Abstract
Background
Globicatella are streptococcus‐like organisms that have been rarely isolated from clinical specimens. Their epidemiology and clinical significance remain largely unknown.
Aims
To describe two cases of Globicatella bacteraemia identified by 16S ribosomal RNA (rRNA) gene sequencing.
Methods
Two unidentified streptococcus‐like bacteria isolated from blood cultures of patients were subject to 16S rRNA gene sequencing.
Results
Two cases of Globicatella bacteraemia were identified by 16S rRNA gene sequencing. In the first case, a gram positive coccus was isolated from the blood culture of an 80 year old woman with diabetes mellitus and nosocomial sepsis, who died the day after developing the bacteraemia. The bacterium was unidentified by conventional phenotypic tests, the Vitek (gram positive identification) and the ATB expression (ID32 Strep) systems. In the second case, a similar bacterium was isolated from the blood culture of a 92 year old woman with polymicrobial acute pyelonephritis complicated by septic shock, who subsequently recovered after antibiotic treatment. 16S rRNA gene sequencing of the two isolates showed 0.5% nucleotide difference from that of G. sulfidifaciens and 0.7% nucleotide difference from that of G. sanguinis, indicating that they were Globicatella species.
Conclusions
Because Globicatella is rarely encountered in clinical microbiology laboratories, it may have been overlooked or misidentified in these cases. 16S rRNA gene sequencing is a useful tool to better characterise the epidemiology and clinical significance of Globicatella.
Keywords: Globicatella , bacteraemia, 16S rRNA gene sequencing
Since the recognition of the 16S ribosomal RNA (rRNA) gene as a new standard for classification and identification of bacteria,1,2 most bacterial species have been subjected to 16S rRNA gene sequence analysis, with reclassification made and new species identified. The taxonomy of the genera Streptococcus, Enterococcus, Lactococcus, and related members have also undergone major revisions.3,4,5 Recently, we reported the use of this technique for identifying and defining the clinical significance of streptococci6,7,8,9,10,11 and streptococcus‐like organisms;12,13,14 and discovery of a novel Streptococcus species.15,16
The genus Globicatella was first described in 1992 when several unidentified streptococcus‐like clinical isolates were characterised in the USA.17 The isolates were isolated from blood cultures of patients with bacteraemia, urine of patients with urinary tract infections, and cerebrospinal fluid of a patient with meningitis, but the clinical details were not described. Based on their unique phenotypic characteristics and phylogenetic position by 16S rRNA gene sequence analysis, they were classified in a new genus Globicatella, as G. sanguis, which was later renamed as G. sanguinis. G. sanguinis was also later isolated from a lamb with meningoencephalitis in Spain.18 Subsequently, a new species of the genus, G. sulfidifaciens, was described, when several animal isolates from Belgium with resemblance to G. sanguinis were studied. They comprised isolates from the lungs of calves and a lamb with purulent lung infections, and joint fluid of a pig and a calf with polyarthritis.19 Although there was 99.2% similarity in their 16S rRNA gene sequences to those of G. sanguinis, they were classified as a new species based on differences in their whole cell protein patterns and biochemical profiles. Since then, there have been no further reports on the isolation of Globicatella from humans and the clinical significance of this rarely encountered genus remains to be determined. In this report, we describe the application of 16S ribosomal RNA gene sequencing in characterising two cases of Globicatella bacteraemia. The difficulties in identifying the two isolates in clinical microbiology laboratories and their clinical significance are also discussed.
METHODS
Microbiological methods
Clinical specimens were collected and handled according to standard protocols. The BACTEC 9240 blood culture system (Becton Dickinson, MD, USA) was used. All isolates were identified by standard conventional biochemical methods,20 the Vitek System (gram positive identification; GPI) (bioMerieux Vitek, Hazelwood, M), USA) and the ATB expression system (ID32 Strep) (bioMerieux Vitek). Antimicrobial susceptibility was tested by disk diffusion and E‐test (AB Biodisk, Solna, Sweden) and results interpreted according to the NCCLS criteria for anaerobic bacteria.21 All tests were performed in triplicate with freshly prepared media on separate occasions.
Extraction of bacterial DNA for 16S ribosomal RNA gene sequencing
Bacterial DNA extraction was performed as described previously.6,15 Briefly, 80 μl of NaOH (0.05 mol/l) was added to 20 μl of bacterial cells suspended in distilled water and the mixture was incubated at 60°C for 45 minutes, followed by addition of 6 μl of Tris‐HCl (pH 7.0), achieving a final pH of 8.0. The resultant mixture was diluted 100×, and 5 μl of the diluted extract was used for PCR.
PCR, gel electrophoresis, and 16S ribosomal RNA gene sequencing
PCR amplification and DNA sequencing of the 16S rRNA gene was performed according to our previous publications.6,12 Briefly, DNase I treated distilled water and PCR master mix (dNTPs, PCR buffer, and Taq polymerase) were used in all PCR reactions by adding 1 U of DNase I (Pharmacia, Sweden) to 40 μl of distilled water or PCR master mix, incubating the mixture at 25°C for 15 minutes, and subsequently at 95°C for 10 minutes to inactivate the DNase I. The bacterial DNA extract and control were amplified with 0.5 μmol/l primers (LPW57 5′‐AGTTTGATCCTGGCTCAG‐3′ and LPW205 5′‐CTTGTTACGACTTCACCC‐3′; Gibco BRL, Rockville, MD, USA). The PCR mixture (50 μl) contained bacterial DNA, PCR buffer (10 mmol/l Tris‐HCl pH 8.3, 50 mmol/l KCl, 2 mmol/l MgCl2, and 0.01% gelatin), 200 μmol/l of each dNTP and 1.0 U Taq polymerase (Boehringer Mannheim, Germany). The mixtures were amplified in in an automated thermal cycler (Perkin‐Elmer Cetus, Gouda, The Netherlands), using 40 cycles of 94°C for 1 minute, 60°C for 1 minute, and 72°C for 2 minutes, with a final extension at 72°C for 10 minutes. DNase I treated distilled water was used as the negative control. A 10 μl sample of each amplified product was electrophoresed in 1.5% (w/v) agarose gel, with a molecular size marker (φX174 HaeIII digest,; Boehringer Mannheim, Germany) in parallel. Electrophoresis in Tris borate EDTA buffer was performed at 100 V for 1.5 hours. The gel was stained with ethidium bromide (0.5 μg/ml) for 15 minutes, rinsed, and photographed under 312 nm ultraviolet light illumination.
The PCR products were gel purified using the QIAquick PCR purification kit (QIAgen, Hilden, Germany). Both strands of the PCR product were sequenced twice with an ABI 377 automated sequencer according to manufacturers' instructions (Perkin‐Elmer, Foster City, CA, USA), using the PCR primers. The sequences of the PCR products were compared with known 16S rRNA gene sequences in GenBank (hwww.ncbi.nlm.nih.gov) by multiple sequence alignment using the Clustal W program.22 Phylogenetic tree construction was performed using Clustal X (version 1.81) 23 and the neighbour joining method with GrowTree (Genetics Computer Group Inc., San Diego, USA). In total, 1360 nucleotide positions were included in the analysis.
RESULTS
Patients and identification of the bacterial strains by conventional methods and commercially available systems
Patient 1
An 80 year old Chinese woman was admitted to hospital because of a 3 month history of chronic diarrhoea. She had been passing loose stool with mucus 2–3 times a day. She had diabetes mellitis, hypertension, and gouty arthritis, and was receiving oral hypoglycaemic and anti‐hypertensive medications. On admission, she was afebrile. Abdominal examination and colonoscopy was unremarkable. She was found to have hypercalcaemia with adjusted serum calcium 3.14 mmol/l and was given intravenous fluid replacement, loop diuretics, and bisphosphonate with gradual normalisation of serum calcium. She developed sudden cardiac arrest 2 weeks after admission and was stabilised after initial resuscitation. Her total leukocyte count was 22×109/l, haemoglobin level 9.8 g/dl, and platelet count 457×109/l. Her serum urea was 19.3 mmol/l, creatinine 211 μmol/l, albumin 30 g/l, globulin 36 g/l, bilirubin 7 μmol/l, alkaline phosphatase 115 IU/l, and alanine aminotransferase 87 IU/l. Blood cultures were performed. However, she continued to deteriorate and succumbed on the next day.
On day 2 post‐incubation, the aerobic blood culture bottle showed a gram positive coccus (isolate 1). It grew on sheep blood agar as greyish white, α‐haemolytic colonies of 0.5 mm in diameter after 24 hours of incubation at 37°C in ambient air. There was enhancement of growth in 5% CO2 and anaerobic environment. It also grew on MacConkey agar but did not grow in 6.5% NaCl. It was positive for pyrolidonyl arylamidase and esculin, and negative for catalase, arginine, or hippurate hydrolysis, bile aesculin, and Voges‐Proskauer tests. It was non‐groupable with Lancefield groups A, B, C, D, F, or G antisera. Both the Vitek and ATB expression systems showed that it was “unidentified” (table 1). It was susceptible to vancomycin (MIC 0.25 μg/ml), intermediately susceptible to penicillin (MIC 2 μg/ml), and resistant to and cefotaxime (MIC 4 μg/ml), erythromycin, clindamycin, and neomycin.
Table 1 Phenotypic characteristics of the two blood culture isolates, Globicatella sulfidifaciens, and Globicatella sanguinis.
| Characteristics | Isolate 1 | Isolate 2 | G. sulfidaciens19 | G. sanguinis17,19 | |||
|---|---|---|---|---|---|---|---|
| Catalase | − | − | − | − | |||
| Lancefield grouping | Non‐group A, B, C, D, F, or G | Non‐group A, B, C, D, F, or G | |||||
| Resistance to bacitracin | − | − | |||||
| Resistance to optochin | − | − | |||||
| Growth in 6.5% NaCl | − | + | + | + | |||
| Growth in 10% bile | − | + | |||||
| Growth in 40% bile | − | − | |||||
| Esculin hydrolysis | + | − | + | + | |||
| Hippurate hydrolysis | − | − | − | + | |||
| Arginine hydrolysis | − | − | − | − | |||
| Urease | − | − | − | − | |||
| Voges−Proskauer test | − | − | − | − | |||
| Tetrazolium reduction | − | − | − | ||||
| Resistance to novobiocin | − | − | |||||
| Production of H2S | − | − | + | − | |||
| Utilisation of: | |||||||
| Hemicellulase | − | − | |||||
| Dextrose | + | + | + | ||||
| Lactose | − | − | − | V | |||
| Mannitol | − | − | − | + | |||
| Raffinose | − | + | + | + | |||
| Salicin | + | + | + | ||||
| Sorbitol | − | − | − | V | |||
| Sucrose | + | + | + | + | |||
| Trehalose | + | + | + | + | |||
| Arabinose | − | − | − | ||||
| Pyruvate | − | − | − | ||||
| Pullulan | − | + | V | ||||
| Inulin | − | + | + | − | |||
| Melibiose | − | + | + | + | |||
| Melezitose | − | − | − | ||||
| Cellobiose | + | + | + | ||||
| Ribose | + | − | − | + | |||
| Xylose | − | − | − | ||||
| Maltose | + | + | + | + | |||
| Glycogen | − | + | + | + | |||
| D‐Arabitol | − | − | |||||
| Methyl‐B‐D‐glucopyranoside | + | + | |||||
| Tagatose | − | − | |||||
| Cyclodextrin | − | − | |||||
| Pyrrolidonylarylamidase | + | + | − | + | |||
| α‐galactosidase | + | + | + | + | |||
| β‐glucuronidase | − | − | + | − | |||
| β‐galactosidase | + | + | V | + | |||
| β‐glucosidase | + | − | |||||
| Alanine‐phenylalanine‐proline arylamidase | + | + | + | ||||
| N‐acetyl‐β‐glucosaminidase | − | − | − | + | |||
| Glycyl‐tryptophane arylamidase | − | − | |||||
| β‐mannosidase | + | − | |||||
| Alkaline phosphatase | − | − | − |
Patient 2
A 92‐year‐old Chinese woman was admitted to hospital because of fever and productive cough with yellow sputum for one day. She had dementia, congestive heart failure, and history of recurrent urinary tract infection. She was bedridden and was put on nasogastric tube feeding. On admission, her oral temperature was 39.5°C. Physical examination did not reveal an obvious focus of infection. Her total leukocyte count was 36.3×109/l, haemoglobin level 11.1 g/dl, and platelet count 95×109/l. Her renal and liver function tests were within normal limits. Blood cultures were performed. She went into septic shock soon after admission and empirical intravenous cefuroxime was administered. Urine microscopy showed the presence of numerous leucocytes and bacteria. Urine culture recovered Pseudomonas aeruginosa with bacterial count >100 000 cfu/ml. Ultrasonography of the kidneys only showed the presence of a renal stone on the right side with no evidence of obstructive uropathy. Antibiotic was switched to intravenous ceftazidime. Her fever responded and she was discharged after 2 weeks of antibiotics.
On day 2 post‐incubation, the aerobic blood culture bottle turned positive with a gram negative bacillus and two gram positive cocci. The gram negative bacillus and one of the gram positive cocci were identified as Pseudomonas aeruginosa and Streptococcus sanguis respectively. The other gram positive coccus (isolate 2) possessed phenotypic characteristics similar to isolate 1 (table 1). It was susceptible to vancomycin (MIC 0.25 μg/ml) and clindamycin, intermediately susceptible to penicillin (MIC 1 μg/ml) and cefotaxime (MIC 2 μg/ml), and resistant to erythromycin and neomycin.
16S ribosomal RNA gene sequencing and phylogenetic analysis
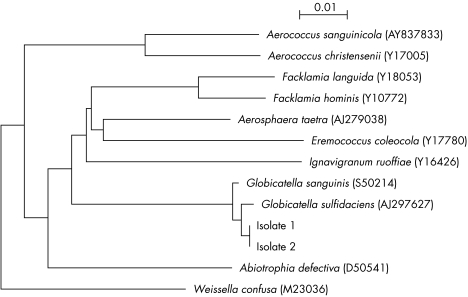
PCR of the 16S rRNA gene of both isolates showed bands at about 1450 bp. The 16S rRNA gene sequences of the two isolates were identical and had 0.5% nucleotide difference from G. sulfidifaciens (GenBank accession no. AJ297627), 0.7% nucleotide difference from G. sanguinis (GenBank accession no. S50214), 3.69% nucleotide difference from Facklamia hominis (GenBank accession no. Y10772), 4.9% difference from Aerosphaera taetra (GenBank accession no. AJ279038), and 5.5% difference from F. languida (GenBank accession no. Y18053), indicating that the two isolates belong to the genus Globicatella (fig 1).
Figure 1 Phylogenetic tree showing the relationships of the two blood culture isolates to Globicatella and members of other related genera. The tree was constructed by using the neighbour joining method. The scale bar indicates the estimated number of substitutions per 100 bases using the Jukes‐Cantor correction. Names and accession numbers are given as cited in the GenBank database.
Based on their phenotypic and genotypic data, the two isolates could not be assigned to a particular species of the genus Globicatella. Phenotypically, they resembled G. sanguinis in the production of pyrrolidonylarylamidase and the negative H2S and β‐glucuronidase production. On the other hand, they resembled G. sulfidifaciens in that they did not use mannitol and did not produce N‐acetyl‐β‐glucosaminidase. However, like G. sanguinis, isolate 1 used ribose, while isolate 2. like G. sulfidifaciens, did not (table 1). Genotypically, the 16S rRNA gene sequences of the two isolates were close to both G. sulfidifaciens and G. sanguinis, which possess highly homologous 16S rRNA sequences with only 0.8% difference. Therefore, the identification of more strains of Globicatella and correlation of their phenotypic characteristics with 16S rRNA gene sequences should help in defining the number of different species within the genus.
DISCUSSION
We describe two cases of Globicatella bacteraemia characterised by 16S rRNA gene sequencing. The two isolates were unidentified by conventional biochemical tests and commercial identification systems, and were only confirmed to be Globicatella species after 16S rRNA gene sequencing. In retrospect, the phenotypic characteristics of the two blood culture isolates actually closely resembled those of Globicatella (table 1). They are facultative anaerobic, catalase negative, α‐haemolytic, gram positive cocci. Similar to G. sanguinis and G. sulfidifaciens, arginine is not hydrolysed, urease not produced, and Voges‐Proskauer test negative for the two isolates. While both G. sanguinis and G. sulfidifaciens grow in 6.5% NaCl, one of our isolates did not. Identification of more isolates of Globicatella would help delineate the phenotypic characteristics and variations within the genus. As Globicatella is rarely encountered in clinical laboratories, most technicians and microbiologists are not familiar with their phenotypic characteristics and identification. As a result, the bacterium may be overlooked when isolated or reported as unidentified streptococcus‐like organisms. 16S rRNA gene sequencing will continue to be useful in the characterisation of rarely encountered bacteria and defining their clinical significance.6,13,24 As PCR and sequencing techniques are becoming more readily available in clinical laboratories, this molecular tool is probably the most practical approach to identify these bacteria.
The clinical significance of the two isolates in our two patients is evident by their isolation from blood in association with the development of fever and neutrophilia. Their infections have been severe because the first patient died soon after the bacteraemia and the second was complicated by septic shock. While the source of the bacteraemia in the first patient could not be identified, the second case probably represents a case of polymicrobial acute pyleonephritis complicated by bacteraemia. S. sanguis and P. aeruginosa, which were concomitantly isolated from the blood of the second patient, are known pathogens of the urinary tract and can reside in the gastrointestinal tract.25 Moreover, G. sanguis has been isolated from the urine of patients with urinary tract infections.17 Therefore, Globicatella is likely to be an uncommon cause of urinary tract infections and may have originated from the gut. However, only P. aeruginosa was recovered in her urine. This may be explained by the slower growth of Globicatella and S. sanguis as opposed to the larger colonies of P. aeruginosa. Nevertheless, as the second patient had mixed infections, the role of Globicatella in this case still remains to be determined. Further studies are required to investigate the role of Globicatella in human infections.
TAKE HOME MESSAGES
Globicatella are streptococcus‐like organisms that have been rarely isolated from clinical specimens, whose epidemiology and clinical significance remain largely unknown.
Using 16S rRNA sequencing, two cases were identified as Globicatella spp.
The infections were severe, leading to death in one case and septic shock in the other.
Further studies are required to investigate the role of Globicatella in human infections.
ACKNOWLEDGEMENTS
This work was partly supported by the University Development Fund, and the Committee of Research and Conference Grant, The University of Hong Kong; Research Fund for the Control of Infectious Diseases of the Health, Welfare and Food Bureau of the Hong Kong SAR Government; and the William Benter Infectious Disease Fund.
Abbreviations
GPI - gram positive identification
rRNA - ribosomal RNA
References
- 1.Relman D A, Loutit J S, Schmidt T M.et al The agent of bacillary angiomatosis. An approach to the identification of uncultured pathogens. N Engl J Med 19903231573–1580. [DOI] [PubMed] [Google Scholar]
- 2.Relman D A, Schmidt T M, MacDermott R P.et al Identification of the uncultured bacillus of Whipple's disease. N Engl J Med 1992327293–301. [DOI] [PubMed] [Google Scholar]
- 3.Collins M D, Ash C, Farrow J A.et al 16S ribosomal ribonucleic acid sequence analyses of lactococci and related taxa. Description of Vagococcus fluvialis gen. nov., sp. nov. J Appl Bacteriol 198967453–460. [DOI] [PubMed] [Google Scholar]
- 4.Bentley R W, Leigh J A, Collins M D. Intrageneric structure of Streptococcus based on comparative analysis of small‐subunit rRNA sequences. Int J Syst Bacteriol 199141487–494. [DOI] [PubMed] [Google Scholar]
- 5.Morse R, Collins M D, O'Hanlon K.et al Analysis of the beta' subunit of DNA‐dependent RNA polymerase does not support the hypothesis inferred from 16S rRNA analysis that Oenococcus oeni (formerly Leuconostoc oenos) is a tachytelic (fast‐evolving) bacterium. Int J Syst Bacteriol 1996461004–1009. [DOI] [PubMed] [Google Scholar]
- 6.Lau S K P, Woo P C Y, Tse H.et al Invasive Streptococcus iniae infections outside North America. J Clin Microbiol 2003411004–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woo P C, Tse H, Chan K M.et al “Streptococcus milleri” endocarditis caused by Streptococcus anginosus. Diagn Microbiol Infect Dis 20044881–88. [DOI] [PubMed] [Google Scholar]
- 8.Woo P C, Fung A M, Lau S K.et al Group G beta‐hemolytic streptococcal bacteremia characterized by 16S ribosomal RNA gene sequencing. J Clin Microbiol 2001393147–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woo P C, Teng J L, Lau S K.et al Analysis of a viridans group strain reveals a case of bacteremia due to Lancefield group G alpha‐hemolytic Streptococcus dysgalactiae subsp. equisimilis in a patient with pyomyositis and reactive arthritis. J Clin Microbiol 200341613–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau S K, Woo P C, Yim T C.et al Molecular characterization of a strain of group a streptococcus isolated from a patient with a psoas abscess. J Clin Microbiol 2003414888–4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee R A, Woo P C, To A P.et al Geographical difference of disease association in Streptococcus bovis bacteraemia. J Med Microbiol 200352903–908. [DOI] [PubMed] [Google Scholar]
- 12.Woo P C, Tam D M, Lau S K.et al Enterococcus cecorum empyema thoracis successfully treated with cefotaxime. J Clin Microbiol 200442919–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woo P C, Lau S K, Fung A M.et al Gemella bacteraemia characterised by 16S ribosomal RNA gene sequencing. J Clin Pathol 200356690–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woo P C, Fung A M, Lau S K.et al Granulicatella adiacens and Abiotrophia defectiva bacteraemia characterized by 16S rRNA gene sequencing. J Med Microbiol 200352137–140. [DOI] [PubMed] [Google Scholar]
- 15.Woo P C Y, Tam D M W, Leung K W.et al Streptococcus sinensis sp. nov., a novel Streptococcus species isolated from a patient with infective endocarditis. J Clin Microbiol 200240805–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woo P C, Teng J L, Leung K W.et al Streptococcus sinensis may react with Lancefield group F antiserum. J Med Microbiol 2004531083–1088. [DOI] [PubMed] [Google Scholar]
- 17.Collins M D, Aguirre M, Facklam R R.et al Globicatella sanguis gen. nov., sp. nov., a new gram‐positive catalase‐negative bacterium from human sources. J Appl Bacteriol 199273433–437. [DOI] [PubMed] [Google Scholar]
- 18.Vela A I, Fernandez E, las Heras A.et al Meningoencephalitis associated with Globicatella sanguinis infection in lambs. J Clin Microbiol 2000384254–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vandamme P, Hommez J, Snauwaert C.et al Globicatella sulfidifaciens sp. nov., isolated from purulent infections in domestic animals. Int J Syst Evol Microbiol 2001511745–1749. [DOI] [PubMed] [Google Scholar]
- 20.Murray P R, Baro E J, Jorgensen J H.et alManual of clinical microbiology. 8th ed. Washington DC: American Society for Microbiology, 2003
- 21.National Committee for Clinical Laboratory Standards M2‐A7, performance standards for antimicrobial disk susceptibility tests. Approved standard. 8th ed. Wayne, PA: National Committee for Clinical Laboratory Standards, 2003
- 22.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position‐specific gap penalties and weight matrix choice. Nucleic Acids Res 1994224673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeanmougin F, Thompson J D, Gouy M.et al Multiple sequence alignment with ClustalX. Trends Biochem Sci 199810403–405. [DOI] [PubMed] [Google Scholar]
- 24.Lau S K, Woo P C, Teng J L.et al Identification by 16S ribosomal RNA gene sequencing of Arcobacter butzleri bacteraemia in a patient with acute gangrenous appendicitis. Mol Pathol 200255182–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christensen J J, Korner B, Kjaergaard H.Aerococcus‐like organism—an unnoticed urinary tract pathogen. APMIS 198997539–546. [DOI] [PubMed] [Google Scholar]