Abstract
We have evaluated the mechanism for sterol-regulated gene expression by the sterol regulatory element binding proteins (SREBPs) in intact cells. We show that activation of SREBPs by sterol depletion results in the increased binding of Sp1 to a site adjacent to SREBP in the promoter for the low density lipoprotein (LDL) receptor gene in vivo. Similarly, sterol depletion resulted in the increased recruitment of two distinct SREBP coregulatory factors, NF-Y and CREB, to the promoter for hydroxymethyl glutaryl CoA reductase, another key gene of intracellular cholesterol homeostasis. Furthermore, increased acetylation of histone H3 but not H4 was also detected in chromatin from both promoters on SREBP activation. Thus, SREBP activation results in the similar selective recruitment of different coregulatory generic transcription factors to two separate cholesterol-regulated promoters. These studies demonstrate the utility of the chromatin immunoprecipitation technique for analyzing the differential action of low-abundance transcription factors in fundamental regulatory events in intact cells. Our results also provide key in vivo support for the mechanism proposed from cell-free experiments, where SREBP increased the binding of Sp1 to the LDL receptor promoter. Finally, our findings also indicate that subtle differences in the pattern of core histone acetylation play a role in selective gene activation.
Feedback regulation of mammalian cholesterol homeostasis is mediated by a positive regulatory mechanism through the sterol regulatory element binding proteins (SREBPs) (1). The SREBPs also regulate key genes of fatty acid metabolism so they control flux into the two major lipid classes in mammalian cells (1, 2). When sterol levels fall in cultured cells, the SREBPs are released from membranes of the endoplasmic reticulum and nuclear envelope through the action of two sequential proteolytic activities (3). The resulting soluble mature transcription factor enters the nucleus, where it activates a set of target genes that are involved in lipid accumulation.
In all promoters for SREBP target genes that have been carefully studied thus far, SREBP-dependent regulation requires an additional generic coregulatory DNA binding factor(s) for efficient expression (4). The identity of the coregulatory factor and the position of its binding site relative to the binding site(s) for the SREBPs differs from promoter to promoter. The common use of SREBP provides a mechanism for coordinate regulation. However, the unique coregulatory factors and subtle differences in promoter architecture provide the opportunity for more subtle promoter-specific regulatory effects that integrate other cellular signaling pathways with simple nutrient sensing to provide optimal control of cellular lipid levels.
To understand the mechanism for coordinate and gene-specific activation by the SREBPs, we have been investigating how they function to activate transcription synergistically with distinct coregulatory factors in different promoters. The gene that encodes the key protein of cholesterol uptake, the low density lipoprotein (LDL) receptor, has a promoter that contains a single SREBP site flanked on either side by a coregulatory site for the generic Sp1 protein (see Fig. 2A). SREBP and Sp1 activate the LDL receptor promoter synergistically in transfected cells (5) and from reconstituted chromatin templates in experiments performed with cell-free extracts (6).
Figure 2.
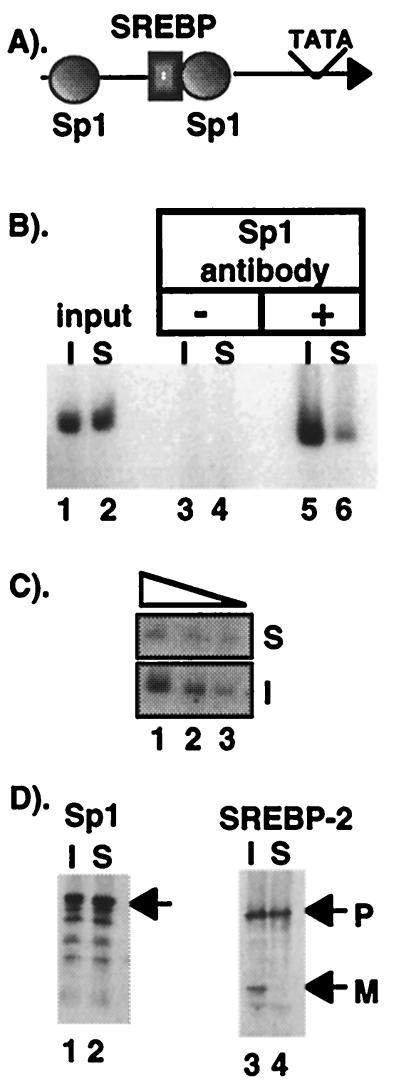
CHIP analysis for Sp1 association with LDL receptor promoter in sterol-depleted CHO-7 cells. (A) A schematic representation of the LDL receptor promoter showing the two required Sp1 sites and the single SREBP site upstream from the TFIID interacting region. All three of these sites are required for sterol regulation (28). (B) PCR analysis of CHIP reactions. Chromatin from cells cultured in the absence (I) or presence (S) of regulatory sterols was analyzed by PCR with LDL receptor promoter-specific primers, as described in Materials and Methods. The starting chromatin from each preparation was diluted 1:1,000, and 2 μl were analyzed in lanes 1 and 2. Samples were processed through the CHIP protocol, and the primary antibody was left out (lanes 3 and 4) or an Sp1 antibody was included (lanes 5 and 6). Twenty-eight cycles of PCR were used. (C) The input material from the induced (I) or suppressed (S) was serially diluted in 3-fold steps in lanes 1–3 to document that the amount of PCR product accurately reflects the amount of template DNA added to the PCR. (D) Equal aliquots of starting chromatin from each sample were subjected to SDS/PAGE and immunoblot analysis with the Sp1- or SREBP-2-specific primary antibodies, as indicated. P and M denote the positions of putative precursor and mature forms, respectively, of SREBP-2.
DNA binding studies with purified proteins in vitro demonstrated that SREBP stimulated the adjacent binding of Sp1 at the LDL receptor promoter approximately 10-fold through stimulating the initial on rate of binding (5, 7). Based on this and other key observations, we proposed a model for sterol regulation of the LDL receptor promoter that is presented in Fig. 1. We wanted to evaluate this model further and address other aspects of SREBP-dependent gene activation in intact cells to establish a working model to account for the ability of SREBPs to function with different coregulatory proteins in stimulating different promoters. Toward this goal, we have used the chromatin immunoprecipitation (CHIP) technique to analyze SREBP coregulatory factor binding to promoters of endogenous sterol-regulated genes in intact cells. The experiments demonstrate that SREBP does indeed recruit Sp1 to the endogenous LDL receptor promoter and further show that a similar mechanism for coregulatory factor recruitment occurs in another cholesterol-regulated promoter, where different generic coregulatory factors function synergistically with SREBP. Additionally, we show that SREBP activation results in an increased level of acetylation of histone H3, but not H4, in chromatin at sterol-regulated promoters.
Figure 1.
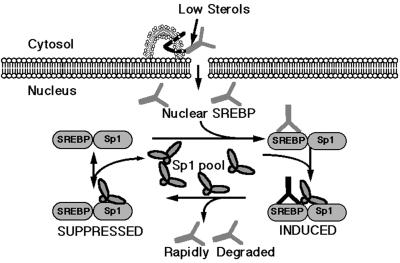
Model for SREBP-dependent gene activation. A schematic representation of key steps in activation of the LDL receptor promoter by SREBP and Sp1. The model depicts full-length SREBP tethered to the endoplasmic reticulum membrane through its two-pass membrane spanning domain. On sterol depletion, the protein is clipped out of the membrane and migrates to the nucleus, where it binds to the SREBP site in the LDL receptor promoter adjacent to an Sp1 site. This stimulates Sp1 to bind to the adjacent site, whereon the DNA bound Sp1 leads to a destablilization of DNA-bound SREBP (5). After it falls off the DNA, SREBP is rapidly degraded by a nuclear calpain type of protease activity (27). Under this model, the activation of the LDL receptor promoter by SREBP is transient and rapidly reversible, unless processing continues and SREBP levels continue to increase because of a chronic low cellular sterol level.
Materials and Methods
Cell Cultures and Media.
Stock flasks of Chinese hamster ovary (CHO)-7 cells (8) were grown in a 50/50 mixture of Ham's F-12 and DMEM (Irvine Scientific) containing 10% (vol/vol) FBS at 37°C and 8% CO2. Twenty to twenty-four 15-cm tissue culture dishes were plated at 2,000,000 cells per dish on day 0 in the above medium. On day 1, the dishes were rinsed twice with 1× PBS, and half of the dishes were fed with either induced media (Ham's F-12/DMEM containing lipoprotein-depleted serum instead of FBS). The other half were fed suppressed media (Ham's F-12/DMEM containing lipoprotein-depleted serum with 10 μg/ml cholesterol and 1 μg/ml of 25-OH-cholesterol). Cells were processed for the CHIP procedure (described below) after an additional 24-h incubation. Lipoprotein-deficient serum was prepared by ultracentrifugation as described (9). Stock solutions of cholesterol and 25-OH-cholesterol (Steraloids, Wilton, NH) were prepared in ethanol.
CHIP Assay.
We used a modification of the technique described by P. Farnham and colleagues (10). Dishes of CHO-7 cells were placed in a fume hood, and formaldeyde was added to the culture medium to a final concentration of 1% followed by a room temperature incubation for 4, 6, 8, or 10 min. Glycine was added to a final concentration of 125 mM, and the dishes were incubated for an additional 5 min at room temperature, medium was removed followed by three rinses with cold 1× PBS. Cells were scraped into 1× PBS and collected by centrifugation, washed once with 1× PBS containing 1 mM PMSF, and the cell pellets were resuspended by gentle pipeting in 5 ml of cell lysis buffer [5 mM Pipes (KOH), pH 8.0/85 mM KCl/0.5% (vol/vol) NP-40] with the protease inhibitors leupeptin, pepstatin, and PMSF all added at 0.2 mM. Samples were incubated 10 min on ice and subjected to 7–10 strokes in a Dounce homogenizer with a B pestle. Nuclei were collected by centrifugation at 2,000 rpm for 5 min in a H-6000A rotor in a Sorvall RC3B, and the supernatant was discarded. The nuclear pellets were resuspended in 3 ml (per six initial 15-cm dishes) of nuclear lysis buffer [50 mM Tris, pH 8.0/10 mM EDTA/1% (wt/vol) SDS plus the protease inhibitors mentioned above], and the samples were resuspended by light sonication. The samples were layered over CsCl step gradients (3.6 ml of 1.33 g/ml; 2.7 ml of 0.8 g/ml; 2.1 ml of 0.44 g/ml) in SW41 tubes and centrifuged at 27,000 rpm for 20 h at 20°C in an SW41 rotor. The chromatin-containing fraction was collected as a flocculent layer, and it was removed and resuspended in nuclear lysis buffer (50 μl/dish of cells), sonicated lightly, and diluted 20-fold with immunoprecipitation dilution buffer [1% (vol/vol) Triton X-100/16.7 mM Tris, pH 8/1.2 mM EDTA/167 mM NaCl plus the protease inhibitors described above], and the sample was sonicated with repeated 10-s bursts until the average size of the resulting DNA was 400–500 bp. The amount of sonication varied based on the time of formaldehyde crosslinking. The samples were subjected to dialysis against two changes of 500 ml of buffer [0.05% (wt/vol) SDS/1% Triton X-100/20 mM Tris, pH 8.0/150 mM NaCl/2 mM EDTA plus protease inhibitors] over a 16-h period. Samples were clarified (10,000 rpm for 10 min in an SS34 rotor) and adjusted to a concentration of 25 A260 units/ml. Equal aliquots (300 μl) were used for individual CHIP reactions, and remaining aliquots were stored frozen at −80°C. The crosslinked chromatin was stable for 4–6 wk at −80°C. The samples were processed from this point on with slight differences, depending on the primary antibody that was to be used. Where indicated below, protein A or G beads (60 μl each; Roche Molecular Biochemicals) or a mixture (30 μl of each) were added to each tube. The beads were pretreated with salmon sperm DNA and BSA to block nonspecific binding as described (10). The samples were incubated with the beads for 1 h at 4°C on a rotating wheel, and beads were collected by centrifugation at 2,000 rpm for 1 min. The supernatant was transferred to a new tube, and the primary antibody (see below) was added and the samples were incubated at 4°C overnight on a rotating wheel. The samples were centrifuged for 10 min at 10,000 rpm, the supernatant was transferred to a new tube, and 60 μl of protein A, G, or a mixture of A and G beads were added and samples were incubated 1–2 h at 4°C on a rotating wheel. The samples were washed four times with 1 ml wash buffer [0.1% (vol/vol) Triton X-100/20 mM Tris, pH 8.0/150 mM NaCl/2 mM EDTA] and eluted by three successive 5-min incubations with 150 μl elution buffer [1% (wt/vol) SDS/50 mM NaHCO3]. The eluates were pooled, 1 μl of RNase (10 mg/ml) was added and NaCl was adjusted to 0.3 M, and the samples were incubated at 65°C for 4 h to reverse the formaldehyde crosslinking. Digestion buffer was added [10 μl 2 M Tris, pH 6.8/10 μl 0.5 M EDTA/2 μl of proteinase K (20 mg/ml)], and the samples were placed at 45°C for 2 h. Samples were extracted with phenol/CHCl3 then CHCl3 only followed by precipitation with ethanol. Samples were resuspended in 50 μl sterile H2O, and 2–4 μl were used in each PCR.
Standard PCR for hamster hydroxymethyl glutaryl (HMG) CoA reductase or LDL receptor promoters were performed with 32P-kinased oligonucleotides and Amplitaq Gold (Perkin–Elmer). The primers for the hamster LDL receptor promoter were designed to hybridize and amplify a 150-bp fragment encompassing the region displayed in Fig. 2A. The HMG CoA reductase primers were designed to hybridize and amplify a ≈230-bp product encompassing the region displayed in Fig. 3A. To provide reactions that were in the linear dose response for the individual samples, we performed test PCR and varied the number of cycles for each promoter and antibody combination, as indicated in the figure legends. Additionally, for each immunoprecipitation experiment, a dilution curve of the starting material was performed to ensure that the final quantity of PCR product was proportional to the input. An example of this is shown for Sp1 binding to the LDL receptor promoter in Fig. 2C. All of the data presented in the figures are representative examples of several different experiments performed for each antibody and PCR combination.
Figure 3.
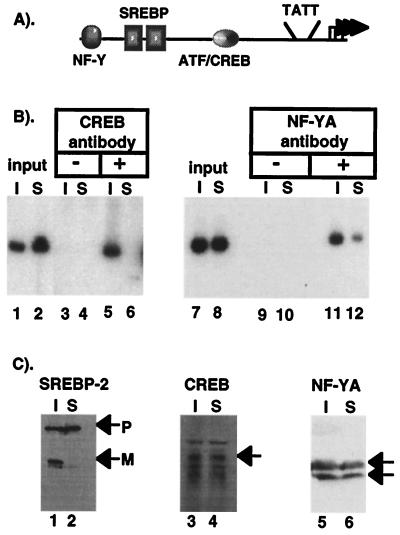
CHIP analysis for CREB and NF-Y association with HMG CoA reductase promoter in sterol-depleted CHO-7 cells (A) A schematic representation of the HMG CoA reductase promoter showing the binding sites for SREBP, NF-Y, and CREB upstream from the TFIID interacting region. All of these sites are required for sterol regulation of HMG CoA reductase (11, 12). (B) PCR analysis of CHIP reactions. Chromatin from sterol-depleted cells (I) or sterol-replete cells (S) was analyzed by PCR with HMG CoA reductase promoter-specific primers. The starting chromatin from each preparation was diluted 1:1,000, and 2 μl were analyzed in lanes 1, 2 and 7, 8. Samples were processed through the CHIP protocol, and the primary antibody was left out (lanes 3, 4, 9, 10) or either a CREB antibody (lanes 5, 6) or NF-Y A antibody (lanes 11, 12) was included as the primary antibody. Thirty cycles were used for the CREB PCR, and 24 were used for NF-Y. PCR analysis performed on serial dilutions of the input and precipitated material demonstrated that the PCR accurately reflected the amount of starting material as shown in Fig. 2 (data not shown). (C) Equal aliquots of starting chromatin from each sample were subjected to SDS/PAGE and immunoblot analysis with the SREBP-2-, CREB-, or NF-Y-specific primary antibodies as indicated. P and M denote the positions of putative precursor and mature forms, respectively, of SREBP-2.
The amounts of primary antibodies and the use of protein A or G beads varied as follows: for antiacetylated H3 and H4 (Upstate Biotechnology 06-599 or 06-866, respectively), 5 μg of antibody and protein A beads were used; for anti-NF-YA(CBF-B) (Santa Cruz Biotechnology sc-7712), 10 μg of antibody and protein G were used; for CREB-1 (Santa Cruz Biotechnology sc-186), 10 μg and a mixture of protein A and G were used; for Sp1 (Santa Cruz Biotechnology sc-59), 30 μg of antibody and a mixture of protein A and G were used.
SDS/PAGE and Immunoblot Analysis.
Equivalent amounts (normalized for A260) of chromatin extracts were analyzed by SDS/PAGE, transferred onto nitrocellulose, and incubated in Tris-buffered saline (TBS; 20 mM Tris, pH 7.6/137 mM NaCl) containing 5% (wt/vol) nonfat dry milk overnight at 4°C. The next day, the filters were washed in TBS with 0.05% (vol/vol) Tween 20 and incubated with the indicated primary antibody for 1 h, washed three times with TBS, and incubated with secondary antibody for 1 h. After three TBS washes, bound antibody was detected by using the Super Signal Kit (Pierce) and autoradiography. The IGG 7D4 mAb (obtained from the American Type Culture Collection) was isolated from the supernatant of cultured hybridoma cells by sequential precipitation with ammonium sulfate and octanoate and used as the primary antibody for analysis of SREBP-2 expression. The polyclonal antibodies used in the CHIP protocol described above for Sp1, the A subunit of NF-Y(CBF-B), and cAMP response element binding protein (CREB) were the primary antibodies used for immunoblotting as well.
Results
Sterol Depletion Results in Sp1 Recruitment to the LDL Receptor Promoter in Vivo.
We adapted the CHIP assay to evaluate the mechanism for synergistic activation of the LDL receptor promoter by SREBP and Sp1 in intact cells. CHO-7 cells were cultured in the absence (induced, I) or presence (suppressed, S) of regulatory sterols and harvested after a brief treatment with formaldeyhde. Isolated chromatin was subjected to sonication followed by immunoprecipitation with an antibody directed against Sp1. Analysis of the material before the immunoprecipitation revealed equal levels of LDL receptor promoter DNA in chromatin from induced or suppressed cells (Fig. 2B, lanes 1 and 2). However, after immunoprecipitation, there was substantially more LDL receptor promoter DNA in the sample prepared from sterol-depleted chromatin (Fig. 2B, lanes 5 and 6). That the level of PCR product accurately reflects a difference in starting material was documented by performing a serial dilution on the input samples (Fig. 2C). A control in which the Sp1 antibody was omitted from the immunoprecipitation reaction resulted in no detectable PCR products (Fig. 2B, lanes 3 and 4). Additionally, the immunoblot in Fig. 2D shows that equal levels of Sp1 were present in the starting chromatin preparations (lanes 1 and 2) and that SREBP-2 processing occurred normally in response to sterol depletion (lanes 3 and 4). Thus, Sp1 was recruited to the LDL receptor in vivo in response to sterol depletion, indicating that it is a key SREBP coregulatory factor and that its DNA binding is stimulated by SREBPs in living cells.
Increased Recruitment of Coregulatory Factors by SREBP at Other Sterol-Regulated Promoters.
Because the SREBP coregulatory protein(s) in different promoters vary, we wanted to evaluate another promoter where SREBP functions synergistically with different coregulators. HMG CoA reductase is the rate-controlling enzyme of cholesterol biosynthesis. The promoter for its gene contains multiple SREBP recognition elements and two different coregulatory sites; one for CAAT box binding factor/nuclear factor Y (CBF/NF-Y; herein referred to as NF-Y) and one for a CREB/ATF family member (Fig. 3A) (11, 12). We used the CHIP technique to evaluate the association of both NF-Y and CREB with the HMG CoA reductase promoter in response to changes in cell cholesterol (Fig. 3B). The PCR results demonstrated that both CREB and NF-Y were bound to the HMG CoA reductase promoter in chromatin prepared from sterol-depleted cells (Fig. 3B, lanes 5 and 11), but the amount of DNA immunoprecipitated from chromatin prepared from sterol-replete cells was significantly lower (Fig. 3B, lanes 6 and 12). Immunoblotting experiments revealed that starting levels for both CREB and NF-Y were similar in both chromatin preparations (Fig. 3C). Thus, analogous to Sp1 in the LDL receptor promoter, these two distinct coregulatory proteins are recruited to the HMG CoA reductase promoter when SREBPs are activated in response to sterol depletion.
SREBP Activation Leads to Increased Acetylation of Chromatin-Associated Histone H3 but Not Histone H4.
Several recently described transcriptional coactivator proteins, such as CREB binding protein (CBP) and P300, have been shown to possess intrinsic histone acetylase (HAT) activity (13) and they interact with P/CAF, which has a distinct HAT activity (14). Because the activation domain of SREBP-1a interacts with CBP (6, 15) and P300 (unpublished observations), we evaluated whether nutrient regulation by the SREBPs also was accompanied by increased acetylation of specific histones in chromatin at sterol-regulated promoters. We used the CHIP technique and antibodies directed against acetylated forms of either histone H3 or H4 to search for cholesterol-dependent changes in the levels of acetylation of these histones at the LDL receptor (Fig. 4A) and HMG CoA reductase (Fig. 4B) promoters. In both cases, acetylated H3 containing DNA was enriched in samples recovered from cholesterol-depleted cells (Fig. 4 A and B, lanes 5 and 6). However, in contrast, the level of histone H4 acetylation was the same under both culture conditions for both promoters (Fig. 4 A and B, lanes 7 and 8). It should be noted that others have used the same H4 antibody used in our studies and showed a preferential association of H4 to the active FMR1 gene (16).
Figure 4.
CHIP analysis for acetylated histone association with LDL receptor and HMG CoA reductase promoters in sterol-depleted CHO-7 cells. Antibodies for acetylated H3 or H4 were used in a CHIP analysis followed by PCR for either the LDL receptor (A) or HMG CoA reductase (B) promoters, respectively. All symbols and notations are similar to Figs. 2 and 3. The starting chromatin was analyzed in lanes 1 and 2, and the primary antibody was left out of the incubations in lanes 3 and 4. Twenty cycles of PCR were used in both A and B. Serial dilutions of the input and precipitated material demonstrated that the PCR accurately reflected the amount of starting material as described in the legend to Fig. 2 (data not shown).
Discussion
Sterol regulation of the LDL receptor promoter requires the concerted action of two proteins: the sterol-regulated SREBP and the generic coregulator Sp1. We previously have demonstrated that SREBP increases the DNA binding of Sp1 to the LDL receptor promoter in vitro (5), and it requires only the DNA binding domain of SREBP and a region encompassing the DNA binding and conserved buttonhead (btd) domains of Sp1 (17, 18). However, other domains in both proteins were additionally required for efficient activation when synergistic activation by SREBP and Sp1 on the LDL receptor promoter was evaluated by a transient DNA transfection assay in Drosophila SL2 cells (17). Thus, we proposed that synergistic activation of the LDL receptor promoter occurs at two steps in the activation process: initially, at the level of DNA binding and also at a subsequent step in the assembly of a transcriptionally active complex once the two proteins are bound to DNA. Recently, the synergistic activation by SREBP and Sp1 has been recapitulated on artificially assembled chromatin templates in vitro and requires a number of coactivators in addition to CBP/P300 (6). In the current studies, we demonstrate that, as sterol levels fall, the increased processing and nuclear accumulation of SREBPs results in an increased association of Sp1 with the LDL receptor promoter DNA in vivo (Fig. 2), which is in support of our prior in vitro DNA binding observations.
We also show that the mechanism for SREBP recruitment of its generic coregulatory factor is not specific for Sp1 in the LDL receptor promoter because similar increased recruitment of the different required coregulatory factors for the HMG CoA reductase promoter, NF-Y and CREB, was also observed on depletion of intracellular cholesterol. In other reports, we have demonstrated that SREBPs interact directly with Sp1, NF-Y, and CREB in solution in the absence of DNA (4, 19, 20). Thus, the increased recruitment of generic transcription factors to sterol-regulated promoters by a direct protein–protein interaction is likely to be a common aspect of regulated gene expression mediated by the SREBP proteins.
The N-terminal activation domain of SREBPs interacts with the CBP and P300 coactivator proteins (refs. 15 and 21, and unpublished observations). These multifunctional non-DNA binding coactivator proteins are brought to promoters through interactions with site-specific DNA binding proteins, and they stimulate transcription both actively through their intrinsic HAT activity and passively by providing a scaffold to assemble other required coactivators at specific promoters (13). One of the key passive roles is the recruitment of the P/CAF protein, which is a distinct HAT enzyme (14).
Because the activation domain of SREBPs interact with CBP and P300, we evaluated the importance of histone acetylation in sterol-regulated gene expression. We used antibodies specific for the acetylated forms of either histone H3 or H4 separately in the CHIP protocol and as shown in Fig. 4, increased acetylation of histone H3 was associated with SREBP activation. However, the level of histone H4 acetylation at both the LDL receptor and HMG CoA reductase promoters was constant and independent of SREBP activation. Thus, acetylation of chromatin, specifically at histone H3, is associated with gene activation by SREBPs. Because the intrinsic HAT activity of P300 readily modifies all four core histones, whereas the P/CAF enzyme shows a strong preference for histone H3 in vitro (22), our results are consistent with a role for the histone acetylase activity of P/CAF in gene activation by SREBPs.
In previous studies, the levels of acetylation for core histones H3 and H4 were evaluated during the activation of specific genes by using similar antibodies in the CHIP technique. In the case of gene activation by nuclear receptors and at the active FMR1 locus, there was a preference for H4 hyperacetylation (16, 23), and, in the activation of interferon β expression by virus infection, both H3 and H4 were hyperacetylated at proximal promoter sites (24). Along with the preferential acetylation of H3 demonstrated for the LDL receptor and HMG CoA reductase genes in our studies, the selective hyperacetylation of H3 was also recently shown to be associated with the cell cycle regulation of the p21waf protein gene (25).
It was not immediately obvious why CBP/P300 would recruit the seemingly redundant P/CAF histone acetylase because CBP and P300 possess intrinsic HAT activity. By analyzing CBP mutants that lack either the intrinsic HAT activity or its separate P/CAF interacting domain, Korzus et al. (26) provided evidence that each distinct HAT activity was involved in the selective activation of different promoters and in response to different signals. Taken together with the differential substrate specificity for the P300 and P/CAF (22), these studies provide indirect evidence that acetylation of specific histones may be associated with selective gene activation. The detection of selective hyperacetylation of histones H3 and H4 by activation of SREBP and nuclear receptors, respectively, directly demonstrate that selective increases in acetylation of specific core histone subtypes is of fundamental importance in defining different pathways for gene activation in eukaryotic genomes.
Boyd et al. (10) demonstrated the usefulness of the CHIP technique in detecting site-specific binding of transcription factors at single copy genes in higher eukaryotic cells. We have adapted the method to analyze the regulated interaction of multiple different low-abundance factors in the activation of two key genes in response to changes in cell cholesterol. With antibodies that specifically interact with single members of transcription factor families, the CHIP method has the potential to identify the individual members involved in a specific regulatory response in vivo. This is important because individual members typically bind to similar DNA sites and with comparable affinity in vitro. In our studies, we have demonstrated that CREB and Sp1 are coregulators for SREBP activation of the HMG CoA reductase and LDL receptor promoters, respectively. However, the isolated CREB DNA binding site from the HMG CoA reductase promoter binds other CREB/ATF family members in vitro (11), and Sp3 can substitute for Sp1 in activating the LDL receptor promoter in transient transfection studies (18). Whether additional CREB/ATF family members are also putative coregulatory factors for HMG CoA reductase and whether Sp3 really can substitute for Sp1 in sterol regulation awaits further investigation by the CHIP procedure.
Acknowledgments
We thank Julie Wells and Peggy Farnham for providing us with a copy of their CHIP protocol. This work was supported in part by grants from the National Institutes of Health (HL48044) and the American Heart Association (96008190).
Abbreviations
- SREBP
sterol regulatory element binding protein
- LDL
low density lipoprotein
- HMG
hydroxymethyl glutaryl
- CHIP
chromatin immunoprecipitation
- CHO
Chinese hamster ovary
- HAT
histone acetylase
- CREB
cAMP response element binding protein
- CBP
CREB binding protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Brown M S, Goldstein J L. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 2.Lopez J M, Bennett M K, Sanchez H B, Rosenfeld J M, Osborne T F. Proc Natl Acad Sci USA. 1996;93:1049–1053. doi: 10.1073/pnas.93.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown M S, Goldstein J L. Proc Natl Acad Sci USA. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dooley K A, Bennett M K, Osborne T F. J Biol Chem. 1999;274:5285–5291. doi: 10.1074/jbc.274.9.5285. [DOI] [PubMed] [Google Scholar]
- 5.Sanchez H B, Yieh L, Osborne T F. J Biol Chem. 1995;270:1161–1169. doi: 10.1074/jbc.270.3.1161. [DOI] [PubMed] [Google Scholar]
- 6.Nåår A M, Beaurang P A, Robinson K M, Oliner J D, Avizonis D, Scheek S, Zwicker J, Kadonaga J T, Tjian R. Genes Dev. 1998;12:3020–3031. doi: 10.1101/gad.12.19.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Osborne T F, LaMorte V J. Methods. 1998;16:42–48. doi: 10.1006/meth.1998.0643. [DOI] [PubMed] [Google Scholar]
- 8.Metherall J E, Goldstein J L, Luskey K L, Brown M S. J Biol Chem. 1989;264:15634–15641. [PubMed] [Google Scholar]
- 9.Goldstein J, Basu S, Brown M. Methods Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- 10.Boyd K E, Wells J, Gutman J, Bartley S M, Farnhman P. Proc Natl Acad Sci USA. 1998;95:13887–13892. doi: 10.1073/pnas.95.23.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osborne T F, Gil G, Goldstein J L, Brown M S. J Biol Chem. 1988;263:3380–3387. [PubMed] [Google Scholar]
- 12.Millinder-Vallett S, Sanchez H B, Rosenfeld J M, Osborne T F. J Biol Chem. 1996;271:12247–12253. doi: 10.1074/jbc.271.21.12247. [DOI] [PubMed] [Google Scholar]
- 13.Ogryzko V V, Schlitz R L, Russanova V, Howard B H, Nakatani Y. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 14.Yang X-J, Ogryzko V V, Nishikawa J-I, Howard B H, Nakatani Y. Nature (London) 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 15.Ericsson J, Edwards P A. J Biol Chem. 1998;273:17865–17870. doi: 10.1074/jbc.273.28.17865. [DOI] [PubMed] [Google Scholar]
- 16.Coffee B, Zhang F, Warren S T, Reins D. Nat Genet. 1999;22:98–101. doi: 10.1038/8807. [DOI] [PubMed] [Google Scholar]
- 17.Yieh L, Sanchez H B, Osborne T F. Proc Natl Acad Sci USA. 1995;92:6102–6106. doi: 10.1073/pnas.92.13.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Athanikar J N, Sanchez H B, Osborne T F. Mol Cell Biol. 1997;17:5193–5200. doi: 10.1128/mcb.17.9.5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bennett M K, Ngo T T, Athanikar J N, Rosenfeld J R, Osborne T F. J Biol Chem. 1999;274:13025–13032. doi: 10.1074/jbc.274.19.13025. [DOI] [PubMed] [Google Scholar]
- 20.Dooley K A, Millinder S, Osborne T F. J Biol Chem. 1998;273:1349–1356. doi: 10.1074/jbc.273.3.1349. [DOI] [PubMed] [Google Scholar]
- 21.Oliner J D, Andresen J M, Hansen S K, Zhou S, Tjian R. Genes Dev. 1996;10:2903–2911. doi: 10.1101/gad.10.22.2903. [DOI] [PubMed] [Google Scholar]
- 22.Schiltz R L, Mizzen C A, Vassilev A, Cook R G, Allis C D, Nakatani Y. J Biol Chem. 1999;274:1189–1192. doi: 10.1074/jbc.274.3.1189. [DOI] [PubMed] [Google Scholar]
- 23.Chen H, Lin R J, Xie W, Wilpitz D, Evans R M. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- 24.Prakesh B S, Maniatis T. Mol Cell. 1999;3:125–129. doi: 10.1016/s1097-2765(00)80181-1. [DOI] [PubMed] [Google Scholar]
- 25.Sambucetti L C, Fischer D D, Zabiudoff S, Kwon P O, Chamberlin H, Trogani N, Xu H, Cohen D. J Biol Chem. 1999;274:34940–34947. doi: 10.1074/jbc.274.49.34940. [DOI] [PubMed] [Google Scholar]
- 26.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T-M, Glass C K, Rosenfeld M G. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 27.Wang X, Sato R, Brown M S, Hua X, Goldstein J L. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 28.Sudhof T, Van der Westhuyzen D, Goldstein J, Brown M, Russell D. J Biol Chem. 1987;262:10773–10779. [PubMed] [Google Scholar]