Abstract
Background
Activated leucocyte cell adhesion molecule (ALCAM, CD166) is a cell surface member of the immunoglobulin superfamily. ALCAM expression has prognostic relevance in prostate and colon cancer.
Objective
To evaluate ALCAM protein expression in breast cancer by immunohistochemistry and to correlate expression levels with clinicopathological data.
Methods
162 primary breast carcinomas with a mean clinical follow up time of 53 months were immunostained using a monoclonal ALCAM antibody. The staining was evaluated as an immunoreactive score (IRS) and grouped into low v high for both membranous and cytoplasmic staining.
Results
Intraductal and invasive carcinomas showed a higher ALCAM expression (median IRS 4 and 6 respectively) than normal breast tissue (IRS 2). In univariate survival analyses a significant association of high cytoplasmic ALCAM expression with shortened patient disease‐free survival (mean (SD) five year non‐progression rate, 69.4 (4.6)% v 49.4 (11.1)%, p = 0.0142) was found. In multivariate analyses of disease‐free survival times, high cytoplasmic ALCAM expression (relative risk (RR) = 2.086, p = 0.026) and nodal status (RR = 2.246, p = 0.035) were significantly associated with earlier disease progression, whereas tumour grading (RR = 1.6, p = 0.052) was of borderline significance.
Conclusions
The data suggest that strong cytoplasmic ALCAM expression in primary breast cancer, as detected by immunohistochemistry, might be a new marker for a more aggressive breast cancer biology.
Keywords: breast cancer, ALCAM, prognostic marker, immunohistochemistry
Breast cancer is the most common malignant tumour of women in the western world. In the USA alone 211 240 new cases were expected for 2005.1 The clinical course is highly variable, so it is important to be able to predict the course in the individual patient to ensure adequate treatment and surveillance. Established conventional prognostic and predictive markers for breast cancer are age, nodal status, tumour grade, tumour size, tumour type, and oestrogen receptor status.2,3 Additionally, molecular markers are being sought to refine the prognosis. Candidate genes of current interest are telomerase, kallikrein 5, urokinase, plasminogen activator (uPA) and its inhibitor (PAI‐1), tissue inhibitor of metalloproteinases 1 (TIMP‐1), Ep‐Cam, c‐erbB2, osteopontin, and CD24.4,5,6,7,8,9,10
Activated leucocyte cell adhesion molecule (ALCAM; MEMD, CD166) is a cell surface immunoglobulin superfamily member involved in cell–cell interactions through homophilic and heterophilic (ALCAM‐CD6) binding.11,12,13,14 ALCAM was identified on thymic epithelial cells and activated leucocytes and later in a wide variety of tissues and cells, including epithelia, lymphoid, and myeloid cells, fibroblasts, neurones, hepatocytes, pancreas acinar and islet cells, and bone marrow.11,15,16,17 In neoplasia, ALCAM has been described in invasive cells of primary and metastasised melanoma and in various carcinoma cell lines including breast, lung, colon, and prostate.12,18,19 ALCAM expression correlates with the aggregation and metastatic capacity of human melanoma cell lines, suggesting a role in melanocytic tumour progression.12 Overexpression of ALCAM and annexin II was also suggested to play a role in chemoresistance.20 Recently, we found ALCAM upregulation in prostate cancer and were able to show that dysregulated ALCAM expression was a marker of disease progression.21,22
We aimed to investigate the expression patterns of ALCAM in a cohort of clinically characterised breast cancers and to correlate these findings with clinicopathological variables including patient survival.
Methods
Patients
Our study included 162 patients diagnosed with primary breast cancer at the Institute of Pathology, Charité University Hospital, Berlin, between 1991 and 1997. Patient age at the time of diagnosis ranged from 30 to 87 years with a median of 59 years (mean 59). The average observation time for overall survival was 57 months for patients still alive at the time of analysis, and ranged from one to 130 months. Twenty seven patients (16.7%) died during follow up, and 51 (31.5%) experienced disease progression defined by either metastatic or local recurrence. Oestrogen receptor status, determined according to Harvey et al, and the expression of c‐erbB2, determined according to Dako Hercep test criteria, were taken from archival pathology reports.23 For statistical analysis we arranged the patients into two groups: the first group received either no or only local therapy/radiotherapy (35 cases), or systemic therapy excluding tamoxifen (31 cases). The second group had received tamoxifen with or without an additional systemic or local therapy (88 cases). For eight patients no data on adjuvant treatment was available. The clinicopathological characteristics of the tumour collection are described in table 1.
Table 1 Clinicopathological variables of the tumour set.
| Variable | Cases (n (%)) |
|---|---|
| Total number | 162 (100%) |
| Histology | |
| Invasive ductal | 144 (88.9%) |
| Invasive lobular | 17 (10.5%) |
| Mixed | 1 (0.6%) |
| pT stage | |
| pT1 | 103 (63.6%) |
| pT2 | 44 (27.7%) |
| pT3 | 7 (4.3%) |
| pT4 | 9 (4.9%) |
| Lymph node status | |
| pN0 | 81 (50.0%) |
| pN1 | 35 (21.6%) |
| pN2 | 20 (12.3%) |
| pN3 | 26 (16.0%) |
| Tumour grade | |
| G1 | 40 (24.7%) |
| G2 | 82 (50.6%) |
| G3 | 40 (24.7%) |
| Age (years) | |
| <60 | 86 (53.1%) |
| > = 60 | 76 (46.9%) |
| Adjuvant treatment (n = 154) | |
| None/radiotherapy | 35 (21.6%) |
| Chemotherapy only | 31 (19.1%) |
| Tamoxifen±chemotherapy | 88 (54.3%) |
| Oestrogen receptor (n = 148) | |
| Negative | 42 (25.9%) |
| Positive | 106 (65.4%) |
| c‐erbB2 (n = 135) | |
| 0, 1+ | 100 (61.7%) |
| 2+, 3+ | 35 (21.6%) |
pT, primary tumour.
Table 3 Relation between low and high cytoplasmic ALCAM expression immunoreactivity scores and various clinicopathological factors.
| Characteristic | All cases | ALCAM IRS 0–6 | ALCAM IRS 7–12 | p Value |
|---|---|---|---|---|
| A. Cytoplasmic | ||||
| All carcinomas | 162 | 134 (82.7%) | 28 (17.3%) | |
| Age at surgery (y) | 0.217 | |||
| ⩽60 | 86 | 68 (79.1%) | 18 (20.9%) | |
| >60 | 76 | 66 (86.8%) | 10 (13.2%) | |
| Histological type | 0.741 | |||
| Ductal carcinoma | 144 | 118 (81.9%) | 26 (18.1%) | |
| Lobular carcinoma | 18 | 16 (88.9%) | 2 (11.1%) | |
| Primary tumour size | 0.487* | |||
| pT1 | 103 | 87 (84.5%) | 16 (15.5%) | |
| pT2 | 44 | 35 (79.5%) | 9 (20.5%) | |
| pT3/4 | 15 | 12 (80.0%) | 3 (20%) | |
| Lymph node status | 0.836 | |||
| pN0 | 79 | 66(83.5%) | 13 (16.5%) | |
| pN1+ | 81 | 66 (81.5%) | 15 (18.5%) | |
| Histological grade | 0.039* | |||
| G1 | 40 | 36 (90.0%) | 4 (10.0%) | |
| G2 | 82 | 69 (84.1%) | 13 (15.9%) | |
| G3 | 40 | 29 (72.5%) | 11 (27.5%) | |
| Oestrogen receptor | 0.812 | |||
| Negative | 42 | 34 (80.9%) | 8 (19.1%) | |
| Positive | 106 | 88 (83.0%) | 18 (17.0%) | |
| c‐erbB2 expression | 0.214 | |||
| 0, 1+ | 100 | 84 (84.0%) | 16 (16.0%) | |
| 2+, 3+ | 35 | 26 (74.3%) | 9 (25.7%) | |
| Adjuvant treatment | 0.834 | |||
| No tamoxifen | 66 | 55 (83.3%) | 11 (16.7%) | |
| Tamoxifen | 88 | 72 (81.8%) | 16 (18.2) | |
| B. Membranous | ||||
| All carcinomas | 162 | 126 (77.8%) | 36 (22.2%) | |
| Age at surgery (y) | 0.060 | |||
| ⩽60 | 86 | 72 (83.7%) | 14 (16.3%) | |
| >60 | 76 | 54 (71.1%) | 22 (28.9%) | |
| Histological type | 0.128 | |||
| Ductal carcinoma | 144 | 115 (79.9%) | 29 (20.1%) | |
| Lobular carcinoma | 18 | 11 (61.1%) | 7 (38.9%) | |
| Primary tumour size | 0.112* | |||
| pT1 | 103 | 82 (79.6%) | 21 (20.4%) | |
| pT2 | 44 | 36 (81.8%) | 8 (18.2%) | |
| pT3/4 | 15 | 8 (53.3%) | 7 (46.7%) | |
| Lymph node status | 0.851 | |||
| pN0 | 79 | 62 (78.5%) | 17 (21.5%) | |
| pN1+ | 81 | 62 (76.5%) | 19 (23.5%) | |
| Histological grade | 0.789* | |||
| G1 | 40 | 30 (75.0%) | 10 (25.0%) | |
| G2 | 82 | 65 (79.3%) | 17 (20.7%) | |
| G3 | 40 | 31 (77.5%) | 9 (22.5%) | |
| Oestrogen receptor | 0.521 | |||
| Negative | 42 | 34 (81.0%) | 8 (19.0%) | |
| Positive | 106 | 79 (74.5%) | 27 (25.5%) | |
| c‐erbB2 expression | 1.000 | |||
| 0, 1+ | 100 | 76 (76.0%) | 24 (24.0%) | |
| 2+, 3+ | 35 | 27 (77.1%) | 8 (22.9%) | |
| Adjuvant treatment | 0.433 | |||
| No tamoxifen | 66 | 54 (81.8%) | 12 (18.2%) | |
| Tamoxifen | 88 | 67 (76.1%) | 21 (23.9%) |
*χ2 test.
ALCAM, activated leucocyte cell adhesion molecule; IRS, immunoreactive score.
Immunohistochemistry
Formalin fixed, paraffin embedded tissue sections were mounted on superfrost slides (Menzel Gläser, Braunschweig, Germany), dewaxed with xylene, and gradually hydrated. Antigen retrieval was achieved by pressure cooking in 0.01 M citrate buffer for five minutes. The primary ALCAM antibody (clone MOG/07, Novocastra Laboratories, Newcastle upon Tyne, UK) was diluted 1:100 using a background reducing dilution buffer (Dako, Hamburg, Germany) and incubated at room temperature for one hour. Detection took place using labelled streptavidin‐biotin (Dako, Hamburg, Germany). Fast‐Red (Sigma‐Aldrich, Munich, Germany) served as chromogen. The slides were briefly counterstained with haematoxylin and aqueously mounted.
Evaluation of the immunohistochemical staining
The immunostainings were independently examined by three clinical pathologists, who were blinded to patient outcome. Membranous and cytoplasmic staining intensity of ALCAM were evaluated separately for invasive carcinoma, ductal carcinoma in situ (DCIS), and adjacent normal tissue on the same sample. An immunoreactive score (IRS) was applied. The IRS is the product of staining intensity (graded between 0 and 3) and the percentage of positive cells (graded between 0 and 4: 0, nil; 1, <10%; 2, 10–50%; 3, 51–80%; 4, >80%).
Statistical analysis
The data was compiled with the software package SPSS, version 12.0. Fisher's exact and χ2 tests were used to assess the statistical significance of the correlation between expression of ALCAM and clinicopathological variables. Univariate survival analysis was undertaken by the Kaplan–Meier method, and differences in survival curves assessed by the log rank test. Multivariate survival analysis was carried out on all variables that were found to be significant on univariate analysis using the Cox regression model. Probability (p) values of <0.05 were considered significant.
Results
ALCAM immunostaining in breast tissue
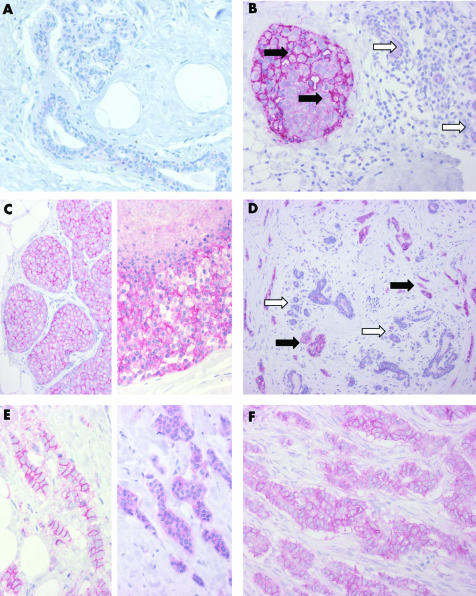
An overview of cytoplasmic and membranous expression of ALCAM in invasive breast carcinoma, adjacent normal breast tissue (n = 137), and ductal carcinoma in situ (DCIS, n = 65) is shown in table 2. In normal breast tissue, ALCAM was weakly expressed in a membranous and cytoplasmic fashion (median IRS = 2; fig 1, panel A). A stronger staining quality was observed in intraductal (median IRS = 4) and invasive carcinomas (median IRS = 6; fig 1, panels B to F; table 2). In invasive carcinomas, the staining qualities (membranous and cytoplasmic) were weakly but significantly correlated (correlation coefficient = 0.164, p = 0.037). Taken both staining qualities together, we found higher overall ALCAM expression in tumour tissue in 103 cases (75.2%), a lower expression in 21 cases (15.3%), and 13 cases (9.5%) showed no differences.
Table 2 Cytoplasmic and membranous expression of ALCAM evaluated as immunoreactivity score in normal breast tissue, ductal carcinoma in situ, and invasive breast carcinoma.
| ALCAM | Normal breast tissue (n = 137) | DCIS (n = 65) | Invasive carcinoma (n = 162) |
|---|---|---|---|
| IRS cytoplasmic | |||
| median | 2 | 4 | 6 |
| mean | 2.7 | 5.1 | 5.4 |
| IRS membranous | |||
| Median | 3 | 4 | 4 |
| Mean | 3.2 | 4.9 | 4.7 |
ALCAM, activated leucocyte cell adhesion molecule; DCIS, ductal carcinoma in situ; IRS, immunoreactive score.
Figure 1 Activated leucocyte cell adhesion molecule (ALCAM) immunohistochemistry of breast glands and breast cancer. (A) Normal breast duct and gland epithelium with minimal ALCAM expression. (B) Ductal carcinoma in situ (black arrows) with strong membranous and moderate cytoplasmic staining; normal breast glands (white arrows) with weak cytoplasmic staining. (C) Lobular carcinoma in situ (on the left) with strong membranous and weak to moderate cytoplasmic staining; high grade ductal carcinoma in situ (on the right) with comedo necrosis and strong membranous and cytoplasmic staining. (D) Invasive ductal carcinoma (black arrows) with strong membranous and cytoplasmic staining; normal breast glands with weak cytoplasmic staining (white arrows). (E) Invasive ductal carcinoma with strong membranous and weak cytoplasmic staining (on the left) and a different case with strong cytoplasmic staining lacking membranous immunoreactivity (on the right). (F) Invasive ductal carcinoma with strong membranous and moderate cytoplasmic staining.
We did not find any significant association of cytoplasmic, membranous, or total ALCAM staining intensity with patient age, tumour type, tumour size (pT), nodal status, oestrogen receptor, or c‐erbB2 expression. Only higher tumour grading was linked to higher rates of cytoplasmic ALCAM positivity (table 3A). No correlation between membranous or total ALCAM immunoreactivity and clinicopathological variables was found (table 3B).
Univariate survival analysis
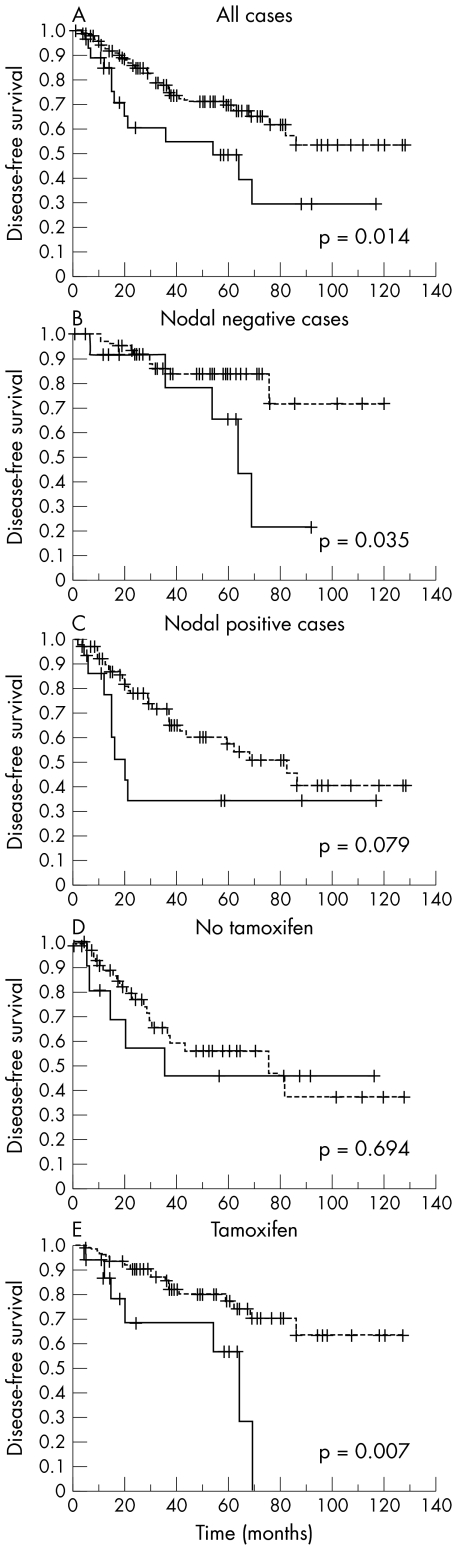
We analysed the impact of all variables on overall survival and disease‐free survival. The conventional prognostic markers of tumour grade, tumour size/pT, and nodal status reached significance for overall and disease‐free survival. High rates of cytoplasmic ALCAM expression were significantly correlated with shortened disease‐free survival (mean 58 v 88 months, p = 0.0142; fig 2A; table 4). Higher rates of membranous ALCAM expression where associated with shorter overall survival, but not disease‐free survival times (table 4). For total ALCAM, no significant differences became apparent for either disease‐free survival (p = 0.865) or overall survival (p = 0.345).
Figure 2 Univariate survival analyses of activated leucocyte cell adhesion molecule (ALCAM) expression in breast cancer. Impact of cytoplasmic ALCAM expression on disease‐free survival (dotted line = low cytoplasmic ALCAM expression, continuous line = high cytoplasmic ALCAM expression): (A) in all 162 cases; (B) in nodal negative cases; (C) in nodal positive cases; (D) in cases either untreated or not treated with tamoxifen; (E) in cases treated with tamoxifen.
Table 4 Univariate survival analysis (Kaplan–Meier): overall and disease‐free survival times of all patients with breast cancer according to clinicopathological factors and ALCAM expression.
| Characteristic | Overall survival | Disease‐free survival | ||||||
|---|---|---|---|---|---|---|---|---|
| No of cases | No of events | 5 year survival (SE) | p Value | No of cases | No of events | 5 year non‐progression (SE) | p Value | |
| Membranous ALCAM expression | 0.031 | 0.779 | ||||||
| Low | 126 | 17 | 88.4 (3.2) | 126 | 40 | 65.9 (5.0) | ||
| High | 36 | 10 | 76.4 (8.0) | 36 | 11 | 66.0 (8.9) | ||
| Cytoplasmic ALCAM expression | 0.067 | 0.014 | ||||||
| Low | 134 | 20 | 88.9 (3.1) | 134 | 38 | 69.4 (4.6) | ||
| High | 28 | 7 | 68.4 (10.1) | 28 | 13 | 49.4 (11.1) | ||
| Age | 0.945 | 0.437 | ||||||
| <60 years | 86 | 14 | 89.1 (3.7) | 86 | 29 | 59.2 (6.4) | ||
| ⩾60 years | 76 | 13 | 82.1 (5.0) | 76 | 22 | 73.8 (5.3) | ||
| Histology | 0.552 | 0.264 | ||||||
| Ductal | 144 | 25 | 84.8 (3.4) | 144 | 48 | 65.1 (4.6) | ||
| Lobular | 18 | 2 | 92.9 (6.9) | 18 | 3 | 73.9 (13.1) | ||
| pT stage | 0.0005 | 0.0005 | ||||||
| pT1 | 103 | 7 | 94.1 (2.6) | 103 | 24 | 78.5 (4.6) | ||
| pT2 | 44 | 14 | 74.0 (7.6) | 44 | 22 | 34.8 (9.4) | ||
| PT3/4 | 15 | 6 | 63.0 (13.3) | 15 | 5 | 63.0 (13.3) | ||
| Nodal status | 0.0064 | 0.0018 | ||||||
| pN0 | 79 | 5 | 93.6 (3.1) | 79 | 15 | 80.5 (5.2) | ||
| pN1+ | 81 | 22 | 77.9 (5.1) | 81 | 34 | 53.7 (6.5) | ||
| UICC stage | 0.0078 | 0.0064 | ||||||
| I | 80 | 5 | 93.7 (3.1) | 80 | 16 | 79.4 (5.2) | ||
| II | 56 | 13 | 84.0 (5.6) | 56 | 25 | 47.8 (8.2) | ||
| III | 26 | 9 | 65.9 (10.0) | 26 | 10 | 61.5 (10.2) | ||
| Histological grade | 0.0114 | 0.0029 | ||||||
| G1 | 40 | 3 | 92.0 (5.4) | 40 | 5 | 84.2 (6.6) | ||
| G2 | 82 | 11 | 89.6 (3.8) | 82 | 28 | 67.0 (5.9) | ||
| G3 | 40 | 13 | 71.4 (7.8) | 40 | 18 | 43.9 (10.4) | ||
| Oestrogen receptor | 0.1639 | 0.1155 | ||||||
| Negative | 42 | 11 | 79.6 (6.5) | 42 | 17 | 59.6 (8.3) | ||
| Positive | 106 | 14 | 87.5 (3.8) | 106 | 29 | 70.4 (5.3) | ||
| c‐erbB2 expression | 0.4172 | 0.4059 | ||||||
| 0, 1+ | 100 | 16 | 86.8 (3.8) | 100 | 29 | 71.3 (5.5) | ||
| 2+, 3+ | 35 | 6 | 81.9 (7.4) | 35 | 12 | 59.27 (9.2) | ||
ALCAM, activated leucocyte cell adhesion molecule; pT, primary tumour; UICC, International Union Against Cancer.
Survival analysis in patient subgroups
In a stratified analysis according to nodal status (fig 2, panels B and C) and adjuvant treatment (fig 2, panels D and E), we found a significant prognostic impact of cytoplasmic ALCAM on disease progression in the group of nodal negative cases (mean 99 v 61 months, p = 0.035) and in the group of patients who had received tamoxifen as adjuvant treatment (mean 97 months v 48 months, p = 0.0068).
Multivariate survival analysis
In the Cox regression we included cytoplasmic ALCAM expression (low v high), tumour grade (G1, G2, and G3), nodal status (negative v positive), and primary tumour (pT) stage (pT1, pT2, and pT3/4). Disease‐free survival time was significantly dependent on nodal status and cytoplasmic ALCAM positivity and borderline significantly dependent on tumour grade (table 5).
Table 5 Cox regression model including conventional variables and cytoplasmic ALCAM.
| Disease‐free survival (51 events) | |||
|---|---|---|---|
| RR | 95% CI | p Value | |
| ALCAM (cytoplasmic) | 2.086 | 1.092 to 3.984 | 0.026 |
| pT stage | 0.884 | 0.544 to 1.436 | 0.618 |
| Nodal status | 2.246 | 1.058 to 4.765 | 0.035 |
| Grading | 1.600 | 0.996 to 2.572 | 0.052 |
ALCAM, activated leucocyte cell adhesion molecule; CI, confidence interval; pT, primary tumour; RR, relative risk.
Discussion
ALCAM is a cell surface immunoglobulin superfamily member involved in cell adhesion.11,12,13,14 In neoplasia, ALCAM expression has been implicated in malignant melanoma, fibrosarcoma cell lines, prostate, colon, and breast cancer, oral and oesophageal squamous cell carcinoma, and hepatocellular carcinoma.12,24,25,26 Our previous studies on ALCAM in prostate cancer showed that ALCAM is upregulated in most tumours compared with normal tissue, although in a minority of high grade tumours reduced ALCAM immunoreactivity was noted.21 In a follow up study using a different antibody which works on formalin fixed paraffin embedded tissues, we could confirm and complement these results by showing that higher levels of cytoplasmic ALCAM were predictive of shorter disease‐free survival times.22 Raised levels of ALCAM protein expression have also been associated with an adverse prognosis and aggressive tumour behaviour in carcinoma of the bladder, colon cancer, oesophageal cancer, and melanoma; in the latter it has been described as a hallmark of the vertical growth phase.18,27,28 High ALCAM levels were also found in vasculogenetic tissue in the lung, which might suggest a role in the vascular proliferation that is necessary for metastatic tumour growth.29 Recent studies have also pointed to a role of ALCAM in chemoresistance, showing ALCAM upregulation in a chemoresistant fibrosarcoma cell line.20
Our findings in breast cancer in the present study underscore the role of ALCAM in the disease progression of human carcinomas. In a previous study using a dot blot analysis we found increased ALCAM transcript levels in five of eight breast cancers in comparison with matching normal tissue.21 This is in concordance with findings of King et al, who reported higher ALCAM transcript levels in invasive breast cancer than in normal tissue.30 Interestingly, they also found that reduced ALCAM transcript levels in primary tumours were indicative of a more aggressive phenotype and worse prognosis. In contrast to our findings, King et al described a weaker immunostaining in malignant tissues compared with normal tissues, which is unfortunately not evident in their published figures, although the same antibody was employed as in our study. Their finding of a reduced ALCAM expression in tumour tissue might be a result of overstaining, which can mask expression differences between normal and malignant tissues; it could also result from methodological differences, and especially the rather high concentration of primary antibody used (1:40). We found that these high concentrations of antibody resulted in a reduction in contrast between normal and tumour staining intensity. Finally, in contrast to our study, they did not show immunostaining of carcinoma with adjacent normal tissue to prove their point. Concerning the general discrepancy between RNA and protein levels, King et al suggested that this might be based on cellularity of the samples. Indeed, after normalisation of the ALCAM transcript levels by cytokeratin 19 transcript levels, ALCAM appeared to be downregulated. However, CK19 itself is differentially expressed in breast tissues with an overexpression in tumour cells compared with normal cells.31 Furthermore, CK19 expression is strongly correlated with tumour grading in breast cancer, showing loss of expression in high grade tumours, which again opposes its use as a gene to normalise for epithelial cellularity.32 As our study did not incorporate RNA data, we could not clarify whether there is a general discrepancy between ALCAM transcript levels and ALCAM protein expression.
Take home message
Strong cytoplasmic ALCAM expression in primary breast cancer can be detected by immunohistochemistry and might be a new marker for more aggressive breast cancer biology
To our knowledge the present study is the first to investigate the role of ALCAM in breast cancer by immunohistochemistry on a large number of clinically characterised cases. We evaluated the two ALCAM staining patterns that have already been described—namely, the membranous and cytoplasmic patterns. Importantly, we found that the cytoplasmic proportion of the ALCAM immunoreactivity was prognostically relevant, which endorses the biological significance of this staining pattern. The mechanism responsible for this association of higher cytoplasmic ALCAM levels with a more aggressive course of the disease is not fully elucidated. High ALCAM levels in the cytoplasm of cells might be the result of aberrant ALCAM protein expression, because physiologically ALCAM shows a membranous staining pattern. Tomita et al investigated prostate cancer cell lines and found that ALCAM was linked to the cytoskeleton by α‐catenin; they were able to show that a loss of α‐catenin led to cytoplasmic localisation of ALCAM and to a more invasive phenotype, whereas membranous ALCAM was indicative of a functioning α‐catenin/E‐cadherin axis.19 The underlying mechanisms appear to be complex, but recently the metastasis associated agent, T lymphoma invasion and metastasis 1 (TIAM1), was found to modulate the localisation of ALCAM in a melanoma cell line by localising ALCAM to the membrane and cell–cell contacts, and it increased proliferation and decreased the migration capabilities of the cell line, suggesting a mechanism similar to cadherin mediated inhibition of migration.33 Thus cytoplasmic localisation of ALCAM might enhance the migratory properties of malignant cells. It is also conceivable that cytoplasmic ALCAM does not function properly as a cell surface sensor for growth saturation and the regulation of cellular signalling and dynamic responses, as has been hypothesised recently for malignant melanoma.34
Tomita et al have noted in their immunostainings a striking similarity between α‐catenin and ALCAM expression. We did not look at α‐catenin expression in our tumour cohort, but it is of interest that Gillett et al could show, in analogy to our finding of the prognostic value of cytoplasmic ALCAM, that higher levels of cytoplasmic α‐catenin were associated with a worse prognosis in breast cancer.35
In summary, we describe ALCAM protein expression as a new prognostic marker in breast cancer. It retains its prognostic impact on disease‐free survival even in a multivariate analysis, and is especially valid in the group of nodal negative patients. The presence of high cytoplasmic ALCAM expression levels in breast cancer could suggest the need for more aggressive treatment. Likewise, the group of patients with a better prognosis according to conventional variables (G1/G2, nodal negative) might be spared unnecessary treatment if the primary tumour shows a low level of cytoplasmic ALCAM expression. It remains to be proven in prospective studies whether ALCAM is a true prognostic marker of breast cancer progression. Additional prospective and functional studies to investigate the role of ALCAM in normal breast tissue and in breast cancer are clearly needed.
Acknowledgements
We are grateful to Britta Beyer for excellent technical assistance, and to A E Neumann and Ilka Olson for helpful discussions.
Abbreviations
ALCAM - activated leucocyte cell adhesion molecule
DCIS - ductal carcinoma in situ
IRS - immunoreactive score
References
- 1.Jemal A, Murray T, Ward E.et al Cancer statistics, 2005. CA Cancer J Clin 20055510–30. [DOI] [PubMed] [Google Scholar]
- 2.Hayes D F, Isaacs C, Stearns V. Prognostic factors in breast cancer: current and new predictors of metastasis. J Mammary Gland Biol Neoplasia 20016375–392. [DOI] [PubMed] [Google Scholar]
- 3.Mori I, Yang Q, Kakudo K. Predictive and prognostic markers for invasive breast cancer. Pathol Int 200252186–194. [DOI] [PubMed] [Google Scholar]
- 4.Poremba C, Heine B, Diallo R.et al Telomerase as a prognostic marker in breast cancer: high‐throughput tissue microarray analysis of hTERT and hTR. J Pathol 2002198181–189. [DOI] [PubMed] [Google Scholar]
- 5.Yousef G M, Borgono C A, Scorilas A.et al Quantitative analysis of human kallikrein gene 14 expression in breast tumours indicates association with poor prognosis. Br J Cancer 2002871287–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duffy M J. Urokinase plasminogen activator and its inhibitor, PAI‐1, as prognostic markers in breast cancer: from pilot to level 1 evidence studies. Clin Chem 2002481194–1197. [PubMed] [Google Scholar]
- 7.Nakopoulou L, Giannopoulou I, Stefanaki K.et al Enhanced mRNA expression of tissue inhibitor of metalloproteinase‐1 (TIMP‐1) in breast carcinomas is correlated with adverse prognosis. J Pathol 2002197307–313. [DOI] [PubMed] [Google Scholar]
- 8.Spizzo G, Obrist P, Ensinger C.et al Prognostic significance of Ep‐CAM AND Her‐2/neu overexpression in invasive breast cancer. Int J Cancer 200298883–888. [DOI] [PubMed] [Google Scholar]
- 9.Rudland P S, Platt‐Higgins A, El‐Tanani M.et al Prognostic significance of the metastasis‐associated protein osteopontin in human breast cancer. Cancer Res 2002623417–3427. [PubMed] [Google Scholar]
- 10.Kristiansen G, Winzer K J, Mayordomo E.et al CD24 expression is a new prognostic marker in breast cancer. Clin Cancer Res 200394906–4913. [PubMed] [Google Scholar]
- 11.Patel D D, Wee S F, Whichard L P.et al Identification and characterization of a 100‐kD ligand for CD6 on human thymic epithelial cells. J Exp Med 19951811563–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Degen W G, van Kempen L C, Gijzen E G.et al MEMD, a new cell adhesion molecule in metastasizing human melanoma cell lines, is identical to ALCAM (activated leukocyte cell adhesion molecule). Am J Pathol 1998152805–813. [PMC free article] [PubMed] [Google Scholar]
- 13.Bowen M A, Aruffo A A, Bajorath J. Cell surface receptors and their ligands: in vitro analysis of CD6‐CD166 interactions. Proteins 200040420–428. [PubMed] [Google Scholar]
- 14.Bowen M A, Bajorath J, Siadak A W.et al The amino‐terminal immunoglobulin‐like domain of activated leukocyte cell adhesion molecule binds specifically to the membrane‐proximal scavenger receptor cysteine‐rich domain of CD6 with a 1:1 stoichiometry. J Biol Chem 199627117390–17396. [DOI] [PubMed] [Google Scholar]
- 15.Bowen M A, Patel D D, Li X.et al Cloning, mapping, and characterization of activated leukocyte‐cell adhesion molecule (ALCAM), a CD6 ligand. J Exp Med 19951812213–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stephan J P, Bald L, Roberts P E.et al Distribution and function of the adhesion molecule BEN during rat development. Dev Biol 1999212264–277. [DOI] [PubMed] [Google Scholar]
- 17.Seshi B, Kumar S, Sellers D. Human bone marrow stromal cell: coexpression of markers specific for multiple mesenchymal cell lineages. Blood Cells Mol Dis 200026234–246. [DOI] [PubMed] [Google Scholar]
- 18.van Kempen L C, van den Oord J J, van Muijen G N.et al Activated leukocyte cell adhesion molecule/CD166, a marker of tumor progression in primary malignant melanoma of the skin. Am J Pathol 2000156769–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomita K, van Bokhoven A, Jansen C F.et al Coordinate recruitment of E‐cadherin and ALCAM to cell‐cell contacts by alpha‐catenin. Biochem Biophys Res Commun 2000267870–874. [DOI] [PubMed] [Google Scholar]
- 20.Choi S, Kobayashi M, Wang J.et al Activated leukocyte cell adhesion molecule (ALCAM) and annexin II are involved in the metastatic progression of tumor cells after chemotherapy with Adriamycin. Clin Exp Metastasis 20001845–50. [DOI] [PubMed] [Google Scholar]
- 21.Kristiansen G, Pilarsky C, Wissmann C.et al ALCAM/CD166 is up‐regulated in low‐grade prostate cancer and progressively lost in high‐grade lesions. Prostate 20035434–43. [DOI] [PubMed] [Google Scholar]
- 22.Kristiansen G, Pilarsky C, Wissmann C.et al Expression profiling of microdissected matched prostate cancer samples reveals CD166/MEMD and CD24 as new prognostic markers for patient survival. J Pathol 2005205359–376. [DOI] [PubMed] [Google Scholar]
- 23.Harvey J M, Clark G M, Osborne C K.et al Estrogen receptor status by immunohistochemistry is superior to the ligand‐binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol 1999171474–1481. [DOI] [PubMed] [Google Scholar]
- 24.Arora S, Matta A, Shukla N K.et al Identification of differentially expressed genes in oral squamous cell carcinoma. Mol Carcinog 20054297–108. [DOI] [PubMed] [Google Scholar]
- 25.Borlak J, Meier T, Halter R.et al Epidermal growth factor‐induced hepatocellular carcinoma: gene expression profiles in precursor lesions, early stage and solitary tumours. Oncogene 2005241809–1819. [DOI] [PubMed] [Google Scholar]
- 26.Verma A, Shukla N K, Deo S V.et al MEMD/ALCAM: a potential marker for tumor invasion and nodal metastasis in esophageal squamous cell carcinoma. Oncology 200568462–470. [DOI] [PubMed] [Google Scholar]
- 27.Tomita K, Van Bokhoven A, Jansen C F.et al Activated leukocyte cell adhesion molecule (ALCAM) expression is associated with a poor prognosis for bladder cancer patients. Urooncology 20033121–129. [Google Scholar]
- 28.Weichert W, Knosel T, Bellach J.et al ALCAM/CD166 is overexpressed in colorectal carcinoma and correlates with shortened patient survival. J Clin Pathol 2004571160–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.King J, Ofori‐Acquah S, Stevens T.et al Potential role for activated leukocyte cell adhesion molecule and neural cadherin in metastasis to the lung microcirculation. Chest 2004125150–1S. [DOI] [PubMed] [Google Scholar]
- 30.King J A, Ofori‐Acquah S F, Stevens T.et al Activated leukocyte cell adhesion molecule in breast cancer: prognostic indicator. Breast Cancer Res 20046R478–R487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitt A O, Specht T, Beckmann G.et al Exhaustive mining of EST libraries for genes differentially expressed in normal and tumour tissues. Nucleic Acids Res 1999274251–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abd El‐Rehim D M, Pinder S E, Paish C E.et al Expression of luminal and basal cytokeratins in human breast carcinoma. J Pathol 2004203661–671. [DOI] [PubMed] [Google Scholar]
- 33.Uhlenbrock K, Eberth A, Herbrand U.et al The RacGEF Tiam1 inhibits migration and invasion of metastatic melanoma via a novel adhesive mechanism. J Cell Sci 20041174863–4871. [DOI] [PubMed] [Google Scholar]
- 34.Swart G W, Lunter P C, Kilsdonk J W.et al Activated leukocyte cell adhesion molecule (ALCAM/CD166): signaling at the divide of melanoma cell clustering and cell migration? Cancer Metastasis Rev 200524223–236. [DOI] [PubMed] [Google Scholar]
- 35.Gillett C E, Miles D W, Ryder K.et al Retention of the expression of E‐cadherin and catenins is associated with shorter survival in grade III ductal carcinoma of the breast. J Pathol 2001193433–441. [DOI] [PubMed] [Google Scholar]