Abstract
Background
Anaerotruncus colihomonis is a newly described bacterial genus and species isolated from the stool specimens of children. Its clinical significance, however, is unknown.
Aims
To describe a case of A colihominis bacteraemia identified by 16S ribosomal RNA (rRNA) gene sequencing and provide an emended description of the species.
Methods
An unidentified anaerobic bacillus (strain HKU19) that stains Gram negative was subjected to characterisation by 16S rRNA gene sequencing, G+C content determination and electron microscopy.
Results
Strain HKU19 was isolated from the blood culture of a 78‐year‐old woman with nosocomial bacteraemia. It was found to be an anaerobic, non‐motile, pleomorphic, thin bacillus that stains Gram negative. It produces Indole and utilises glucose and mannose. Identifying the strain to the species level was not possible by conventional phenotypic tests and commercial identification systems. The G+C content of strain HKU19 was found to be 53.43 mol%. A similarity of 99.3% nucleotide identities was found between the 16S rRNA gene sequence of strain HKU19 and that of A colihominis WAL 14 565T, which was isolated from a human faecal specimen. In contrast with the original description of A colihominis, HKU19 was found to produce occasional oval, terminal spores, although the other phenotypic characteristics matched. Spores were also occasionally observed when the two previously reported strains were re‐examined.
Conclusions
Although the source of the bacteraemia in the patient cannot be determined, this report suggests that A colihominis is of clinical significance. Spore formation is proposed as an emended description of A colihominis.
The identification of anaerobic Gram‐positive bacilli is often difficult in clinical microbiology laboratories. Firstly, spore formation is sometimes not obvious in bacterial isolates recovered directly from clinical specimens. Secondly, identification of bacilli by commercial kits, or analysis of cell wall fatty acids and metabolic end products by gas–liquid chromatogaphy, is difficult because of the limited database and a lack of special equipment and expertise. Thirdly, despite having Gram‐positive cell walls, certain species, including many Clostridium species, often stain as Gram negative. As a result, the epidemiology and pathogenic potential of many members of this group of bacteria have been poorly understood. Since the recognition of the 16S ribosomal RNA (rRNA) gene as a new standard for classification and identification of bacteria,1,2 their classification has been revised and many new species identified.3,4,5,6
Anaerotruncus is a newly described bacterial genus in which A colihominis is the only species.7 The bacterium was first identified by phenotypic and phylogenetic studies on two isolates of an unidentified Gram‐positive, anaerobic bacillus isolated from the stool specimens of two children. Owing to its loose phylogenetic association with, and phenotypic difference to, its closest relatives within the C leptum rRNA cluster, and, more importantly, the absence of spore formation, it was classified as a distinct genus and species. It is not known, however, whether the bacterium is only part of the human gut flora or whether it can be associated with disease. In this study, we describe the identification of an anaerobic Gram‐positive bacillus associated with nosocomial bacteraemia by a combination of phenotypic and genotypic tests, including 16S rRNA gene analysis. Although the bacterium was able to produce spores, it is most likely to be a strain of A colihominis by phylogenetic analysis. On the basis of our findings, we propose an emended description of the species.
Methods
Microbiological methods
Clinical data were collected prospectively. Clinical specimens were collected and handled according to standard protocols. The BACTEC 9240 blood culture system (Becton Dickinson, Maryland, USA) was used. The isolate was identified by a combination of standard conventional biochemical methods8,9 and the Vitek (ANI; bioMerieux Vitek, North Carolina, USA), API (20A; bioMerieux Vitek, Hazelwood, Missouri, USA) and ATB Expression (rapid ID32A; bioMerieux Vitek, USA) systems. Antimicrobial susceptibility was tested by the E‐test (AB Biodisk, Solna, Sweden) on Brucella blood agar plates and the results were interpreted according to the NCCLS criteria for anaerobic bacteria.10 All tests were carried out in triplicate, with freshly prepared media on separate occasions.
Scanning electron microscopy
Scanning electron microscopy (SEM) was carried out as described in our previous publications.11,12 Briefly, bacterial cells were settled onto a polycarbonate membrane (Nucleopore) for fixing in 2.5% glutaraldehyde. Critical dried material was coated with palladium in a BAL‐TEC SCD 005 SEM coating system and examined under a Leica Cambridge Stereoscan 440 scanning electron microscope operating at 12 kV.
16S rRNA gene sequencing and phylogenetic characterisation
Bacterial DNA extraction, PCR amplification and DNA sequencing of the 16S rRNA gene of HKU19 were carried out with polymerase chain reaction (PCR) primers (LPW57 5′‐AGTTTGATCCTGGCTCAG‐3′ and LPW205 5′‐CTTGTTACGACTTCACCC‐3′) and additional sequencing primers (LPW104 5′‐ACTCCTACGGGAGGCAGCAGTA‐3′ and LPW306 5′‐TGAGATGTTGGGTTAAGT‐3′; Gibco BRL, Rockville, Maryland, USA), as described previously.13,14 The sequences of the PCR products were compared with known 16S rRNA gene sequences in the GenBank (http://www.ncbi.nlm.nih.gov) by multiple sequence alignment with the Clustal W program.15 The phylogenetic tree was constructed using Clustal X V.1.8116 and the neighbour‐joining method with GrowTree (Genetics Computer Group, San Diego, USA).
G+C content determination
The genomic DNA G+C content was determined by thermal denaturation according to previously published protocols.12,17 Briefly, the temperature of the genomic DNA in standard sodium citrate (0.15 M NaCl with 0.015 M sodium citrate) buffer (25 μg/ml) was increased slowly (0.5°C/min) from 25°C and the absorbance of the solution at 260 nm was monitored continuously against a blank containing only standard sodium citrate buffer. The melting temperature (Tm) of the DNA is defined as the temperature at 50% hyperchromicity. The G+C content of the genomic DNA was calculated by the formula (G+C)% = 2.44Tm−169.18
Nucleotide sequence accession number
The 16S rRNA gene sequence of HKU19 has been lodged in the GenBank sequence database under the accession number DQ002932.
Results and discussion
Patient
A 78‐year‐old woman was admitted to hospital because of acute exacerbation of chronic obstructive airway disease. She also had ischaemic heart disease, hypertension and gout. She was put on prolonged mechanical ventilation because of respiratory failure and secondary pneumothorax, which resolved gradually with chest drain. During the fifth week of hospitalisation, she developed fever, with no localised symptoms or signs. Her total leucocyte count was 17.18×109/l (neutrophils 14.11×109/l, lymphocytes 1.42×109/l, monocytes 1.11×109/l and basophils 0.15×109/l), haemoglobin level 10 g/dl and platelet count 419×109/l. The renal and liver function tests were within normal limits. Blood culture was performed. Empirical intravenous cefoperazone/sulbactam and metronidazole were started. On day 2 post‐incubation, the anaerobic blood culture bottle turned positive with a pure culture of a pleomorphic, thin bacillus that stained Gram negative (strain HKU19). Subsequently, no bacteria were grown from repeated blood cultures. No infection was evident and the patient responded to antibiotics. Her condition was subsequently complicated by episodes of nosocomial pneumonia and gastrointestinal bleeding associated with steroid treatment over the next few weeks. The patient ultimately survived after 3 months of hospitalisation.
Phenotypic characteristics
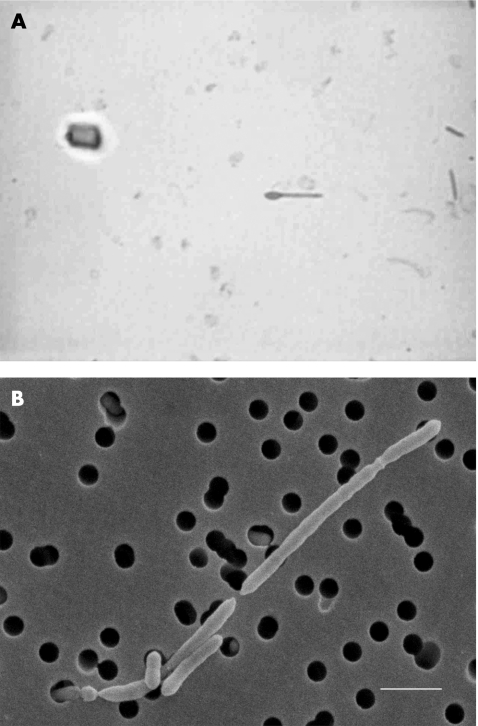
Strain HKU19 grew on sheep blood agar as non‐haemolytic, irregularly edged, grey colonies 2–3 mm in diameter after 48 h of incubation at 37°C in an anaerobic environment. It did not grow in ambient air, ambient air supplemented with 5% CO2 or microaerophilic conditions. Cells formed occasional oval terminal endospores, producing a drumstick‐like appearance (fig 1A). The strain was non‐motile. It did not produce catalase or reduce nitrate and failed to hydrolyse aesculin, gelatin or urea. It produced indole and β‐gluocosidase, and used glucose and mannose but not arabinose, cellobiose, glycerol, lactose, maltose, mannitol, melezitose, raffinose, rhamnose, salicin, sorbitol, sucrose, trehalose or xylose. Glucose was fermented, with the production of acid and gas. Lysine arylamidase was detected by the Vitek ANI system. The Vitek ANI system showed that it was “unidentified”, whereas the API system (20A) (code profile 50404003) showed a match of 83.9% to Closfridium cadaveris and 15.5% to C bifermentans, and the ATB Expression system (ID32A; code profile 0010200000) showed a 75% match to C bifermentans, 17% to Fusobacterium nucleatum and 6% to Clostridium tetani. Analysis of metabolic end products by gas–liquid chromatogaphy from peptone–yeast–glucose broth yielded only acetic and butyric acids, but no organic acids. The isolate was sensitive to vancomycin (5 μg), but resistant to colistin (10 μg) identification discs. The minimum inhibitory concentrations of penicillin, cefotaxime, vancomycin and metronidazole were 0.094, 0.047, 0.5 and <0.016 μg/ml, respectively.
Figure 1 Spore stain of HKU19 (A) showing terminal spores (green) with a drumstick‐like appearance. Scanning electron micrograph of HKU19 (B) (size bar 2 μm). Bacterial cells were bacilli, which multiplied by longitudinal division.
SEM
SEM showed that bacterial cells of HKU19 were bacilli (0.5×2−5 μm), which multiplied by longitudinal division (fig 1B).
16S ribosomal RNA gene sequencing, G+C content and phylogenetic characterisation
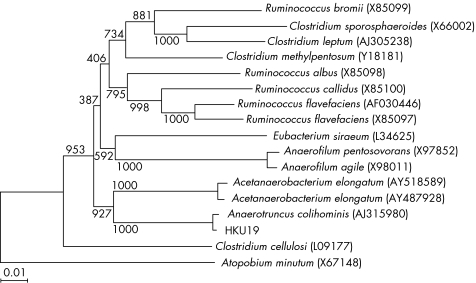
As conventional phenotypic tests and commercial identification kits failed to ascertain the identity of HKU19, its 16S rRNA gene was amplified and sequenced. PCR of the 16S rRNA gene showed a band at about 1400 bp. The 16S rRNA gene sequence of strain HKU19 had a 0.7% nucleotide difference from that of A colihominis (GenBank accession number AJ315980); a 10.2% nucleotide difference from that of Acetanaerobacterium elongatum (GenBank accession number AY487928); a 10.5% difference from that of A elongatum (GenBank accession number AY518589); a 11.5% difference from that of C methylpentosum (GenBank accession number Y18181); and a 12.5% difference from that of C cellulosi (GenBank accession number L09177; fig 2). The G+C content of strain HKU19 (mean (SD)) was 53.43 (1.09) mol%. On the basis of phylogenetic affiliation, HKU19 is a strain of A colihominis, which belongs to the C leptum suprageneric rRNA cluster. Table 1 summarises a comparison between the characteristics of HKU19 and the type strain of A colihominis, WAL 14 565T.
Figure 2 Phylogenetic tree showing the relationships of strain HKU19 to Anaerotruncus colihominis (Genbank accession number AJ315980) and members of other related genera. The tree was constructed by the neighbour‐joining method and bootstrap values calculated from 1000 trees. A total of 1419 nucleotide positions were included in the analysis. The scale bar indicates the estimated number of substitutions per 100 bases using the Jukes–Cantor correction. Names and accession numbers are given as cited in the GenBank database.
Table 1 Comparison between strain HKU19 and Anaerotruncus colihominis7.
| Biochemical reactions/enzymes/substrates | A colihominis | HKU19 |
|---|---|---|
| Spore formation | − | + |
| Catalase | − | − |
| Aesculin hydrolysis | − | − |
| Gelatin hydrolysis | − | − |
| Arginine dehydrogenase | − | − |
| Alkaline phosphatase | − | − |
| Glutamic acid decarboxylase | − | − |
| Indole production | + | + |
| Phosphate choline | − | |
| Urease | − | − |
| Reduction of nitrate | − | − |
| Reduction of triphenyl tetrazolium | − | |
| Oxidation/fermentation of | ||
| Arabinose | − | − |
| Cellobiose | − | − |
| Glucose | + | + |
| Glycerol | − | |
| Lactose | − | − |
| Maltose | − | − |
| Mannitol | − | − |
| Mannose | + | + |
| Melezitose | − | − |
| Raffinose | − | − |
| Rhamnose | − | − |
| Salicin | − | − |
| Sorbitol | − | − |
| Sucrose | − | |
| Trehalose | − | |
| Xylose | − | − |
| α‐Arabinosidase | − | − |
| α‐Fucosidase | − | − |
| β‐Fucosidase | − | |
| α‐Galactosidase | − | − |
| β‐Galactosidase | − | − |
| β‐Galactosidase‐6‐phosphate | − | − |
| α‐Glucosidase | − | − |
| β‐Glucosidase | − | + |
| β‐Glucuronidase | − | − |
| α‐Mannosidase | − | − |
| β‐Lactosidase | − | |
| β‐Xylosidase | ‐ | |
| N‐acetyl‐glucosaminidase | − | − |
| Alanine arylamidase | − | − |
| Arginine arylamidase | − | − |
| Benzoyl‐arginine arylamidase | − | |
| Glutamyl glutamic acid arylamidase | − | − |
| Glycine arylamidase | − | − |
| Histidine arylamidase | − | − |
| Leucine arylamidase | − | − |
| Leucyl glycine arylamidase | − | − |
| Lysine arylamidase | + | |
| Phenylalanine arylamidase | − | − |
| Proline arylamidase | − | − |
| Pyroglutamic acid arylamidase | − | − |
| Serine arylamidase | − | − |
| Tyrosine arylamidase | − | − |
| DNA G+C content (mol%) | 54 | 53 |
The present isolate was probably more clinically important than a contaminant because of its association with an acute febrile illness, leucocytosis, the absence of other infective foci and the patient's prompt response to antibiotics. The source of the bacteraemia was, however, unknown. The patient did not have any gastrointestinal tract disease nor had she undergone gut instrumentation. Further studies with 16S rRNA gene sequencing to characterise more clinical isolates of unidentified anaerobic Gram‐positive bacilli may help better understand the epidemiology and pathogenic potential of this bacterium.
Spore formation was also evident in the two A colihominis strains reported previously.7 Unlike HKU19, the originally described A colihominis strains were non‐spore forming. After the recognition of spore formation in the strain in this study, the two recently reported strains of A colihominis were re‐examined by both malachite green stain and phase contrast microscopy. They were also found to form occasional spores after incubation on anaerobic blood agar for 5 days. As a result, we have written an emended description of this species.
Emended description of A colihominis
This description is based on three strains, including the type strains WAL 14 565T and HKU19.7 Cells are Gram‐positive, thin rods of approximately 0.5×2−5 μm, that form oval, terminal spores. After 48 h of anaerobic incubation at 37°C under an N2/CO2 (80:20, v/v) gas phase, colonies were 2–3 mm in diameter, grey, entire‐edged, irregularly shaped, low pyramidal in profile and translucent. The pH range for growth is 5.5–11 and the growth temperature range is 36–40°C. The cells are catalase negative; nitrate is not reduced to nitrite, but indole is produced. The end products of metabolism from peptone–yeast broth are acetic and butyric acids. Aesculin, gelatin and urea are not hydrolysed. They are able to grow in peptone–yeast broth supplemented with 1% glucose, fructose, mannose or cellobiose, but they do not grow in peptone–yeast broth alone or peptone–yeast broth supplemented with arabinose, inositol, lactose, maltose, mannitol, melezitose, melibiose, raffinose, rhamnose, ribose, salicin, sorbitol, starch or xylose. With the Biolog system, N‐acetyl‐d‐glucosamine, N‐acetyl‐β‐d‐mannosamine, arbutin, d‐cellobiose, dextrin, d‐fructose, d‐galactose, d‐galacturonic acid, α‐d‐glucose, maltose, maltotriose, d‐mannose, methyl‐3‐d‐glucose, methyl‐β‐d‐glucose, methyl‐β‐d‐galactoside, methyl‐β‐d‐glucoside, palatinose, d‐trehalose and turanose are used. α‐Ketobutyric acid, α‐ketovaleric acid, l‐malic acid, pyruvic acid and pyruvic acid methyl ester are also used. Serine, l‐valine, 2′‐deoxyadenosine, inosine, thymidine and uridine are used for growth. With the API ZYM and Rapid ID 32A systems, acid phosphatase and indole activities are detected, but N‐acetyl‐β‐glucosaminidase, alanine arylamidase, alkaline phosphatase, arginine arylamidase, arginine dehydrolase, α‐arabinosidase, ester lipase C8, α‐galactosidase, β‐galactosidase, α‐glucosidase, β‐glucuronidase, glutamyl glutamic acid arylamidase, α‐mannosidase, α‐fucosidase, chymotrypsin, alanine arylamidase, leucyl glycine arylamidase, phosphoamidase, trypsin, cystine arylamidase, esterase C4, β‐galactosidase‐6‐phosphate, glutamic acid decarboxylase, glycine arylamidase, histidine arylamidase, lipase C14, leucine arylamidase, phenylalanine arylamidase, proline arylamidase, pyroglutamic acid arylamidase, serine arylamidase, tyrosine arylamidase and valine arylamidase activities are not detected. Cells are sensitive to vancomycin (5 µg) and kanamycin (1000 µg), but resistant to colistin sulphate (10 µg) identification discs. DNA G+C content is 53–54 mol%. The organism is isolated from human clinical specimens. The type strain is WAL 14 565T = CCUG 45 055T = CIP 107 765T.
Acknowledgements
This work was partly supported by the University Development Fund and University Research Grant Council Grant (HKU 7236/02M), The University of Hong Kong and the William Benter Infectious Disease Fund.
Abbreviations
rRNA - ribosomal RNA
SEM - scanning electron microscopy
Tm - melting temperature
PCR - polymerase chain reaction
rRNA - ribosomal RNA
Footnotes
Competing interests: None.
References
- 1.Relman D A, Loutit J S, Schmidt T M.et al The agent of bacillary angiomatosis. An approach to the identification of uncultured pathogens. N Engl J Med 19903231573–1580. [DOI] [PubMed] [Google Scholar]
- 2.Relman D A, Schmidt T M, MacDermott R P.et al Identification of the uncultured bacillus of Whipple's disease. N Engl J Med 1992327293–301. [DOI] [PubMed] [Google Scholar]
- 3.Collins M D, Lawson P A, Willems A.et al The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol 199444812–826. [DOI] [PubMed] [Google Scholar]
- 4.Spring S, Merkhoffer B, Weiss N.et al Characterization of novel psychrophilic clostridia from an Antarctic microbial mat: description of Clostridium frigoris sp. nov., Clostridium lacusfryxellense sp. nov., Clostridium bowmanii sp. nov. and Clostridium psychrophilum sp. nov. and reclassification of Clostridium laramiense as Clostridium estertheticum subsp. laramiense subsp. nov. Int J Syst Evol Microbiol 2003531019–1029. [DOI] [PubMed] [Google Scholar]
- 5.Lau S K P, Woo P C Y, Woo G K S.et alEggerthella hongkongensis sp. nov. and Eggerthella sinensis sp. nov., two novel Eggerthella species, account for half of the cases of Eggerthella bacteraemia. Diagn Microbiol Infect Dis 200449255–263. [DOI] [PubMed] [Google Scholar]
- 6.Woo P C Y, Fung A M Y, Lau S K P.et alActinomyces hongkongensis sp. nov. A novel Actinomyces species isolated from a patient with pelvic actinomycosis. Syst Appl Microbiol 200326518–522. [DOI] [PubMed] [Google Scholar]
- 7.Lawson P A, Song Y, Liu C.et alAnaerotruncus colihominis gen. nov., sp. nov., from human faeces. Int J Syst Evol Microbiol 200454413–417. [DOI] [PubMed] [Google Scholar]
- 8.Jousimies‐Somer H R, Summanen P, Citron D M.et alWadsworth anaerobic bacteriology manual. 6th edn. Belmont, CA: Star Publishing, 2002
- 9.Murray P R, Baro E J, Jorgensen J H.et alManual of clinical microbiology. 8th edn. Washington, DC: American Society for Microbiology, 2003
- 10.National Committee for Clinical Laboratory Standards Methods for antimicrobial susceptibility testing of anaerobic bacteria. Approved standard M11‐A5. 5th edn. Wayne, PA: National Committee for Clinical Laboratory Standards, 2003
- 11.Woo P C Y, Tam D M W, Leung K W.et alStreptococcus sinensis sp. nov., a novel Streptococcus species isolated from a patient with infective endocarditis. J Clin Microbiol 200240805–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuen K Y, Woo P C Y, Teng J L L.et alLaribacter hongkongensis gen. nov., sp. nov., a novel Gram‐negative bacterium isolated from a cirrhotic patient with bacteraemia and empyema. J Clin Microbiol 2001394227–4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woo P C Y, Fung A M Y, Lau S K P.et al Identification by 16S ribosomal RNA gene sequencing of Lactobacillus salivarius bacteremic cholecystitis. J Clin Microbiol 200240265–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woo P C Y, Fung A M Y, Lau S K P.et al Diagnosis of pelvic actinomycosis by 16S ribosomal RNA gene sequencing and its clinical significance. Diagn Microbiol Infect Dis 200243113–118. [DOI] [PubMed] [Google Scholar]
- 15.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position‐specific gap penalties and weight matrix choice. Nucleic Acids Res 1994224673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeanmougin F, Thompson J D, Gouy M.et al Multiple sequence alignment with ClustalX. Trends Biochem Sci 199810403–405. [DOI] [PubMed] [Google Scholar]
- 17.Ausubel F M, Brent R, Kingston R E. eds. Preparation of genomic DNA from bacteria. In: Current protocols in molecular biology. New York: Wiley, 19982.4.1–32.42.
- 18.Goodfellow M.Chemical methods in bacterial systematics. London: Academic Press, 198567–93.