Abstract
Background
Basophils are highly specialised granulocytes that express a unique profile of antigens and increase in myeloproliferative disorders (MPD). In chronic myeloid leukaemia (CML), basophilia is a diagnostic and prognostic determinant. So far, however, no reliable approach for routine detection and enumeration of bone marrow basophils has become available.
Objective
To detect and enumerate basophils in bone marrow sections in patients with CML and other MPD
Methods
The anti‐basophil antibody 2D7 was applied to paraffin embedded bone marrow sections from normal/reactive subjects (n = 31), patients with CML (chronic phase, n = 37; accelerated phase, n = 9), and other MPD (chronic idiopathic myelofibrosis (CIMF), n = 20; polycythaemia vera (PV), n = 20; essential thrombocythaemia (ET), n = 20; indolent systemic mastocytosis (ISM), n = 7).
Results
As assessed by serial section staining, 2D7+ cells were found to co‐express myeloperoxidase, histidine decarboxylase, CD9, and CD43, but did not express B cell or T cell restricted antigens. 2D7+ bone marrow cells were found to increase in CML compared with normal/reactive bone marrow and other MPD (median numbers of 2D7+ cells/mm2: CML, 33; normal/reactive bone marrow, 6; CIMF, 10; PV, 6; ET, 5; ISM, 3; p<0.05). The highest basophil counts were recorded in accelerated phase CML (115/mm2).
Conclusions
A novel immunohistochemical procedure has been established for basophil detection in normal bone marrow and MPD. This approach should help in the quantification of bone marrow basophils at diagnosis and during anti‐leukaemic treatment.
Keywords: basophil, chronic myeloid leukaemia, immunohistochemistry, myeloproliferative diseases
Basophils are multifunctional haematopoietic cells that store histamine and other proinflammatory mediators in their granules.1,2,3,4,5 Under physiological conditions, basophils are primarily produced in the bone marrow.6,7,8,9 After maturation, they typically reside in the circulation until recruited into tissue sites of inflammation. Basophils express a unique composition of cytoplasmic mediators and cell surface membrane antigens.10,11,12,13,14,15 Although mast cells and basophils produce several mediators and surface antigens in common, they can clearly be discriminated from each other by their unique profile of granular enzymes and CD molecules.12,13,14,15,16,17 During the past two decades, the phenotype of human basophils has been analysed extensively using antibodies directed against various leucocyte differentiation antigens.14,15,16,17,18 Most of these studies have been performed on normal peripheral blood basophils.14,15,16,17 By contrast, little is known about the phenotype of bone marrow basophils.18
Basophilia is detectable in various haematopoietic neoplasms including chronic myeloid leukaemia (CML) and other myeloproliferative disorders (MPD).19,20,21,22 Especially in CML, basophilia is a typical finding.19,20,21 In addition, an increased percentage of basophils in the bone marrow and in the peripheral blood in CML is associated with a poor prognosis.23,24,25,26,27,28,29 Thus basophilia is an independent adverse prognostic variable in CML.27,28,29 Moreover, basophils typically increase in number in the bone marrow as well as in the blood during disease acceleration.23,24,25,26,27 Basophils originate from the malignant clone in patients with CML.30,31 However, whereas basophilia can readily be quantified in the peripheral blood and on bone marrow smears, no generally accepted and easily applicable staining protocol for the identification and quantification of basophils in bone marrow biopsy sections has become available so far. In fact, basophils are difficult to identify by routine histochemistry, and no reliable immunohistochemical marker for basophil detection in the bone marrow has been described.
Recently, several basophil specific antibodies have been generated and used for the immunohistochemical detection of basophils in tissue sections.32,33 One of these granule associated antigens is recognised by a murine monoclonal antibody called 2D7.33 In the present study, 2D7 was used to detect and enumerate basophils in bone marrow sections in patients with CML and other MPD. The results of our study show that 2D7 staining is a highly sensitive and reliable approach for the detection and quantification of bone marrow basophils in these patients.
Methods
Bone marrow sampling and patients' characteristics
Bone marrow biopsy material (iliac crest) was examined in 46 patients with CML (chronic phase, n = 37; accelerated phase, n = 9), 20 with chronic idiopathic myelofibrosis (CIMF), 20 with polycythaemia vera, 20 with essential thrombocythaemia, seven with indolent systemic mastocytosis, and 31 control cases (normal/reactive marrow = no neoplasm involving the bone marrow). Diagnoses were established according to published criteria.34,35 The patients' characteristics are shown in table 1. Informed consent was obtained in all cases before bone marrow or blood was obtained.
Table 1 Patient characteristics.
| Diagnosis | Patients (n) | Median | F:M ratio | Median | Blood basophils |
|---|---|---|---|---|---|
| age (y) | WBC (× 109/l) | (median %) | |||
| CML (all) | 46 | 57.5 | 1:2.1 | 110 | 5 |
| CML‐CP | 37 | 57 | 1:2.1 | 103.5 | 4 |
| CML‐AP | 9 | 58 | 1:2 | 258.4 | 18 |
| PV | 20 | 66.5 | 1:1.2 | 10.05 | 1 |
| ET | 20 | 58 | 1:1 | 9.5 | 2 |
| CIMF | 20 | 71 | 1:1.2 | 11.35 | 1 |
| ISM | 7 | 41 | 1:0.75 | 6.8 | 0 |
AP, accelerated phase; CIMF, chronic idiopathic myelofibrosis; CML, chronic myeloid leukaemia; CP, chronic phase; ET, essential thrombocythaemia; ISM, indolent systemic mastocytosis, PV, polycythaemia vera; y, years.
In 23 patients with CML, whole blood histamine levels were measured by a commercial radioimmunoassay (RIA) (Immunotech) as an indirect (objective) index of basophil lineage cells.
Antibodies
Various monoclonal and polyclonal antibodies were used to determine the phenotype of basophils in bone marrow sections. The monoclonal antibodies AB75 (CD2), PS1 (CD3), 1F6 (CD4), 4C7 (CD5), CD7–272 (CD7), 72F6 (CD9), 56C6 (CD8), 38C12 (CD13), 7 (CD14), 2H7 (CD16), FPC1 (CD22), 1B12 (CD23), 4C9 (CD25), and 1B6 (CD56) were purchased from Novocastra (Newcastle upon Tyne, UK); C8/144B (CD8), L26 (CD20), 1F8 (CD21), BER‐H2 (CD30), JC/70A (CD31), DF1485 (CD44), UCHL1 (CD45), PG‐M1 (CD68), KP‐1 (CD68R), JCB117 (CD79a), CR3/43 (anti‐HLA‐DR), and polyclonal antibodies against KIT (CD117) and myeloperoxidase were from Dako (Copenhagen, Denmark); the CD15 antibody DT07+BC97 from BioCare (Walnut Creek, California, USA); the monoclonal antibodies QBEND10 (CD34) and 97A6 (CD203c) from Immunotech (Marseille, France); MT1 (CD43) from Biotest (Frankfurt, Germany); and HNK‐1 (CD57) from Becton Dickinson (San José, California, USA). The monoclonal antibodies B‐B4 (CD138) and CCI (anti‐chymase) were from SeroTec (Kidlington, Oxford, UK), a polyclonal anti‐VEGF antibody from Santa Cruz (Santa Cruz, California, USA), the monoclonal antibody G3 (anti‐tryptase) from Chemicon (Temecula, California, USA); and a polyclonal antibody against histidine decarboxylase from Progen (Heidelberg, Germany). The monoclonal antibody 2D7 was prepared as described.33
Immunohistochemistry
Immunohistochemistry was performed on serial (2 μm) sections prepared from paraffin embedded, formalin fixed bone marrow specimens using the indirect immunoperoxidase staining technique as described previously.36,37,38,39 Endogenous peroxidase was blocked by methanol/H2O2. For basophil detection, the monoclonal antibody 2D733 was applied. Other monoclonal antibodies were used to confirm the presence of basophils and to determine their phenotype. A specification of monoclonal antibodies and the staining techniques applied are shown in table 2. In general, the antibodies were diluted in 0.05 M Tris‐buffered saline (TBS, pH 7.5) plus 1% bovine serum albumin (BSA) (Sigma, St Louis, Missouri, USA). After washing, slides were incubated with biotinylated horse anti‐mouse IgG or goat anti‐rabbit IgG for 30 minutes, washed, and then exposed to avidin‐biotin‐peroxidase or streptavidin‐biotin‐peroxidase complex for 30 minutes. Diaminobenzidene hydrochloride (DAB) or 3‐amino‐9‐ethylcarbazole (AEC) were used as chromogen. Slides were counterstained in Mayer's Haemalaun. Expression of antigens in basophils was determined by comparing cell reactivity in serial sections stained with 2D7 (every second consecutive section stained) and other antibodies.
Table 2 Specification of antibodies and staining techniques.
| CD | Antibody | Reactive | Ig class | Source | Dilution | Retrieval |
|---|---|---|---|---|---|---|
| (clone*) | antigen | technique† | ||||
| CD2 | AB75 | LFA‐2 | lgG1 | Mouse | 1:40 | Aut |
| CD3 | PS1 | TCR | IgG2a | Mouse | 1:50 | Aut |
| CD4 | 1F6 | T4 | IgG1 | Mouse | 1:10 | Aut |
| CD5 | 4C7 | gp67 | IgG1 | Mouse | 1:20 | Aut |
| CD7 | CD7‐272 | gp40 | IgG1 | Mouse | 1:40 | MW |
| CD8 | C8/144B | T8 | IgG1 | Mouse | 1:20 | Aut |
| CD9 | 72F6 | MRP‐1 | IgG1 | Mouse | 1:200 | MW |
| CD10 | 56C6 | CALLA | IgG1 | Mouse | 1:20 | Aut |
| CD13 | 38C12 | APN | IgG1 | Mouse | 1:80 | Aut |
| CD14 | 7 | LPS‐R | IgG2a | Mouse | 1:100 | MW |
| CD15 | DT07+BC97 | LewisX | IgMκ | Mouse | 1:40 | MW |
| CD16 | 2H7 | FcγRIII | IgG2a | Mouse | 1:40 | Aut |
| CD20 | L26 | B1 | IgG2a | Mouse | 1:200 | MW |
| CD21 | 1F8 | C3dR | IgG1 | Mouse | 1:20 | Prot |
| CD22 | FPC1 | BL‐CAM | IgG1 | Mouse | 1:40 | Aut |
| CD23 | 1B12 | FcεRII | IgG1 | Mouse | 1:40 | Aut |
| CD25 | 4C9 | IL‐2Rα | IgG2b | Mouse | 1:40 | Aut |
| CD30 | BER‐H2 | Ki‐1 | IgG1 | Mouse | 1:40 | MW |
| CD31 | JC/70A | PECAM‐1 | IgG1 | Mouse | 1:80 | MW |
| CD34 | QBEND10 | HPCA‐1 | IgG1 | Mouse | 1:100 | MW |
| CD43 | MT1 | Leukosialin | IgG1 | Mouse | 1:400 | MW |
| CD44 | DF1485 | Pgp‐1 | IgG1 | Mouse | 1:50 | MW |
| CD45 | UCHL1 | UCHL‐1 | IgG2a | Mouse | 1:100 | MW |
| CD56 | 1B6 | NCAM | IgG1 | Mouse | 1:80 | Aut |
| CD57 | HNK‐1 | HNK1 | IgM | Mouse | 1:10 | – |
| CD68 | PG‐M1 | Macrosialin | IgG3 | Mouse | 1:100 | Prot |
| CD68R | KP‐1 | Macrosialin | IgG1 | Mouse | 1:100 | Prot |
| CD79a | JCB117 | Igα/MB1 | IgG1 | Mouse | 1:25 | MW |
| CD117 | αKIT | KIT/SCFR | Poly | Rabbit | 1:150 | MW |
| CD138 | B‐B4 | Syndecan1 | IgG1 | Mouse | 1:40 | MW |
| NC | CR3/43 | HLA‐DR | IgG1 | Mouse | 1:200 | MW |
| NC | Polyclonal | MPO | Poly | Rabbit | 1:400 | MW |
| NC | Polyclonal | VEGF | Poly | Rabbit | 1:50 | MW |
| NC | Polyclonal | HDC | Poly | Rabbit | 1:1000 | MW |
| NC | G3 | Tryptase | IgG1 | Mouse | 1:5000 | – |
| NC | CCI | Chymase | IgG1 | Mouse | 1:1000 | Prot |
| NC | 2D7 | 2D7 | IgG1 | Mouse | 1:500 | Prot |
*Clone of monoclonal antibody.
†Retrieval techniques: Aut, autoclave; MW, microwave oven; Prot, proteinase type XXIV.
HPCA‐1, haemopoietic progenitor cell antigen‐1; IL2Rα, interleukin 2 receptor α; LFA‐2, lymphocyte function associated antigen‐2; NC, not yet clustered; SCF‐R, stem cell factor receptor.
Determination of basophil counts in bone marrow sections
The numbers of basophils in bone marrow sections were determined on 2D7 stained slides in all patients. Basophil counting was done using an objective lens (×20) and an ocular lens (×10) containing an optical grid (original magnification ×200). The ocular system was adapted to define a scanned area of 0.425 mm2. In each section, at least 10 areas were examined. The numbers of 2D7+ cells were expressed as cells/mm2.
Separation of basophils by flow cytometry
In two CML donors with excessive basophilia (51% and 30% basophils in differential blood counts, respectively), peripheral blood leucocytes were separated by Ficoll and subjected to flow cytometry to enrich or to deplete basophils. For this purpose, cells were split and incubated either with the basophil specific monoclonal antibody 97A6 (CD203c) or with a monoclonal antibody against CD15, a broadly expressed myeloid antigen that usually is not detectable on basophils and therefore is widely used to enrich these cells by negative selection.10,16,17 Both cell fractions were subjected to flow cytometry and both populations were purified by cell sorting on a high speed sorter (FACSAria, Becton Dickinson). After purification, cells were spun on cytospin slides and subjected to Wright Giemsa staining. CD15 negative cells (enriched basophils) were subjected to immunocytochemical staining using the 2D7 antibody as well as antibodies against CD9, CD13, CD15, CD25, CD68, CD68R, CD117, tryptase, chymase, myeloperoxidase, VEGF, and histidine decarboxylase. For comparison of antigen expression in primary CML cells, the CML derived basophil cell line KU812 was also stained with these antibodies. CD203c negative cells (basophil depleted cell fractions) were stained with monoclonal antibody 2D7 to reconfirm the specificity of 2D7 for basophils. Immunocytochemistry was undertaken as described previously.33,40
Statistical analysis
To determine whether differences among basophil numbers between the various groups of patients were significant, standard statistical tests including the paired Student's t test and analysis of variance, were applied. Differences were considered to be significant when the p value was <0.05. To define the relations of basophil associated markers, linear correlations were applied and the correlation coefficients (R) determined.
Results
Demonstration that 2D7+ CML cells are basophils
In a first step, we reconfirmed the specificity of 2D7 using Ficoll separated primary peripheral blood leucocytes obtained from patients with CML showing basophilia. These cells were subjected to flow cytometry to obtain a CD203c negative (basophil depleted) fraction and a CD15 negative (basophil enriched) fraction. As expected, the CD203c negative fraction contained fewer than 1% basophils whereas the CD15 negative fractions contained >80% basophils. The monoclonal antibody 2D7 was found to react with enriched CML basophils, but did not react with bone marrow cells depleted of basophils (CD203c negative cells) (fig 1). Identical results were obtained in two CML donors. These data suggest that 2D7 is a basophil specific antigen in patients with CML but is not expressed in other leucocytes, confirming result obtained with normal blood basophils.33
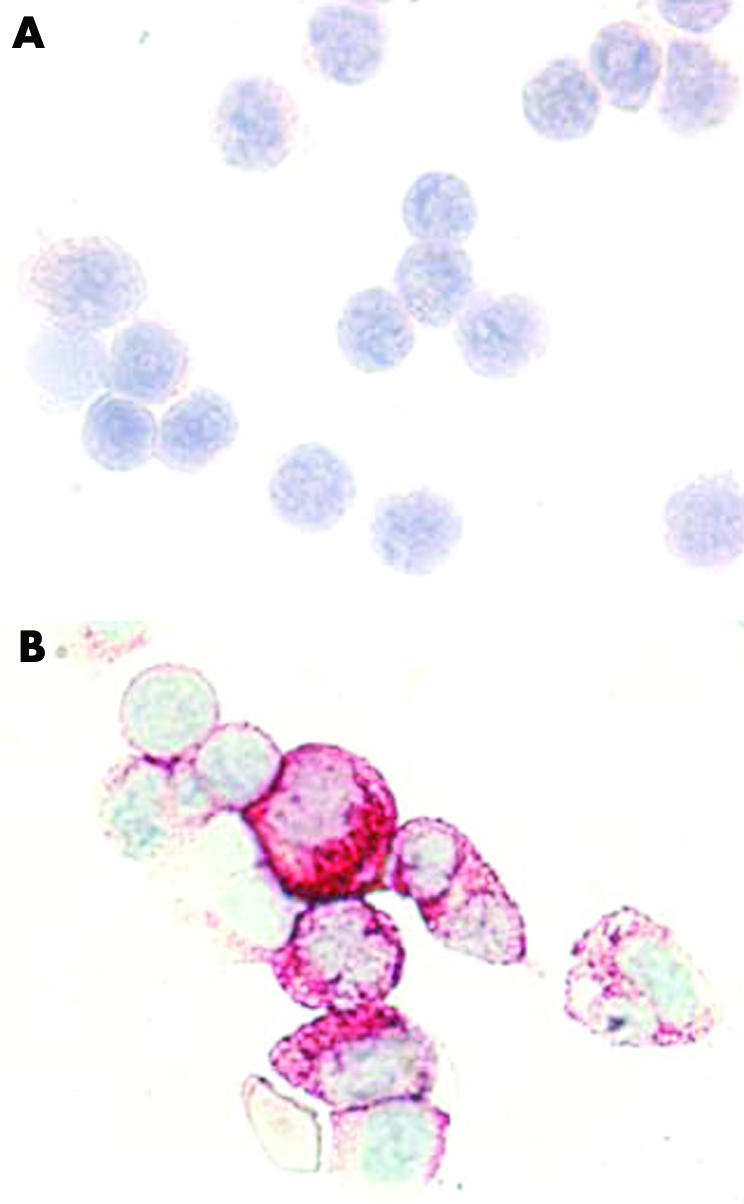
Figure 1 Specificity of 2D7 for basophils in chronic myeloid leukaemia (CML). Primary blood leucocytes obtained from a patient with CML were negatively sorted by flow cytometry using antibodies against CD203c (A) or an antibody against CD15 (B), and then were spun on cytospin slides. The basophil depleted (CD203c negative) cells did not react with the 2D7 antibody (A), whereas virtually all of the basophil enriched cells were labelled by 2D7 (B). Original magnification ×100.
Immunophenotypic characterisation of bone marrow basophils in CML
In a next step, we established the immunophenotype of leukaemic basophils using serial bone marrow sections obtained from patients with CML. Using 2D7 antibody, basophils were easily detectable in all patients, although the numbers varied among donors (fig 2). As assessed by serial section staining, the 2D7+ CML basophils were found to co‐express myeloperoxidase, tryptase, and histidine decarboxylase. In addition, basophils expressed CD9, CD13, CD31, CD43, CD45, and CD68. By contrast, basophils did not react with monoclonal antibodies against CD2, CD3, CD5, CD7, CD8, CD10, CD14, CD16, CD20, CD21, CD22, CD23, CD30, CD56, CD57, CD79a, or CD138 (table 3). This phenotype corresponds to the well established surface marker profile of blood basophils.10,14,15,34 An interesting finding was that basophils express low levels of CD15, a marker antigen that is usually not detectable on the surface of normal or CML derived basophils unless these cells are exposed to neuraminidase.10,14,15
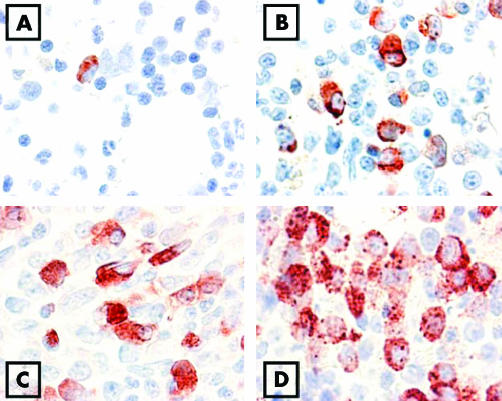
Figure 2 Immunohistochemical detection of basophils by 2D7 antibody in the bone marrow. Bone marrow sections were obtained from a patient without haematological neoplasm (control marrow) (A), one with chronic phase chronic myeloid leukaemia (CML) (B), one with accelerated phase CML (C), and one with accelerated CML and an excess of basophils (D). Slides were stained by the indirect immunoperoxidase technique.
Table 3 Expression of differentiation antigens in basophils.
| Antigen | CD | Reactivity of antibodies with | Surface antigens phenotype* of | |||
|---|---|---|---|---|---|---|
| 2D7+ cells in | Isolated CML | KU812, | CML | KU812 | ||
| CML bm, IHC | basophils, ICC | ICC | Basophils | |||
| LFA‐2 | CD2 | − | NT | NT | − | − |
| TCR | CD3 | − | NT | NT | − | − |
| T4 | CD4 | +/− | NT | NT | −† | +/− |
| Tp67 | CD5 | − | NT | NT | − | − |
| gp40 | CD7 | − | NT | NT | − | − |
| T8 | CD8 | − | NT | NT | − | − |
| MRP‐1 | CD9 | + | + | + | + | + |
| CALLA | CD10 | − | NT | NT | − | − |
| APN | CD13 | + | + | + | + | + |
| LPS‐R | CD14 | − | − | − | − | − |
| LewisX | CD15 | + | +/− | + | − | − |
| FcγRIII | CD16 | − | NT | NT | − | − |
| B1 | CD20 | − | NT | NT | − | − |
| C3dR | CD21 | − | NT | NT | − | − |
| BL‐CAM | CD22 | − | NT | NT | − | − |
| FcεRII | CD23 | − | NT | NT | − | − |
| IL‐2Rα | CD25 | +/− | + | + | + | + |
| Ki‐1 | CD30 | − | NT | NT | − | − |
| PECAM‐1 | CD31 | +/− | + | + | + | + |
| HPCA‐1 | CD34 | − | − | − | − | − |
| Leukosialin | CD43 | + | NT | NT | + | + |
| Pgp‐1 | CD44 | + | + | + | + | + |
| UCHL‐1 | CD45 | + | NT | NT | + | + |
| NCAM | CD56 | − | NT | NT | − | − |
| HNK1 | CD57 | − | NT | NT | − | − |
| Macrosialin | CD68 | + | +/− | + | NT | NT |
| Macrosialin | CD68R | + | + | + | NT | NT |
| Igα/MB1 | CD79a | − | NT | NT | − | − |
| KIT/SCFR | CD117 | +/− | +/− | +/− | − | +/− |
| Syndecan1 | CD138 | − | NT | NT | − | − |
| HLA‐DR | NC | +/− | +/− | +/− | −† | +/− |
| MPO | NC | + | + | + | NT | NT |
| VEGF | NC | + | + | + | NT | NT |
| HDC | NC | + | + | + | NT | NT |
| Tryptase | NC | +/− | + | + | NT | NT |
| Chymase | NC | − | − | NT | NT | NT |
Score of percentage of 2D7+ cells that were labelled: +, 50–100% of all cells reactive; +/−, 10–49% cells reactive; −, <10% of all cells stained.
*Data refer to published results.14,15,16
†After short term culture, CML basophils express surface CD4 and surface HLA‐DR.10
bm, bone marrow; CML, chronic myeloid leukaemia; ICC, immunocytochemistry; IHC, immunohistochemistry; NC, not yet clustered; NT, not tested.
The phenotype of basophils could also be confirmed by immunocytochemistry using enriched CML basophils (sorted CD15 negative cells) and KU812 cells spun on cytospin slides. In line with the data obtained by immunohistochemistry, CML basophils and KU812 cells were found to react with antibodies against 2D7 (fig 1) and myeloperoxidase (fig 3), as well as with antibodies against CD9, CD13, CD15, CD25, CD31, CD44, CD68, histidine decarboxylase, VEGF, and tryptase. A summary of all staining results obtained with CML basophils and KU812 cells is shown in table 3.
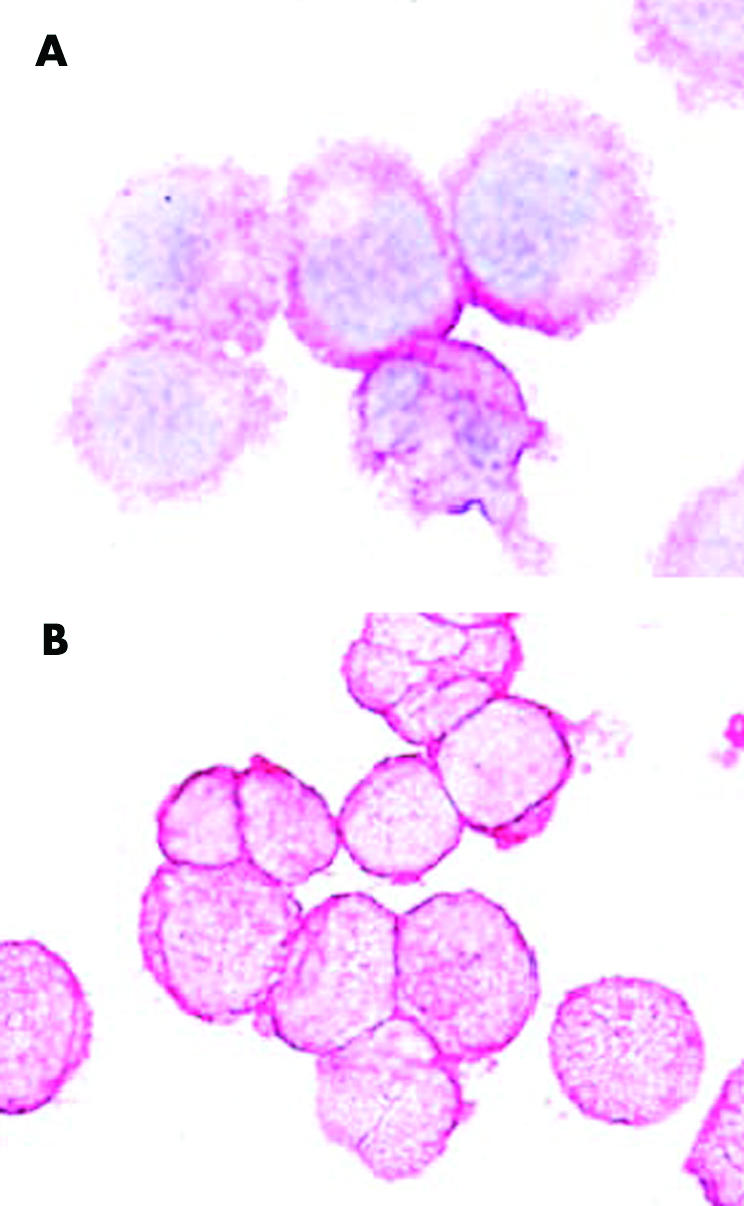
Figure 3 Immunocytochemical detection of myeloperoxidase in chronic myeloid leukaemia (CML) basophils. The CML derived basophil cell line KU812 (A) and enriched FACS sorted primary CML basophils (B) were spun on cytospin slides and stained with an antibody against myeloperoxidase (MPO). Both types of cells reacted with the anti‐MPO antibody. The antibody did not label lymphocytes (not shown).
Numbers of 2D7+ cells in patients with various myeloproliferative disorders
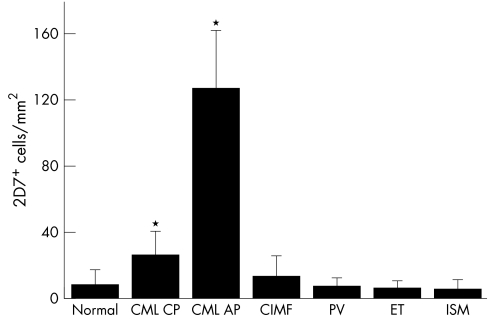
The numbers of 2D7+ cells (basophils) were determined in bone marrow sections in patients with CML, patients with other MPD, and in normal/reactive bone marrow. In normal/reactive bone marrow, the number of 2D7+ (mean (SD)) was 8 (5) cells/mm2 (median 6; range 0 to 18) (fig 2A). In patients with CML, larger numbers of 2D7+ cells (basophils) were found (all CML patients: mean 46 (44) cells/mm2, median 33, range 2 to 211; p<0.05; CML‐CP: mean 26 (14) cells/mm2, median 28, range 2 to 57, p<0.05) (fig 2, panels B, C, and D, and fig 4). The number of 2D7+ cells in patients with CIMF was 13 (12) cells/mm2 (median 10; range 2 to 45). In patients with polycythaemia vera, the number of 2D7+ cells was 7 (5) cells/mm2 (median 6; range 1 to 19), in essential thrombocythaemia 6 (4) (median: 5; range: 1–15 cells/mm2), and in indolent systemic mastocytosis, the number of 2D7+ cells was 5 (4) cells/mm2 (median 3; range 2 to 14). Figure 4 shows the numbers of 2D7+ cells (mean and SD) in all groups of patients. The largest numbers of 2D7+ cells (basophils) were found in patients with accelerated phase CML (mean 126 (33) cells/mm2(median 115, range 103 to 211; p<0.05) (fig 2, panels C and D, and fig 4). Figure 2D shows 2D7+ cells from a patient with accelerated phase CML in whom a massive increase in basophils was found. In this patient, the final diagnosis was “secondary basophilic leukaemia”.
Figure 4 Numbers of 2D7+ cells in the bone marrow of patients with MPD. Numbers of 2D7+ cells in the bone marrow sections were determined by morphometry using an adapted microscope (see text). The figure shows the mean numbers of 2D7+ cells (basophils) per mm2 in each group of patients. Results represent the mean and SD of all donors examined in each group. *p<0.05 v normal bone marrow.
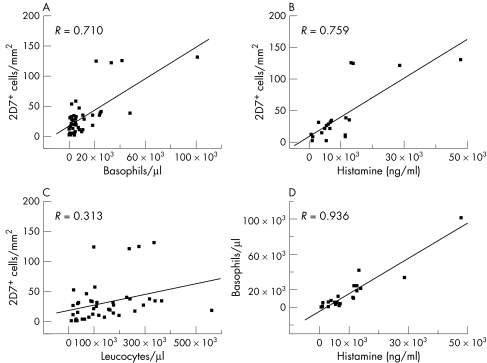
Correlation between 2D7+ cells and other haematological indices in CML
In patients with CML, the (median) number of 2D7+ cells was correlated with various haematological indices including white blood count, peripheral blood basophils, and whole blood histamine levels. In these analyses, significant correlations were found between the numbers of 2D7+ bone marrow cells and numbers of peripheral blood basophils (fig 5A), and between the numbers of 2D7+ cells and the total blood histamine levels (fig 5B). By contrast, there was no correlation between 2D7+ bone marrow cells and peripheral blood leucocytes (fig 5C). As expected, a highly significant correlation between the numbers of peripheral blood basophils and whole blood histamine levels was found (fig 5D). Interestingly, in patients with other MPD, the correlation between 2D7+ bone marrow cells and peripheral blood basophils (and the other variables analysed) was less pronounced than the correlations obtained in patients with CML (not shown).
Figure 5 Correlation between the numbers of 2D7+ cells and other indices in chronic myeloid leukaemia (CML). Correlations were determined between the median numbers of 2D7+ cells in bone marrow sections (2D7+ cells per mm2) and absolute numbers of peripheral blood basophils (calculated from total leucocyte counts and the percentage of basophils in differential counts) (A), between the numbers of 2D7+ bone marrow cells and whole blood histamine levels (measured by radioimmunoassay) (B), between the numbers of 2D7+ bone marrow cells and blood leucocytes (C), and between absolute numbers of blood basophils and whole blood histamine levels (D). The correlation coefficient (R) is shown in each case. All correlations except that between the numbers of 2D7+ bone marrow cells and leucocytes were statistically significant (p<0.05).
Discussion
Basophilia is an important feature of MPD, often a diagnostic finding, and sometimes even of prognostic significance.20,22,23,24,25,26,27,28 However, so far no reliable immunohistochemical marker for basophil detection and enumeration in MPD has become available. We have established 2D7 as a powerful new marker for the immunohistochemical analysis of basophils in patients with MPD. The results of our study show that 2D7 is specific for basophils in normal bone marrow as well as bone marrow in patients with MPD, and that the numbers of basophils can easily be quantified (or estimated) in 2D7 stained bone marrow sections. The 2D7 antibody should therefore be considered a novel helpful marker for basophil detection in haematopathology.
Although the 2D7 antibody is known to label basophils specifically in inflamed tissues41,42,43 and in normal peripheral blood,33 it was of pivotal importance to reconfirm the specificity of 2D7 for basophils in patients with MPD. To address this issue, two different approaches were used. First, purified CML basophils and CML cells depleted of basophils, were analysed. In these experiments, we were able to show that 2D7 is selectively expressed in CML basophils, but is not expressed in basophil depleted cell fractions. In consecutive experiments, serial section staining was carried out in order to reconfirm that the 2D7 stained bone marrow cells express the established phenotype of human basophils.14,15,16
Various previous studies have shown that basophils express a unique cell surface marker profile, including myeloid determinants as well as activation linked cell surface antigens.10,11,14,15,16,34,44 This phenotype has been described for normal blood basophils and for CML derived basophils.10,14,15,16 However, no systematic immunohistochemical analysis of bone marrow basophils has been presented so far. We now show that 2D7+ basophils in the bone marrow in patients with CML express a unique phenotype which corresponds largely to the cell surface phenotype of (normal and CML) basophils. However, some of the antigens detected in CML basophils by immunohistochemistry, like CD4 or CD15, are usually not detectable on the surface of normal blood basophils.10,14,15,16,34 With regard to CD15, this antigen is indeed expressed on the surface of basophils, but is masked by gangliosides and is thus detectable only after treatment with neuraminidase.10 In the case of CD4, the discrepancy may have several explanations. First, the antibody used may cross react with other antigens or may show non‐specific binding to basophils. An alternative explanation may be that CML basophils express very small amounts of CD4 on their cell surface. In this regard it is noteworthy that CML basophils can react with CD4 antibodies in cell surface staining experiments when these cells are kept in culture for several days.10 Whether CML basophils indeed express functional CD4 and thereby can interact with class II HLA antigen or the HIV virus remains presently unknown.
In CML, the numbers of basophils are almost invariably increased and correspond to the phase of disease.20,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45 Especially in the accelerated phase of CML, many patients present with highly upregulated numbers of basophils.22,23,24,25,26,45 This phenomenon was confirmed in the present study by 2D7 staining. In fact, the largest numbers of 2D7+ cells were recorded in patients with accelerated phase CML. Thus the 2D7 antibody is a reliable marker and helpful to enumerate basophil counts in various phases of CML. In this regard, it is noteworthy that disease acceleration in CML is even defined by bone marrow basophilia.28,35,36 In addition, 2D7 may be helpful for monitoring of basophils—that is, their decrease during the first months of treatment with tyrosine kinase inhibitors.
In a few CML patients, basophilia is excessive and results in the clinical (pathological) picture of (secondary) basophilic leukaemia.37,38 Such excessive basophilia was also found in one of the patients examined in the current study. In this particular patient, more than 30% of all nucleated bone marrow cells appeared to react with the 2D7 monoclonal antibody. Thus the 2D7 antibody may also be a helpful marker to confirm the presence of basophilic leukaemia.
Basophils are increased not only in CML, but also in most other subtypes of MPD.19,21,22 Therefore we extended our analyses using 2D7 monoclonal antibody to various groups of patients with MPD. However, interestingly, the numbers of 2D7+ bone marrow basophils were found to be in the same range in patients with other MPD compared with normal/reactive bone marrow. An interesting finding was that bone marrow basophils are not increased in patients with indolent systemic mastocytosis. In these patients, the mast cell infiltrates were clearly composed of 2D7– cells, and the remaining normal appearing marrow did not contain increased numbers of 2D7+ cells when compared with normal/reactive bone marrow. This observation provides further evidence that basophils and mast cells represent two different haematopoietic cell lineages.15 No evidence for the previously reported basophil/mast cell hybrids46 was found in the current study, in agreement with the observations of Foster et al.47
An important question addressed in our study was whether the numbers of 2D7+ bone marrow cells correlate with other basophil related or disease related indices. Indeed, the numbers of bone marrow basophils did correlate with the numbers (percentages) of peripheral blood basophils, histamine levels, and to a lesser degree, the leucocyte counts. These findings further document the value of the 2D7 monoclonal antibody in the immunohistochemical quantification of basophilia in patients with MPD. On the other hand, we quantified 2D7+ cells on a “per mm2” basis, so a possible influence of variation in cellularity between patients and disease categories needs to be taken into account. However, the cellularity among MPD patients did not differ extensively, so that such an influence may not change the overall message of this report. In this regard it should also be pointed out that in the non‐CML MPDs, the numbers of 2D7+ cells did not vary significantly from each other and not much from controls, so that the stain may not be helpful in separating non‐CML MPD from reactive basophilia.
In summary, we have established a novel immunohistochemical staining technique for the detection and enumeration of basophils in routinely processed bone marrow trephine biopsy sections. This may be a helpful new approach in haematopathology.
Acknowledgements
We wish to thank Ilona Schliephake, Agnes Dummer, Mariola Erdmann‐Jensko, Tanja Oeltermann, Katharina Vogel, and Christoph Kroll (University of Lübeck) as well as Susanne Herndlhofer and Hans Semper (Medical University of Vienna, Department of Internal Medicine I) for skilful technical assistance. This study was supported by the Fonds zur Förderung der Wissenschaftlichen Forschung in Österreich (FWF) grant No P17205‐B14, FWF grant No SFB‐018/09, FWF‐Charlotte Bühler grant No H185‐B04, as well as grant No AI20487 from the National Institutes of Health, Bethesda.
Abbreviations
CIMF - chronic idiopathic myelofibrosis
CML - chronic myeloid leukaemia
MPD - myeloproliferative disorder
VEGF - vascular endothelial growth factor
References
- 1.Valent P, Bettelheim P. The human basophil. Crit Rev Oncol Hematol 199010327–352. [DOI] [PubMed] [Google Scholar]
- 2.Dvorak A M. Cell biology of the basophil. Int Rev Cytol 199818087–236. [DOI] [PubMed] [Google Scholar]
- 3.Falcone F H, Haas H, Gibbs B F. The human basophil: a new appreciation of its role in immune responses. Blood 2000964028–4038. [PubMed] [Google Scholar]
- 4.Schroeder J T, MacGlashan D W, Lichtenstein L M. Human basophils: mediator release and cytokine production. Adv Immunol 20017793–122. [DOI] [PubMed] [Google Scholar]
- 5.Dvorak A M. Histamine content and secretion in basophils and mast cells. Prog Histochem Cytochem 199833169–320. [DOI] [PubMed] [Google Scholar]
- 6.Denburg J A, Davison M, Bienenstock J. Basophil production. J Clin Invest 198065390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leary A G, Ogawa M. Identification of pure and mixed basophil colonies in culture of human peripheral blood and marrow cells. Blood 19846478–83. [PubMed] [Google Scholar]
- 8.Saito H, Hatake K, Dvorak A M.et al Selective differentiation and proliferation of hematopoietic cells induced by recombinant human interleukins. Proc Natl Acad Sci USA 1988852288–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valent P, Schmidt G, Besemer J.et al Interleukin‐3 is a differentiation factor for human basophils. Blood 1989731763–1769. [PubMed] [Google Scholar]
- 10.Stain C, Stockinger H, Scharf M.et al Human blood basophils display a unique phenotype including activation linked membrane structures. Blood 1998701872–1879. [PubMed] [Google Scholar]
- 11.Bodger M P, Newton L A. The purification of human basophils: their immunophenotype and cytochemistry. Br J Haematol 198767281–284. [DOI] [PubMed] [Google Scholar]
- 12.MacGlashan D W, Schleimer R P, Peters S P.et al Comparative studies of human basophils and mast cells. Fed Proc 1983422504–2509. [PubMed] [Google Scholar]
- 13.de Boer M, Roos D. Metabolic comparison between basophils and other leukocytes from human blood. J Immunol 19861363447–3454. [PubMed] [Google Scholar]
- 14.Valent P, Majdic O, Maurer D.et al Further characterization of surface membrane structures expressed on human basophils and mast cells. Int Arch Allergy Appl Immunol 199091198–203. [DOI] [PubMed] [Google Scholar]
- 15.Valent P, Bettelheim P. Cell surface structures on human basophils and mast cells: biochemical and functional characterization. Adv Immunol 199252333–423. [DOI] [PubMed] [Google Scholar]
- 16.Agis H, Füreder W, Bankl H C.et al Comparative immunophenotypic analysis of human mast cells, blood basophils and monocytes. Immunology 199687535–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agis H, Beil W J, Bankl H C.et al Mast cell‐lineage versus basophil lineage involvement in myeloproliferative and myelodysplastic syndromes: diagnostic role of cell‐immunophenotyping. Leuk Lymphoma 199622187–204. [DOI] [PubMed] [Google Scholar]
- 18.Bühring H J, Simmons P J, Pudney M.et al The monoclonal antibody 97A6 defines a novel surface antigen expressed on human basophils and their multipotent and unipotent progenitors. Blood 1999942343–2356. [PubMed] [Google Scholar]
- 19.Juhlin L. Basophil and eosinophil leukocytes in various internal disorders. Acta Med Scand 1963174249–254. [DOI] [PubMed] [Google Scholar]
- 20.Spiers A S, Bain B J, Turner J E. The peripheral blood in chronic granulocytic leukaemia. Study of 50 untreated Philadelphia‐positive cases. Scand J Haematol 19771825–38. [PubMed] [Google Scholar]
- 21.Gilbert H S. Myelofibrosis revisited: characterization and classification of myelofibrosis in the setting of myeloproliferative disease. Prog Clin Biol Res 19841543–17. [PubMed] [Google Scholar]
- 22.Arnalich F, Lahoz C, Larrocha C.et al Incidence and clinical significance of peripheral and bone marrow basophilia. J Med 198718293–303. [PubMed] [Google Scholar]
- 23.Gomez G A, Sokal J E, Walsh D. Prognostic features at diagnosis of chronic myelocytic leukemia. Cancer 1981472470–2477. [DOI] [PubMed] [Google Scholar]
- 24.Denburg J A, Wilson W E, Bienenstock J. Basophil production in myeloproliferative disorders: increases during acute blastic transformation of chronic myeloid leukemia. Blood 198260113–120. [PubMed] [Google Scholar]
- 25.Mühleck S D, McKenna R W, Arthur D C.et al Transformation of chronic myelogenous leukemia: clinical, morphologic, and cytogenetic features. Am J Clin Pathol 1984821–14. [DOI] [PubMed] [Google Scholar]
- 26.Kantarjian H M, Dixon D, Keating M J.et al Characteristics of accelerated disease in chronic myelogenous leukemia. Cancer 1988611441–1446. [DOI] [PubMed] [Google Scholar]
- 27.Braga G W, Chauffaille M L, Moncau J E.et al Chronic myeloid leukemia (CML): prognostic factors and survival analysis. Rev Paul Med 19961141083–1090. [DOI] [PubMed] [Google Scholar]
- 28.Hasford J, Pfirrmann M, Hehlmann R.et al A new prognostic score for survival of patients with chronic myeloid leukemia treated with interferon alfa. Writing Committee for the Collaborative CML Prognostic Factors Project Group. J Natl Cancer Inst 199890850–858. [DOI] [PubMed] [Google Scholar]
- 29.Steegmann J L, Odriozola J, Rodriguez‐Salvanes F.et al Stage, percentage of basophils at diagnosis, hematologic response within six months, cytogenetic response in the first year: the main prognostic variables affecting outcome in patients with chronic myeloid leukemia in chronic phase treated with interferon‐alpha. Results of the CML89 trial of the Spanish Collaborative Group on interferon‐alpha2a and CML. Haematologica 199984978–987. [PubMed] [Google Scholar]
- 30.Bosma T J, Kennedy M A, Bodger M P.et al Basophils exhibit rearrangement of the bcr gene in Philadelphia chromosome‐positive chronic myeloid leukemia. Leukemia 19982141–143. [PubMed] [Google Scholar]
- 31.Bodger M P, Morris C M, Kennedy M A.et al Basophils (Bsp‐1+) derive from the leukemic clone in human myeloid leukemias involving the chromosome breakpoint 9q34. Blood 198973777–781. [PubMed] [Google Scholar]
- 32.McEuen A R, Buckley M G, Compton S J.et al Development and characterization of a monoclonal antibody specific for human basophils and the identification of a unique secretory product of basophil activation. Lab Invest 19997927–38. [PubMed] [Google Scholar]
- 33.Kepley C L, Craig S S, Schwartz LB: Identification and partial characterization of a unique marker for human basophils J Immunol. 1995;154:6548–6555. [PubMed] [Google Scholar]
- 34.Vardiman J W, Harris N L, Brunning R D. The World Health Organization (WHO) classification of the myeloid neoplasms. Blood 20021002292–2302. [DOI] [PubMed] [Google Scholar]
- 35.Jaffe E S, Harris N L, Stein H.et al In: WHO classification of tumours. Pathology and genetics of tumours of haematopoietic and lymphoid tissues. Lyon: IARC Press, 2001
- 36.Hsu S M, Raine L, Fanger H. Use of avidin‐biotin‐peroxidase complex (ABC) in immunoperoxidase techniques: a comparison between ABC and unlabeled antibody (PAP) procedures. J Histochem Cytochem 198129577–580. [DOI] [PubMed] [Google Scholar]
- 37.Cattoretti G, Pileri S, Parravicini C.et al Antigen unmasking on formalin‐fixed, paraffin‐embedded tissue sections. J Pathol 199317183–98. [DOI] [PubMed] [Google Scholar]
- 38.Horny H P, Sillaber C, Menke D.et al Diagnostic value of immunostaining for tryptase in patients with mastocytosis. Am J Surg Pathol 1998221132–1140. [DOI] [PubMed] [Google Scholar]
- 39.Jordan J H, Walchshofer S, Jurecka W.et al Immunohistochemical properties of bone marrow mast cells in systemic mastocytosis: evidence for expression of CD2, CD117/Kit, and bcl‐x(L). Hum Pathol 200132545–552. [DOI] [PubMed] [Google Scholar]
- 40.Schernthaner G H, Jordan J H, Ghannadan M.et al Expression, epitope analysis, and functional role of the LFA‐2 antigen detectable on neoplastic mast cells. Blood 2001983784–3792. [DOI] [PubMed] [Google Scholar]
- 41.Irani A M, Huang C, Xia H Z.et al Immunohistochemical detection of human basophils in late‐phase skin reactions. J Allergy Clin Immunol 1998101354–362. [DOI] [PubMed] [Google Scholar]
- 42.Nouri‐Aria K T, Irani A M, Jacobson M R.et al Basophil recruitment and IL‐4 production during human allergen‐induced late asthma. J Allergy Clin Immunol 2001108205–211. [DOI] [PubMed] [Google Scholar]
- 43.Wilson D R, Irani A M, Walker S M.et al Grass pollen immunotherapy inhibits seasonal increases in basophils and eosinophils in the nasal epithelium. Clin Exp Allergy 2001311705–1713. [DOI] [PubMed] [Google Scholar]
- 44.Füreder W, Agis H, Sperr W R.et al The surface membrane antigen phenotype of human blood basophils. Allergy 199449861–865. [DOI] [PubMed] [Google Scholar]
- 45.Vardiman J D, Pierre R, Thiele J.et al Chronic myelogenous leukaemia. In: WHO classification of tumours. Pathology and genetics of tumours of haematopoietic and lymphoid tissues. Lyon: IARC Press, 200120–26.
- 46.Li Y, Li L, Wadley R.et al Mast cells/basophils in the peripheral blood of allergic individuals who are HIV‐1 susceptible due to their surface expression of CD4 and the chemokine receptors CCR3, CCR5, and CXCR4. Blood 2001973484–3490. [DOI] [PubMed] [Google Scholar]
- 47.Foster B, Schwartz L B, Devouassoux G.et al Characterization of mast‐cell tryptase‐expressing peripheral blood cells as basophils. J Allergy Clin Immunol 2002109287–293. [DOI] [PubMed] [Google Scholar]