) into the BamHI/SalI sites of pLex202-PL. p51234GADNOT (encoding Gal4AD-51L234A) was made by insertion of p51 from p6HprotL234A into the BamHI-SalI sites of pGADNOT. Second-site mutations restoring dimerization to lexA202-Ala-66L234A were generated by propagation of p66Ala234Lex202 in XL1-Red (Stratagene). Two independent pools were prepared. CTY10-5d was cotransformed with the mutagenized library and either p51234GADNOT or p51GADNOT. Blue colonies were picked from β-gal colony lift assays and were clonally purified. p66 DNA from isolated plasmids was recloned into a nonmutated pLex202-PL backbone and was reintroduced into CTY10-5d to confirm the phenotype. Mutations present in p66 were determined by automated nucleotide sequencing.
Site-Directed Mutagenesis.
p66 with genotype D110G was prepared from a p66 clone containing both D110G and L234A obtained by random mutagenesis by back-mutation of codon 234 to wild-type. p66 with either the W402R or W406R substitutions were prepared by subcloning a Bsp1286I-SalI fragment (600 bp) from the clones obtained by random mutagenesis of L234A with wild-type BamHI-Bsp1286I fragment (1,080 bp) from p66HXAlaLex202 into pLex202-PL.
In Vitro Heterodimerization.
Plasmids expressing wild-type and p66 mutants with a histidine tag at the C terminus (p66-His) were constructed by cloning the p66 coding region into the SphI-BglII site of pQE-70 (Qiagen, Chatsworth, CA). The C-terminal tag was appended as described [clone 3 (27)]. Glutathione S-transferase-tagged p51 (GST-p51) was prepared by subcloning the BamHI-SalI fragment from p51HXGADNOT into pGEX5X-3 (Amersham Pharmacia). Cells were induced and then lysed by the addition of 1 mg/ml of lysozyme to 1 ml of lysis buffer [50 mM sodium phosphate buffer (pH 7.8), 500 mM NaCl, 0.5% Nonidet P-40, 5 mM DTT, and 1 μg/ml each of pepstatin A, aprotinin, and leupeptin] and were clarified. Lysates were combined and incubated for 16 h at 4°C. The heterodimer was captured on Glutathione Sepharose 4B beads, and unbound subunits were removed by washing with lysis buffer. Heterodimer bound to beads were resolved by SDS/PAGE. For quantification of RT activity, dimers were eluted from beads with 10 mM reduced glutathione in 50 mM Tris (pH 8.0). Samples were assayed for DNA polymerase activity on homopolymeric template-primers for various times, and the activity was determined from the initial slope of the linear phase of the time course. Western blot analysis confirmed equal recovery of GST-51 protein in each sample.
Results
Expression of RT Fusion Proteins and RT Activity.
The stable expression of p66 was tested in several contexts, as either Gal4BD or LexA fusions, and using a six alanine linker to separate p66 from its fusion partner. p66 fused to the C terminus of lexA87, the C or N termini of lexA202 (with or without a six alanine spacer), and in a variety of contexts to the Gal4AD were all stably expressed (Fig. 1). In contrast, p66 fused to the C terminus of the Gal4BD (Gal4BD-66) was not expressed in yeast at detectable levels (Fig. 1). The smaller RT subunit, p51, was well expressed as fusions with the Gal4BD, Gal4AD, and both lexA87 and lexA202.
We examined whether the bait fusions encoded by p66SH2-1, p66AlaLex202, and p66NLexA exhibited RT activity in yeast. All three fusion proteins demonstrated high levels of RT activity compared with protein lysates from yeast transformed with an empty vector (data not shown). These data suggest that the p66 fusion proteins are functional and in a conformation consistent with measurable catalytic activity.
Heteromeric Interactions of p66 and p51 by Transactivation in the Two-Hybrid System.
To test whether the Y2H system could detect the interaction of the p66/p51 heterodimer, we cotransformed yeast reporter strains with plasmids expressing p66DNA BD and p51DNA AD fusion proteins (Table 1). β-gal activity expressed in yeast, which indicates the strength of the interaction between the fusion proteins, was assessed by both qualitative and quantitative assays. The p66 bait fusions expressed from p66SH2-1, p66AlaLex202, and p66NLexA interacted with Gal4AD-p51 domain fusions (Table 1) but not with Gal4AD alone (Table 1). The strongest interactions were observed with p66 baits lexA202-Ala-66 and 66-lexA202. Moreover, p51 expressed in pACTII gave a stronger signal than p51GADNOT when coexpressed with p66 fusion baits. Despite the stable expression of the p66 fusion protein, lexA202-66, no significant interaction with p51 was detected (Fig. 1). Moreover, lexA202-66 yielded the same weak signal with the empty Gal4AD vector, pGADNOT, indicating that this version of p66 is weakly self-activating even without a partner.
Table 1.
Interaction of p66 binding domain fusions with p51 activation domain fusions in the Y2H system
| Constructs | Operator | β-gal activity
|
|
|---|---|---|---|
| Colony* | Liquid† | ||
| p66SH2-1 + pGADNOT | lexA | − | ND |
| p66SH2-1 + pACTII | lexA | − | 0.02 |
| p66SH2-1 + p51GADNOT | lexA | ++ | 0.5 |
| p66SH2-1 + p51ACTII | lexA | +++ | 3.5 |
| p66AlaLex202 + pGADNOT | lexA | − | ND |
| p66AlaLex202 + pACTII | lexA | − | 0.04 |
| p66AlaLex202 + p51GADNOT | lexA | +++ | 1.6 |
| p66AlaLex202 + p51ACTII | lexA | +++ | 7.7 |
| p66NLexA + pGADNOT | lexA | − | 0.06 |
| p66NLexA + pACTII | lexA | − | 0.04 |
| p66NLexA + p51GADNOT | lexA | +++ | 6.6 |
| p66NLexA + p51ACTII | lexA | +++ | 25.0 |
| p66Lex202 + pGADNOT | lexA | +/− | ND |
| p66Lex202 + p51GADNOT | lexA | +/− | ND |
| p66GBT9 + pGADNOT‡ | UASG | − | ND |
| p66GBT9 + p51GADNOT‡ | UASG | − | ND |
Yeast strain CTY10-5d or ‡HF7c were transformed with plasmids encoding p66 bait and p51 prey fusions. Fusion proteins encoded by plasmids are described in Materials and Methods and Fig. 1.
Transformants were lifted onto nitrocellulose and subjected to the β-gal colony lift assay to determine intensities of blue color produced; +++, strong blue in 1 h; ++, blue in 1 h; +/−, weak blue in 3 h; −, white; ND, not done.
† Numbers represent β-gal activity in Miller units.
We also showed that heteromeric interactions between p66 and p51 could be detected in the reciprocal configuration with p51 as either a LexA or Gal4BD fusion and p66 as a Gal4AD fusion (Table 2). The demonstration of heteromeric dimerization of p66 and p51 in different contexts strongly suggests that the interaction is specific. Tests for interaction with five unrelated proteins showed no signal (data not shown), providing further evidence for the specificity of RT heterodimerization in yeast.
Table 2.
Interaction of p51 binding domain fusions with p66 activation domain fusion in the Y2H system
| Constructs | Operator | β-gal activity
|
|
|---|---|---|---|
| Colony* | Liquid† | ||
| p51SH2-1 + pGADNOT | lexA | − | 0.06 |
| p51SH2-1 + pACTII | lexA | − | 0.05 |
| p51SH2-1 + p66AlaACTII | lexA | ++ | 1.2 |
| p51Lex202 + pACTII | lexA | − | 0.05 |
| p51Lex202 + p66AlaACTII | lexA | +++ | 3.2 |
| p51AS2-1 + pACTII‡ | UASG | − | ND |
| p51AS2-1 + p66AlaACTII‡ | UASG | ++ | ND |
Homomeric Interactions.
The interaction of the RT heterodimer p66/p51 has a dissociation constant of 10−9 M whereas the dissociation constants for the p66 and p51 homodimers are 10−6 and 10−5 M, respectively (9). We were unable to detect p51 homodimerization when CTY10-5d was cotransformed with either p51SH2-1 or p51Lex202 baits and p51ACTII prey (data not shown). In contrast, p66 homodimerization could be detected when yeast was cotransformed with p66NlexA bait and p66AlaACTII prey (β-gal activity 0.3 Miller units). p66 homodimerization of these two constructs was 100-fold weaker compared with the interaction of p66NlexA with p51ACTII (Table 1). The strength of the interactions observed in vivo are consistent with biochemical data.
p66 Domains that Interact with p51.
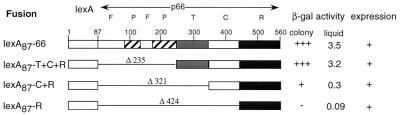
We used the Y2H RT dimerization assay to map the regions of p66 required for binding to p51 (Fig. 2). A series of mutants with sequential deletions in the polymerase subdomains were prepared as C-terminal fusions with lexA87. Deletion of the fingers and palm subdomains (lexA87-T+C+R) did not significantly affect binding to Gal4AD-HA-51. A further deletion of the thumb subdomain (lexA87-C+R) resulted in reduced β-gal activity (Fig. 2). Expression of the RNase H domain alone was not sufficient for interaction with p51. This lack of interaction was not attributable to an aberrant RNase H conformation, as lexA87-R also interacted as strongly as lexA87-66 with a cellular protein, diaphorase, that we find interacts with the RNase H domain of RT in the Y2H system (results not shown). None of the bait fusions demonstrated activation of the lacZ reporter gene when coexpressed with Gal4AD-HA alone, excluding the possibility of nonspecific self-activation by the bait fusions (results not shown). These data suggest that the connection and RNase H subdomains of p66 are sufficient for interaction with p51.
Figure 2.
Interaction of p66 deletion mutants with Gal4AD-HA-51 fusion protein. p66 domains were fused to the C terminus of lexA87 in pSH2-1. CTY10-5d was cotransformed with the appropriate constructs. Transformants were lifted onto nitrocellulose and were subjected to β-gal colony lift assay to determine intensities of blue color produced as defined in Tables 1 and 2. β-gal activity from liquid assays is expressed in Miller units. Expression in CTY10-5d of p66 fusion proteins was detected by using anti-lexA polyclonal antibodies. Expression levels are as defined as in the legend for Fig. 1.
The C Terminus of p51 Is Required for Interaction with p66.
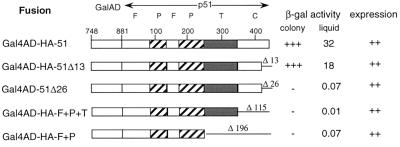
It has previously been shown biochemically that deletion of as little as 25 amino acids from the C terminus of p51 can prevent dimerization to p66 (15). To ascertain whether this effect could be observed under physiological conditions in the Y2H system, we constructed a series of C-terminal deletion mutants of p51 prey fusions and assayed interaction with p66 bait. Deletion of 13 amino acids from the C terminus of p51 had little effect (1.8-fold decrease) on dimerization with p66 (Fig. 3). However, deletions of 26 amino acids and greater abrogated RT dimerization, indicating the importance of the C-terminal 26 amino acids of p51 in these interactions. These results also suggest that the system faithfully recapitulates the behavior of the enzyme as studied in vitro.
Figure 3.
Interaction of C-terminal deletion mutants of p51 with lexA202-Ala-66. p51 domains were fused to the C terminus of the Gal4AD in pACTII. Deletions at the C terminus are denoted by the number of amino acids missing from the end of p51. β-gal activity was determined as described in the legend of Fig. 2. Expression of p51 fusion proteins in CTY10-5d was detected by using anti-GAL4AD antibodies, and expression levels are as denoted in the legend for Fig. 1.
L234A in p66 Subunit Inhibits RT Dimerization.
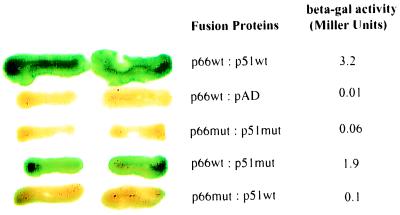
The Y2H RT dimerization assay would be most useful if it could be applied to the analysis of single amino acid substitutions that affect heteromeric interactions. To test the system, we introduced the L234A primer grip mutation, previously shown biochemically to inhibit p66/p51 association (10), into both RT subunits. The presence of L234A in both p66 and p51 totally inhibited RT dimerization as observed by a 53-fold decrease in the β-gal signal compared with wild-type proteins (Fig. 4). To assess the effect of L234A in individual subunits, CTY10d-5 was cotransformed with constructs expressing either p66 mutant bait and wild-type p51 prey, or p66 wild-type bait with p51 mutant prey. Less than a two-fold decrease in the signal compared with wild-type fusions was observed when the L234A mutant p51 (Gal4AD-51L234A) was coexpressed with the wild-type fusion lexA202-Ala-66 (Fig. 4). However, a 32-fold inhibition was observed for the interaction of the mutant lex202-Ala-66L234A with wild-type Gal4AD-51. These data suggest that L234A affects dimerization predominantly through p66, as has been previously reported (10). Analysis of fusion protein expression in yeast by Western blot analysis revealed that all fusion proteins, including the L234A mutants, were stably expressed (results not shown).
Figure 4.
L234A inhibits RT dimerization in the Y2H assay. CTY10-5d was cotransformed with expression constructs, and yeast patches were subjected to both the β-gal colony lift and liquid assays. The green is hydrolyzed X-gal and reflects β-gal activity. p66wt and p51wt denote wild-type lex202-Ala-HX66 and Gal4AD-HX51 fusion proteins, respectively. pAD denotes pGADNOT. p66mut and p51mut denote RT fusion proteins lex202-Ala-66-L234A and Gal4AD-51-L234A, respectively.
Second-Site Mutations that Restore Heterodimerization and RT Activity to the p66L234A Mutant.
To gain insight into the mechanism of inhibition of RT dimerization by L234A, we attempted to select for second-site suppressor mutations in p66 that restore dimerization with p51. To select for p66 mutants with restored dimerization, CTY10-5d was cotransformed with a library generated by mutagenesis of p66AlaL234ALex202 and a plasmid expressing either Gal4AD-51 or the Gal4AD-51-L234A mutant. A total of 25,000 colonies from each of two independently mutated libraries were screened. Six and five blue colonies were obtained when lex202-Ala-66L234A was cotransformed with Gal4AD-51 and Gal4AD-51-L234A, respectively. CTY10-5d was retransformed with each isolated library plasmid and with either p51HXGADNOT or p51L234AGADNOT; the recovered clones showed restored binding activity with both p51 fusion proteins. Five types of mutations were observed (Table 3). Single amino acid changes in the clones that retained the L234A change included D110G, D186V, W402R, and W406R. The remaining three clones had reverted to wild-type at codon 234 (Table 3).
Table 3.
Second site mutations in lexA20266HXL234A that restore dimerization to p51
| Genotype | No. of clones | β-gal activity
|
|
|---|---|---|---|
| Colony* | Liquid† | ||
| Wild type | NA | +++ | 3.2 |
| L234A | NA | − | 0.1 |
| D110G | NA | +++ | 6.9 |
| W402R | NA | +++ | 7.1 |
| W406R | NA | +++ | 5.8 |
| L234A, D110G | 3 | +++ | 1.5 |
| L234A; D186V | 1 | +/− | 0.2 |
| L234A; W402R | 3 | +++ | 7.1 |
| L234A; W406R | 1 | +++ | 6.1 |
| L234 | 3 | +++ | 4.7 |
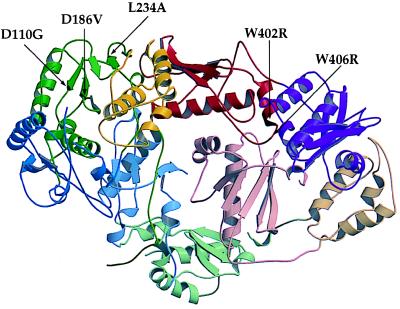
Two of the changes are at the catalytically essential aspartyl residues D110 and D186. These residues are not located at the dimer interface, and mutations at these residues result in an inactive RT (28) (Fig. 5). A variant p66 containing D110G alone, without L234A, gave a 2-fold stronger β-gal signal than wild-type p66 for heterodimerization and was 4.6-fold stronger compared with clones containing both L234A and D110G. Partial restoration of dimerization by D110G suggests that conformational changes at the active site compensate for structural changes mediated by L234A.
Figure 5.
Ribbon diagram of unliganded HIV-1 RT showing position of L234A primer grip mutation and locations of suppressors (shaded black). The figure was generated by molscript (38) and raster3d (39) with coordinates (2) retrieved from the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB) (http://www.rcsb.org/pdb; PDB ID code 1HMV). Domains are defined as in ref. 3: fingers, blue; palm, green; thumb, yellow; connection, red; RNase H, purple. Domains in p66 are in fully saturated colors whereas in p51 they have decreased saturation. Secondary structure was assigned by using dssp (40). Spirals represent α-helices; arrows denote β-strands.
The second set of mutations, W402R and W406R, are located at the dimer interface (Fig. 5) in a tryptophan repeat region that is highly conserved among HIV-1, HIV-2, and closely related simian immunodeficiency virus RTs (29). In the L234A genetic background, these mutations resulted in a dramatic increase in the β-gal signal over the parent and yielded a 2-fold higher signal for heterodimerization compared with wild-type RT fusions (Table 3). W402R and W406R in a wild-type genetic background had the same enhanced β-gal activity as the restored mutants (Table 3). Therefore, the mutations in the tryptophan repeat motif may enhance the interaction with GAL4AD-51 independently of the L234A mediated defect.
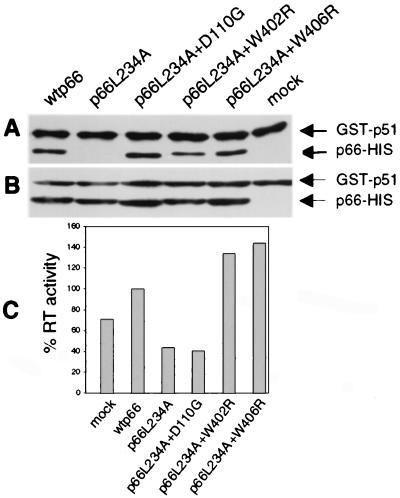
To confirm that the second-site mutations could restore heterodimerization to the L234A parent in an alternative assay, we examined the binding of these p66 mutants to p51 in vitro. Bacterial lysates containing GST-p51 or wild-type and mutant p66-His were incubated together, and heterodimers were captured on Glutathione Sepharose 4B beads. As expected, wild-type p66 dimerized with GST-p51 whereas the p66L234A mutant did not (Fig. 6A). Restoration of dimerization by D110G, W402R, or W406R in the L234A parent was observed (Fig. 6A), thus confirming our observations in the Y2H assay.
Figure 6.
In vitro assay for binding of GST-p51 and p66 to form active RT heterodimers. (A) Bacterial lysates containing GST-p51 and various p66 proteins as indicated were incubated overnight and were captured on glutathione beads. The complexes were eluted, were resolved by SDS/PAGE, were blotted to membrane, and were detected by monoclonal anti-RT antibodies. Mock, GST-p51 alone. (B) An aliquot of each incubation mix, reflecting input protein, was directly analyzed by SDS/PAGE and Western blot analysis as in A. (C) Bound proteins were eluted with glutathione and were assayed for RT activity with homopolymeric template-primer. Values are normalized to the wild-type control.
To determine whether restoration of heterodimerization was associated with enhanced DNA polymerase activity, heterodimers eluted from beads were assayed for RT activity (Fig. 6C). GST-p51 had significant background activity compared with wild-type enzyme. The enzyme resulting from incubation with p66L234A had the same background activity. As expected, heterodimers comprising p66L234A containing the active site mutation D110G also had only background activity. Interestingly, both W402R and W406R mutations not only restored heterodimerization to the L234A parent but also increased RT activity, even above levels of the wild-type control (Fig. 6C).
Discussion
In this study, we have shown that fusions of p66 and p51 can be stably expressed in yeast and can heterodimerize in reciprocal configurations. The presence of spacers in the form of alanine or an HA tag may have been an important aspect for stronger interactions in the Y2H assay. Moreover, we have validated the Y2H assay by comparing previously described effects of p51 deletions and the L234A substitution on heterodimerization. We have also shown how this assay can further the study of the HIV-1 RT structure-function by the identification of second-site mutations that restore RT dimerization.
The palm, connection, and RNase H domains of p66 make major contacts with p51. An indication that the palm region is important is the destabilization of the p66/p51 heterodimer by the nonnucleoside RT inhibitor 2′,5′-bis-O-(tert-butyldimethylsilyl)-3′-spiro-5"-4"-amino-1",2"-oxothiole-2",2"-dioxide)]-β-d-pentofuranosyl (TSAO) by its interaction between the palm subdomain of p66 and the β7-β8 loop in the fingers subdomain of p51 (30, 31). Preliminary tests of the addition of TSAO to our in vitro binding assays confirm the ability of the drug to reduce heterodimerization (data not shown). Tests of the related drug TSAOe3T showed a more modest destabilization only detectable in the presence of denaturants (31). Deletion mapping of the p66 domains required for interaction with p51 suggests that the presence of the connection and RNase H domains are sufficient for interaction with p51 in the Y2H system. It is surprising that the deletion of the palm domain had little effect on binding to p51 as this p66 subdomain provides a major contact with p51 (9); however, the connection and RNase H domains may provide a sufficient surface for saturating the signal in yeast.
Truncation of the C terminus of p51 revealed that a 13-amino acid deletion had little effect on dimerization with p66, but a deletion of 26 amino acids abrogated heterodimerization as seen in the Y2H assay. These data are consistent with previous in vitro studies (15). All C-terminal truncation mutants were stably expressed in yeast, excluding the possibility of decreased expression affecting the signal. It is possible that these C-terminal residues may have a direct role in dimerization; or the deletion of these residues may effect the structural integrity or correct positioning of the structural elements α-L and β-20 (5, 15). These elements contain the tryptophan repeat motif, which has been proposed to play an important role in HIV-1 dimerization (29, 32).
We have shown that the L234A substitution inhibits RT dimerization in yeast most dramatically when present on the p66 subunit of HIV-1 RT, as previously seen in vitro (10). L234A is located in the primer grip region of p66 (5) and is highly conserved among avian, primate, and murine RTs (33). To help determine the mechanism by which L234A affects heterodimerization, we selected for second-site mutations restoring p66/p51 association. Aside from clones that had reverted to the wild-type L234, we observed two classes of mutants: those with alterations either in the tryptophan repeat or in the polymerase active site (Fig. 5). Both classes of suppressors were also shown to restore binding of the mutant p66 subunit to p51 as measured in an in vitro binding assay (Fig. 6A). L234A is not at the dimer interface, and it has been proposed that it affects dimerization by indirectly affecting contacts between P95 in the palm of p66 with residues in the β7-β8 loop of p51 (11). The mutations W402R and W406R are distant from this region, being located in the connection subdomain that contacts the p51 connection domain in the heterodimer. The appearance of a basic residue in both codon 402 and 406 suggests a charge interaction with an acidic residue in p51 or, alternatively, an increase in electrostatic potential between the surfaces at the connection domain interface.
The recovery of second-site suppressor mutations at the catalytically essential aspartyl residues suggests that there is a relationship between dimerization and active site residues. Neither D186V nor D110G make obvious contacts with L234A, although both are in the same palm subdomain (Fig. 5) (2). Interaction between the NNRTI binding site, which includes L234, and the RT catalytic site has been suggested by both structural and enzymatic data explaining the mechanism of resistance to NNRTIs (34, 35). The D110G or D186V changes would probably result in loss of one of the two magnesium ions bound to the active site (36). A loss of chelated magnesium in addition to a glycine change at 110 may lead to increased flexibility in that region, thus affecting dimerization. Determination of the crystal structure of the D110G RT mutant will help resolve these issues.
Heterodimerization of HIV-1 has been suggested as a target for chemotherapeutic intervention (7). To date, there are no HIV-1 RT dimerization inhibitors being used in the clinic. Nevertheless, there are several reports of HIV-1 and HIV-2 RT dimerization inhibitors based on peptides representing the conserved tryptophan repeat region of RT (32, 37). These peptides have been shown to prevent the association of p66/p51 (32) and have demonstrable in vitro anti-HIV-1 activity (37). TSAO has been shown to destabilize the p66/p51 heterodimer and may represent a nonpeptide RT dimerization inhibitor (30). In preliminary tests of this drug for its effects on heterodimerization in the Y2H system, we saw no inhibition of β-gal activity (data not shown). However, we cannot rule out the possibility that the drug is not taken up by yeast. The availability of a Y2H assay for RT dimerization will facilitate the screening for other such inhibitors of this process.
Acknowledgments
We thank Drs. Wayne A. Hendrickson and Marianna Orlova for helpful discussions, Dr. Vinayaka Prasad for providing the p6HprotL234A construct, and Dr. Sergei Kuchin for advice on yeast protocols. G.T. was supported in part by C. J. Martin Fellowship 977375 awarded by the National Health and Medical Research Council of Australia. S.P.G. is an Investigator of the Howard Hughes Medical Institute. H.-E.G.A. was supported by National Institutes of Health Grant JM34102-16 to W. A. Hendrickson.
Abbreviations
- RT
reverse transcriptase
- Y2H
yeast two-hybrid
- β-gal
β-galactosidase
- HA
hemagglutinin
- GST-p51
glutathione S-transferase-tagged p51
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.di Marzo Veronese F, Copeland T D, DeVico A L, Rahman R, Oroszlan S, Gallo R C, Sarngadharan M G. Science. 1986;231:1289–1291. doi: 10.1126/science.2418504. [DOI] [PubMed] [Google Scholar]
- 2.Rodgers D W, Gamblin S J, Harris B A, Ray S, Culp J S, Hellmig B, Woolf D J, Debouck C, Harrison S C. Proc Natl Acad Sci USA. 1995;92:1222–1226. doi: 10.1073/pnas.92.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ren J, Esnouf R, Garman E, Somers D, Ross C, Kirby I, Keeling J, Darby G, Jones Y, Stuart D, et al. Nat Struct Biol. 1995;2:293–302. doi: 10.1038/nsb0495-293. [DOI] [PubMed] [Google Scholar]
- 4.Huang H, Chopra R, Verdine G L, Harrison S C. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 5.Jacobo-Molina A, Ding J, Nanni R G, Clark A D, Jr, Lu X, Tantillo C, Williams R L, Kamer G, Ferris A L, Clark P, et al. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 7.Restle T, Muller B, Goody R S. J Biol Chem. 1990;265:8986–8988. [PubMed] [Google Scholar]
- 8.Becerra S P, Kumar A, Lewis M S, Widen S G, Abbotts J, Karawya E M, Hughes S H, Shiloach J, Wilson S H. Biochemistry. 1991;30:11707–11719. doi: 10.1021/bi00114a015. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Smerdon S J, Jager J, Kohlstaedt L A, Rice P A, Friedman J M, Steitz T A. Proc Natl Acad Sci USA. 1994;91:7242–7246. doi: 10.1073/pnas.91.15.7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh M, Jacques P S, Rodgers D W, Ottman M, Darlix J L, Le Grice S F. Biochemistry. 1996;35:8553–8562. doi: 10.1021/bi952773j. [DOI] [PubMed] [Google Scholar]
- 11.Wohrl B M, Krebs R, Thrall S H, Le Grice S F J, Scheidig A J, Goody R S. J Biol Chem. 1997;272:17581–17587. doi: 10.1074/jbc.272.28.17581. [DOI] [PubMed] [Google Scholar]
- 12.Goel R, Beard W A, Kumar A, Casas-Finet J R, Strub M P, Stahl S J, Lewis M S, Bebenek K, Becerra S P, Kunkel T A, et al. Biochemistry. 1993;32:13012–13018. doi: 10.1021/bi00211a009. [DOI] [PubMed] [Google Scholar]
- 13.Divita G, Restle T, Goody R S. FEBS Lett. 1993;324:153–158. doi: 10.1016/0014-5793(93)81383-b. [DOI] [PubMed] [Google Scholar]
- 14.Debyser Z, De Clercq E. Protein Sci. 1996;5:278–286. doi: 10.1002/pro.5560050212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacques P S, Wohrl B M, Howard K J, Le Grice S F. J Biol Chem. 1994;269:1388–1393. [PubMed] [Google Scholar]
- 16.Fields S, Song O. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 17.Kalpana G V, Goff S P. Proc Natl Acad Sci USA. 1993;90:10593–10597. doi: 10.1073/pnas.90.22.10593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luban J, Bossolt K L, Franke E K, Kalpana G V, Goff S P. Cell. 1993;73:1067–1078. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- 19.Rose M D, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. [Google Scholar]
- 20.Szilvay A M, Nornes S, Haugan I R, Olsen L, Prasad V R, Endresen C, Goff S P, Helland D E. J Acquired Immune Defic Syndr. 1992;5:647–657. [PubMed] [Google Scholar]
- 21.Telesnitsky A, Blain S, Goff S P. Methods Enzymol. 1995;262:347–362. doi: 10.1016/0076-6879(95)62029-x. [DOI] [PubMed] [Google Scholar]
- 22.Hanes S D, Brent R. Cell. 1989;57:1275–1283. doi: 10.1016/0092-8674(89)90063-9. [DOI] [PubMed] [Google Scholar]
- 23.Ruden D M, Ma J, Li Y, Wood K, Ptashne M. Nature (London) 1991;350:250–252. doi: 10.1038/350250a0. [DOI] [PubMed] [Google Scholar]
- 24.Legrain P, Dokhelar M C, Transy C. Nucleic Acids Res. 1994;22:3241–3242. doi: 10.1093/nar/22.15.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maldarelli F, Martin M A, Strebel K. J Virol. 1991;65:5732–5743. doi: 10.1128/jvi.65.11.5732-5743.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ratner L, Haseltine W, Patarca R, Livak K J, Starcich B, Josephs S F, Doran E R, Rafalski J A, Whitehorn E A, Baumeister K, et al. Nature (London) 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 27.Maier G, Dietrich U, Panhans B, Schroder B, Rubsamen-Waigmann H, Cellai L, Hermann T, Heumann H. Eur J Biochem. 1999;261:10–18. doi: 10.1046/j.1432-1327.1999.00304.x. [DOI] [PubMed] [Google Scholar]
- 28.Larder B A, Purifoy D J, Powell K L, Darby G. Nature (London) 1987;327:716–717. doi: 10.1038/327716a0. [DOI] [PubMed] [Google Scholar]
- 29.Baillon J G, Nashed N T, Kumar A, Wilson S H, Jerina D M. New Biol. 1991;3:1015–1019. [PubMed] [Google Scholar]
- 30.Harris D, Lee R, Misra H S, Pandey P K, Pandey V N. Biochemistry. 1998;37:5903–5908. doi: 10.1021/bi9728452. [DOI] [PubMed] [Google Scholar]
- 31.Sluis-Cremer N, Dmitrienko G I, Balzarini J, Camarasa M J, Parniak M A. Biochemistry. 2000;39:1427–1433. doi: 10.1021/bi991682+. [DOI] [PubMed] [Google Scholar]
- 32.Divita G, Restle T, Goody R S, Chermann J C, Baillon J G. J Biol Chem. 1994;269:13080–13083. [PubMed] [Google Scholar]
- 33.Georgiadis M M, Jessen S M, Ogata C M, Telesnitsky A, Goff S P, Hendrickson W A. Structure (London) 1995;3:879–892. doi: 10.1016/S0969-2126(01)00223-4. [DOI] [PubMed] [Google Scholar]
- 34.Esnouf R, Ren J, Ross C, Jones Y, Stammers D, Stuart D. Nat Struct Biol. 1995;2:303–308. doi: 10.1038/nsb0495-303. [DOI] [PubMed] [Google Scholar]
- 35.Spence R A, Kati W M, Anderson K S, Johnson K A. Science. 1995;267:988–993. doi: 10.1126/science.7532321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel P H, Jacobo-Molina A, Ding J, Tantillo C, Clark A D, Jr, Raag R, Nanni R G, Hughes S H, Arnold E. Biochemistry. 1995;34:5351–5363. doi: 10.1021/bi00016a006. [DOI] [PubMed] [Google Scholar]
- 37.Morris M C, Robert-Hebmann V, Chaloin L, Mery J, Heitz F, Devaux C, Goody R S, Divita G. J Biol Chem. 1999;274:24941–24946. doi: 10.1074/jbc.274.35.24941. [DOI] [PubMed] [Google Scholar]
- 38.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 39.Merrit E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 40.Kabsch W, Sander C. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]