Abstract
Background
INK4 (p15, p16, p18 and p19) and CIP/KIP (p21, p27 and p57) are two families of cyclin‐dependent kinase inhibitors (CKI) targeting CDK4/6 and CDK2, respectively.
Aim
To study the role of methylation in the inactivation of CKI in chronic lymphocytic leukaemia (CLL).
Materials and methods
Methylation‐specific polymerase chain reaction was carried out on DNA obtained from the bone marrow of 56 newly diagnosed patients with CLL.
Results
Similar demographic features and clinical outcome were observed in our patients when compared with Caucasian patients, including an indolent clinical course (10‐year overall survival 51%) and advanced Rai stage (p = 0.006), and a high‐risk karyotype such as trisomy 12 and complex aberrations (p = 0.03). In the INK4 family, methylation in p15 and p16 occurred in 20 (35.7%) and 8 (14.3%) patients, respectively. In all, 5 (8.9%) CLL samples harboured concurrent methylation of both p15 and p16. Apart from an association of p16 methylation with higher presenting leucocyte count (64.5×109/l in methylated p16 and 16.0×109/l in unmethylated p16 patients; p = 0.016), there was no association between p15 and p16 methylation and age, sex and Rai stage. No difference was observed in the overall survival for patients with and without p15 and p16 methylation. By contrast, p18 and Rb were unmethylated in all samples. In the CIP/KIP family, apart from infrequent methylation of p57 in 4 (7.1%) patients, methylation of p21 and p27 was uniformly absent.
Conclusion
p15 and, less frequently, p16 of the INK4 family of CKI, instead of the CIP or KIP family, were targeted by methylation in CLL. p16 methylation was associated with a higher lymphocyte count at presentation. This is the first comprehensive study of the epigenetic dysregulation of the INK4 and CIP/KIP families of CKI in Chinese patients with CLL.
Cellular proliferation is mediated by progression through the cell cycle, where two cell cycle checkpoints are located at G1S and G2M.1 Quiescent cells in G0 phase contain hypophosphorylated retinoblastoma (RB), which sequesters the transcription factor E2F. On activation by mitogens, up regulation of D‐type cyclins results in the activation of cyclin‐dependent kinases 4 and 6 (CDK4/6), leading to phosphorylation of RB.1 Hyperphosphorylated RB results in release of E2F, which activates transcription of S1‐specific genes. At the same time, the CIP/KIP family of cyclin‐dependent kinase inhibitors (CKIs) will detach from the CDK2 or cyclin E complex and bind to CDK4/6 instead, resulting in de‐repression (and thus activation) of CDK2 and activation of CDK4/6, and thus irreversible commitment of the cell to transit the G1S cell cycle checkpoint.1 Therefore, the cell cycle is triggered by the binding of cyclin D to and activation of cyclin‐dependent kinase 4/6 (CDK4/6), and further potentiated by the subsequent activation of CDK2, but is negatively regulated by the INK4 (p15, p16, p18 and p19) and the CIP/KIP (p21CIP, p27KIP1and p57KIP2) families of CKIs.2 The INK4 family of CKIs binds to and inhibits CDK4/6.2 In contrast, the CIP/KIP family of CKIs may bind to both CDK2 and CDK4/6, with inhibition of CDK2 and activation of CDK4/6 on binding.2
Chronic lymphocytic leukaemia (CLL) is the most common leukaemia in the Western population, but not in Asians.3 For example, the age‐adjusted incidence of CLL in the US in 2000 was 3.4/100 000. On the other hand, the age‐adjusted incidence of CLL in Hong Kong in 2000 was 0.45/100 000, as estimated from the Hong Kong Cancer Registry. Patients are usually elderly people and present with lymphocytosis, lymphadenopathy and hepatospenomegaly.4 The disease runs an indolent clinical course, but may be complicated by development of autoimmune disorders, marrow failure and Richter transformation.
DNA methylation, catalysed by DNA methyltransferase, includes the addition of a methyl group to the carbon 5 position of the cytosine ring in the CpG dinucleotide, converting it to methylcytosine.5,6 In many cancers and haematological malignancies, the CpG islands of selected genes are aberrantly methylated (hypermethylated), resulting in transcriptional repression. This may serve as an alternative mechanism of gene inactivation.5,6 Indeed, epigenetic dysregulation of cell cycle control has been shown in various haematological malignancies.7,8,9,10 In CLL, the importance of cell cycle dysregulation has been suggested by the effective induction of apoptosis of B‐CLL cells by CDI.11 Despite the frequency of the disease, data on methylation in CLL, especially with regard to a defined cellular pathway, are surprisingly scanty. Therefore, we hypothesised that epigenetic inactivation of the INK4 and CIP/KIP families of CKI may be associated with CLL. p19 of the INK4 family was not included in the analysis, as tumour suppressor activity has not been shown in transgenic mice deficient in p19.12 This is the first comprehensive study of the epigenetic dysregulation of the INK4 and CIP/KIP families of CKI in Chinese patients with CLL.
Patients and methods
Patients and treatment
The diagnosis of CLL was made according to standard criteria,13 and staged according to the Rai staging system.13 In all, there were 44 men and 12 women, with a median age of 64.5 (range: 37–91) years. The median lymphocyte count was 17×109/l. Apart from two patients with an uncertain Rai stage at diagnosis, there were 22 (40.7%) and 32 (59.3%) patients with Rai stage >2 and ⩽2, respectively (table 1). Treatment would be given for B symptoms, symptomatic organomegaly, extreme lymphocytosis, immune cytopenia or a rapid rise in lymphocyte count. The treatment included prednisolone, fludarabine or chlorambucil, or combination chemotherapy such as COPP (cyclophosphamide, vincristine, prednisolone and procabazine), CVP (cyclophosphamide, vincristine and prednisolone) or FND (fludarabine, mitoxantrone and dexamethasone).14
Table 1 Demographic data of the patients.
| n | % | |
|---|---|---|
| Patients | 56 | 100 |
| Sex | ||
| Men | 44 | 78.6 |
| Women | 12 | 21.4 |
| Median age (years) | 37–91 (64.4) | |
| Presenting lymphocyte count (×109/l) | 2–236 (17.5) | |
| Rai stage | ||
| >2 | 22 | 40.7 |
| ⩽2 | 32 | 59.3 |
| Unknown | 2 | |
| Karyotypic abnormalities | 39 | |
| del 13q14 | 3 | 7.7 |
| Trisomy 12 | 5 | 12.8 |
| Complex | 11 | 28.2 |
| Normal | 20 | 51.3 |
High‐molecular‐weight genomic DNA was isolated by standard protocols from diagnostic bone marrow aspirates of 56 patients with CLL and 12 normal bone marrow donors in addition to DNA from the peripheral blood of 12 healthy blood donors from the Hong Kong Red Cross Association.15 Cytogenetic data were available in 39 patients.16 Previous studies showed that trisomy 12 in CLL is associated with atypical morphology, progressive disease and poor survival, whereas del(13q) seems to indicate a good prognosis.17 Therefore, in this study, patients with poor‐risk cytogenetic aberrations were those with trisomy 12 and complex abnormalities, and patients with standard‐risk cytogenetic aberration included those with normal karyotype and isolated deletion of 13q14.
Methylation‐specific polymerase chain reaction
The methylation‐specific polymerase chain reaction (MSP) for gene promoter methylation was carried out as described in detail previously.7,17 Briefly, treatment of DNA with bisulphite for conversion of unmethylated, but not methylated, cytosine to uracil was carried out with a commercially available kit (CpGenome DNA modification kit, Intergen, New York, New York, USA) according to the manufacturer's instructions. Table 2 shows the primers for the methylated (M‐MSP) and unmethylated (U‐MSP) gene promoter regions for p15, p16, p18, Rb, p21, p27 and p57.18 DNA from eight normal donors was used as negative control, whereas methylated‐control DNA (CpGenome Universal Methylated DNA, Intergen) was used as positive control in all the experiments. MSP was carried out in a thermal cycler (9700, PE Biosystems, Foster City, California, USA). The polymerase chain reaction (PCR) mixture contained 50 ng of bisulphite‐treated DNA, 0.2 mM deoxynucleoside triphosphates, 2 mM magnesium chloride, 10 pmol of each primer, 1 × PCR buffer II and 2.5 units of AmpliTaq Gold (Perkin‐Elmer Biosystems, Wellesley, Massachusetts, USA) in a final volume of 50 μl.
Table 2 MSP primers and PCR conditions.
| Gene | Forward primer (5′–3′) | Reverse primer (5′–3′) | Tm and cycles | Size in bp | Locus | Reference |
|---|---|---|---|---|---|---|
| p15 | ||||||
| M‐MSP | TGAGGATTTCG CGACGCGTTC | CGTACAATAA CCGAACGAC CGATCG | 63°C/35 | 162 | 9p21 | 15 |
| U‐MSP | TGAGGATTTTG TGATGTGTTT | CATACAATAA CCAAACAAC CAATCA | 162 | |||
| p16 | ||||||
| M‐MSP | TTA TTA GAG GGT GGG GCG GAT CGC | GAC CCC GAA CCG CGA CCG TAA | 65°C/35 | 150 | 9p21 | 15 |
| U‐MSP | TTA TTA GAG GGT GGG GTG GAT TGT | CAA CCC CAA ACC ACA ACC ATA A | 151 | |||
| p18 | ||||||
| M‐MSP | TTATCGAATTG TTATTTTCGTT CG | CGTCTCGCCG AAAAAATAA TC | 64°C/35 | 93 | 1p32 | 15 |
| U‐MSP | GGGTTATTGAA TTGTTATTTTT GTTTG | CATCCATCTC ACCAAAAAAA TAATC | 95 | |||
| RB | ||||||
| M‐MSP | GGGAGTTTCGC GGACGTGAC | ACGTCGAAAC ACGCCCCG | 66°C/35 | 152 | 13q14 | 15 |
| U‐MSP | GGGAGTTTTGT GGATGTGAT | ACATCAAAAC ACACCCCA | 152 | |||
| p21 | ||||||
| M‐MSP | TTG GGC GCG GAT TCG TC | CTA AAC CGC CGA CCC GA | 62°C/35 | 100 | 6p21 | 25 |
| U‐MSP | TTA GTT TTT TGT GGA GTT G | CTC AAC TCT AAA CCA CAA | 120 | |||
| p27 | ||||||
| M‐MSP | AAG AGG CGA GTT AGC GT | AAA ACG CCG CCG AAC GA | 66°C/35 | 195 | 12p13 | 25 |
| U‐MSP | ATG GAA GAG GTG AGT TAG T | AAA ACC CCA ATT AAA ACA | 212 | |||
| p57 | ||||||
| M‐MSP | TCG GTT AGG TTT GAG CGA GC | TAC GTA TAC GAA AAA CGC GAC GAC | 62°C/35 | 137 | 11p15 | 26 |
| U‐MSP | TTG GTT AGG TTT GAG TGA GTG A | TCT ACA TAT ACA AAA AAC ACA ACA A | 139 | — | ||
PCR, polymerase chain reaction; M, methylated; U, unmethylated; MSP, methylation‐specific PCR.
DNA sequencing
The identity of the methylated and unmethylated sequences was confirmed by automated DNA sequencing. PCR products were gel purified, sequenced bi‐directionally (DYEnamic ET Terminator Cycle Sequencing kit, Amersham Biosciences, Piscataway, New Jersey, USA), and analysed on an automated DNA sequence analyser (ABI Prism 3700 DNA analyser, Applied Biosystem, Foster City, California, USA). M‐MSP of methylated positive control and selected patients were sequenced.
Statistics
Correlations between p15 and p16 methylation and continuous variables (median age, median lymphocyte counts) and categorical variables (sex and Rai staging) were studied by Mann–Whitney test and χ2 test, respectively. Overall survival is defined as the time from diagnosis to the time of death or last follow‐up. Overall survivals of patients with limited Rai stage (stages 0, I and II) were compared with those with advanced stage (stage III and IV). The effect of karyotype was studied by comparing overall survival in patients with standard‐risk and poor‐risk cytogenetic changes. Survival curves are plotted by the Kaplan–Meier method, and compared by the log rank test. All p values were two‐sided.
Results
Patient outcome
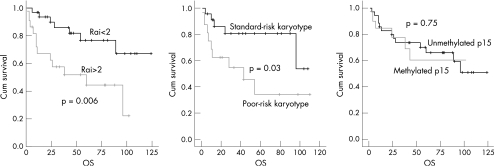
Overall survival for the whole group was 64.4% at 5 years and 51.2% at 10 years. Five‐year overall survival in patients with advanced (>2) and limited (⩽2) Rai stage was 44.4% and 76.7% (p = 0.006), respectively. Poor‐risk cytogenetic changes (trisomy 12 and complex karyotypes) were found in 16 of 39 patients (41.0%; table 1). Overall survival for patients with poor‐risk and standard‐risk karyotype was 84.5% and 32.2%, respectively (p = 0.03).
Controls
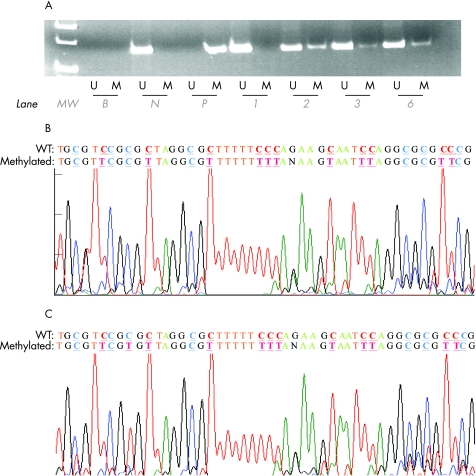
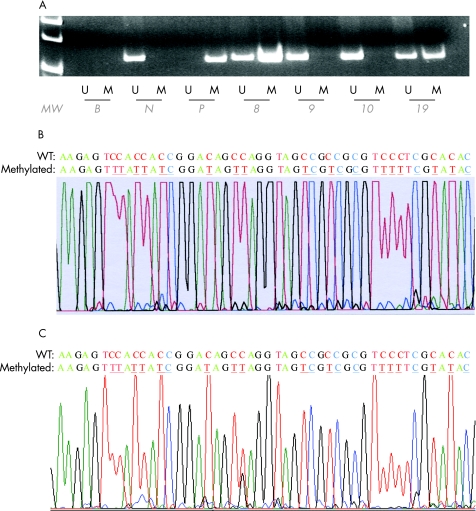
None of the seven genes tested were methylated in 12 normal bone marrow and 12 normal peripheral blood samples. The positive and negative controls showed the expected MSP results (normal DNA: U‐MSP positive/M‐MSP negative; methylated DNA: U‐MSP negative/M‐MSP positive; figs 1A and 2A). Authenticity of MSP was confirmed by sequencing of samples from the M‐MSP products from methylated positive control (figs 1B and 2B).
Figure 1 (A) Methylated methylation‐specific polymerase chain reaction (M‐MSP) and unmethylated methylation‐specific poylmerase chain reaction (U‐MSP) of p15 in primary chronic lymphocytic leukaemia (CLL) samples. B, reagent blank; N, normal marrow DNA controls; P, methylated positive control; patients 1, 2, 3 and 6, primary CLL marrow samples. (B) Sequence of positive control M‐MSP product and (C) sequence of M‐MSP of the primary CLL marrow sample. Wildtype (WT) cytosine (C) residues that remained unchanged in methylated CpG are coloured blue and underlined, whereas those that were changed to thymidine (T) are coloured red and underlined. U‐MSP shows that the methylated control (M) was totally methylated.
Figure 2 (A) Methylated methylation‐specific polymerase chain reaction (M‐MSP) and unmethylated methylation‐specific polymerase chain reaction (U‐MSP) of p57 in primary chronic lymphocytic leukaemia (CLL) samples. B, reagent blank; N, normal marrow DNA controls; P, methylated positive control; patients 8, 9, 10 and 19, primary CLL marrow samples. (B) Sequence of positive control M‐MSP amplification. (C) M‐MSP sequence of primary CLL marrow sample. Wildtype (WT) cytosine and (C) residues that remained unchanged in methylated CpG were coloured blue and underlined, whereas those that were changed to thymidine (T) are coloured red and underlined. U‐MSP shows that the methylated control (M) was totally methylated.
MSP in primary CLL marrow samples
In the INK4 family, methylation of p15 and p16 occurred in 20 (35.7%) and 8 (14.3%) patients, respectively (table 3). Of them, 5 (8.9%) patients with CLL harboured concurrent methylation of both p15 and p16. In contrast, p18 and Rb are unmethylated. In the CIP or KIP family, apart from infrequent methylation of p57 in 4 (7.1%) patients, methylation of p21 and p27 are absent. Authenticity of MSP was confirmed by sequencing samples from the M‐MSP products from methylated samples (figs 1C and 2C).
Table 3 Association between gene methylation and demographic characteristics.
| p15 | p | p16 | p | p57 | p | ||||
|---|---|---|---|---|---|---|---|---|---|
| Unmethylated | Methylated | Unmethylated | Methylated | Unmethylated | Methylated | ||||
| Median age | 63.5 | 68.5 | 0.19 | 64.5 | 65.5 | 0.72 | 64.5 | 67.5 | 0.47 |
| Median lymphocyte count | 17.0 | 21.5 | 0.25 | 16.0 | 64.5 | 0.016 | 16.0 | 31.5 | 0.17 |
| Sex | |||||||||
| Men | 30 | 14 | 0.31 | 38 | 6 | 0.99 | 41 | 3 | 0.99 |
| Women | 6 | 6 | 10 | 2 | 11 | 1 | |||
| Rai stage | |||||||||
| ⩽2 | 21 | 10 | 0.77 | 27 | 5 | 0.99 | 30 | 2 | 0.99 |
| >2 | 13 | 9 | 19 | 3 | 20 | 2 | |||
| Poor‐risk karyotype | |||||||||
| Yes | 15 | 8 | 0.74 | 21 | 2 | 0.63 | 21 | 2 | 0.50 |
| No | 9 | 7 | 13 | 3 | 16 | 0 | |||
Association between gene methylation and age and lymphocyte count was analysed by the Mann–Whitney test, and association with sex, Rai stage and presence of poor‐risk karyotype was analysed by the χ2 or Fisher's exact test.
Association of gene methylation with demographic data and overall survival
No association was found between p15 methylation and age (p = 0.19) and diagnostic Rai stage (p = 0.77), presence of poor‐risk karyotype (p = 0.74) and median lymphocyte count at presentation. No correlation was observed between p16 methylation and sex (p = 0.99), Rai stage (p = 0.99), poor‐risk karyotype (p = 0.63) and age (p = 0.72), but p16 methylation was associated with a high median lymphocyte count at diagnosis (p = 0.016). Similarly, no correlation was identified between p57 methylation and sex (p = 0.99), Rai stage (p = 0.99), poor‐risk karyotype (p = 0.50), age (p = 0.47) and presenting lymphocyte count (p = 0.17). Projected 5‐year overall survival for patients with and without p15 methylation was 66% and 59%, respectively (p = 0.75; fig 3).
Figure 3 Overall survival (OS) for patients (A) with and without advanced Rai stage disease, (B) poor‐risk karyotype and (C) p15 methylation.
Discussion
Ideally, MSP status of the neoplastic lymphocytes will only be elucidated if marrow cells have been sorted for CD5 and CD23 dually positive cells. Here, MSP of the genes has been first validated in normal control DNA (both normal bone marrow and normal peripheral blood DNA) by showing a lack of methylation and further verified by sequencing. This confirms that gene methylation is an aberration that does not exist in normal cells, be it marrow or peripheral blood cells. Given that methylation detected by MSP is a positive signal with a high sensitivity (up to 1×105 for p15 gene),7 our results are still valid without sorting for lymphocytes. The presence of amplification in U‐MSP in some methylated primary samples was due to the presence of the unmethylated gene in normal marrow cells.
Although global hypomethylation has been first shown in CLL,19 a recent genome‐wide methylation study showed that 2.5–8% of the CpG islands are aberrantly methylated in a non‐random manner in CLL.20 Moreover, although CLL has conventionally been described to be a disease with impaired apoptosis, the role for cell cycle dysregulation may be less important. However, a subpopulation of cycling B cells,21 especially those in the pseudofollicles, may be important in disease progression.
Although there are recent reports of p53 and ZAP‐70 gene methylation in CLL, with the inherent effect on prognosis,22,23 data on methylation of p15 and p16 in CLL are surprisingly scanty. Two studies reporting on the frequency of p15 and p16 methylation in Caucasian patients were available. One study showed infrequent p15 (5% of patients) and p16 (9%) methylation, whereas another showed frequent methylation of p15 (50%) and less frequently of p16 (17%), in Caucasian patients with CLL.24,25 Our study on Chinese patients for methylation of p15 and p16 showed a rate of 37.5% and 14.3%, respectively, which is comparable to that in the Caucasians. It is noteworthy that deletion or mutation associated with p15 and p16 is extremely rare in CLL.26 Therefore, methylation of p15 and p16 may be the major mechanism of p15 and p16 gene inactivation in CLL. Compared with the high frequency (>70%) of p15 methylation in acute leukaemia,7,8 the lower frequency of p15 methylation in CLL suggested a less important role for cell cycle dysregulation in CLL. Interestingly, a similar rate of p15 methylation has also been reported in multiple myeloma, a disease also characterised by a mature B cell phenotype and impaired apoptosis instead of cell proliferation.9 Our data also showed that p16 methylation was associated with a higher leucocyte count at presentation (64.5×109/l in patients with methylated p16 and 16.0×109/l in patients with unmethylated p16; p = 0.016), suggesting the activation of the cell cycle.
A tumour suppressor property of p18 gene has been demonstrated in mice.27 However, p18 methylation was absent in our patients in this and other studies.25,26 A literature review indicates that p18 methylation is probably not targeted in cell cycle dysregulation in CLL.28,29 Although 13q deletions are common in CLL,30Rb is not associated with the minimally deleted region, and thus genes other than Rb are targeted.30 Our results showed that Rb was not methylated in CLL.
We have also shown the infrequent p57, and absence of p21 and p27, methylation in CLL. p21CIP1/WAF1/CDKN1A is a tumour suppressor gene inducible by wildtype p53 in the presence of DNA damage, leading to cell cycle arrest. This is the first report of the absence of p21 methylation in CLL. p21 methylation has been shown to be rare in other lymphoid malignancies.31 Infrequent p27KIP1/CDKN1B methylation has been reported to occur in solid cancers,32 but has not been studied in haematological cancers. However, recent studies showed that cellular p27 level may be down regulated by post‐translational ubiquitination and proteasomal degradation.33 Moreover, functional inactivation of p27 may also be mediated by the phosphorylation of the nuclear localisation domain, resulting in the cytoplasmic mislocalisation of p27, thus precluding p27 from its negative regulation of CDK2.33 Therefore, although p27 methylation may not have an important role in its inactivation, post‐translational modification by ubiquitination or phosphorylation may still be important in oncogenesis. Therefore, our data suggest that epigenetic inactivation of the cell cycle control primarily targets the CDK4/INK4 complex and does not require additional inactivation of the CDK2/CIP/KIP. Indeed, a recent study showed that CDK2 (target of inhibition by KIP proteins), but not CDK4, is dispensable for cell proliferation,34 underscoring the importance of CDK4 in cell proliferation and thus the potential importance of INK4 targeting by methylation.
In summary, p15 and, less frequently, p16 of the INK4 family, instead of the CIP or KIP families, are targets of methylation in CLL, and thus might be important for its pathogenesis, posing potentially important targets for 5‐azacytidine demethylation.
Acknowledgements
We thank Professor LC Chan and Dr Clarence Lam in the Department of Pathology for the diagnosis, and Miss Chan YY and nursing staff of K20N, Division of Haematology, Queen Mary Hospital, Hong Kong, for their clinical management of the patients.
Abbreviations
CDK4/6 - cyclin‐dependent kinases 4 and 6
CKI - cyclin‐dependent kinase inhibitors
CLL - chronic lymphocytic leukaemia
MSP - methylation‐specific polymerase chain reaction
M‐MSP - methylated MSP
U‐MSP - unmethylated MSP
PCR - polymerase chain reaction
Footnotes
Competing interests: None declared.
References
- 1.Malumbres M, Barbacid M. To cycle or not to cycle: a critical decision in cancer. Nat Cancer Rev 20011222–231. [DOI] [PubMed] [Google Scholar]
- 2.Sherr C J, Roberts J M. CDK inhibitors: positive and negative regulators of G1‐phase progression. Genes Dev 1999131501–1512. [DOI] [PubMed] [Google Scholar]
- 3.Parker S L, Tong T, Bolden S.et al Cancer statistics. CA Cancer J Clin 1997475. [DOI] [PubMed] [Google Scholar]
- 4.Keating M J, Chiorazzi N, Messmer B.et al Biology and treatment of chronic lymphocytic leukemia. Hematology. Am Soc Hematol Educ Program 2003153–175. [DOI] [PubMed]
- 5.Herman J G, Baylin S B. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 20033492042–2054. [DOI] [PubMed] [Google Scholar]
- 6.Chim C S, Liang R, Kwong Y L. Gene promoter hypermethylation in hematologic malignancies. Hematol Oncol 200220167–176. [DOI] [PubMed] [Google Scholar]
- 7.Chim C S, Liang R, Tam C.et al P15 and P16 promoter methylation in acute promyelocytic leukemia. J Clin Oncol 2001192033–2040. [DOI] [PubMed] [Google Scholar]
- 8.Chim C S, Tam C, Liang R.et al P15 and P16 gene methylation in adult acute leukemia: lack of prognostic significance. Cancer 2001912222–2229. [PubMed] [Google Scholar]
- 9.Chim C S, Fung T K, Liang R. Disruption of INK4/CDK/Rb cell cycle pathway by gene hypermethylation in multiple myeloma and MGUS. Leukemia 2003172533–2535. [DOI] [PubMed] [Google Scholar]
- 10.Chim C S, Wong A S Y, Kwong Y L. Epigenetic inactivation of INK4/CDK/RB cell cycle pathway in acute leukemias. Ann Haematol 200382738–742. [DOI] [PubMed] [Google Scholar]
- 11.Hahntow I N, Schneller F, Oelsner M.et al Cyclin‐dependent kinase inhibitor Roscovitine induces apoptosis in chronic lymphocytic leukemia cells. Leukemia 200418747–755. [DOI] [PubMed] [Google Scholar]
- 12.Zindy F, van Deursen J, Grosveld G.et al INK4d‐deficient mice are fertile despite testicular atrophy. Mol Cell Biol 200020372–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheson B D, Bennett J M, Grever M.et al National Cancer Institute‐sponsored Working Group guidelines for chronic lymphocytic leukemia: revised guidelines for diagnosis and treatment. Blood 1996874990–4997. [PubMed] [Google Scholar]
- 14.Ma S Y, Au W Y, Chim C S.et al Fludarabine, mitoxantrone and dexamethasone in the treatment of indolent B‐ and T‐cell lymphoid malignancies in Chinese patients. Br J Haematol 2004124754–761. [DOI] [PubMed] [Google Scholar]
- 15.Chim C S, Fung T K, Cheung J.et al SOCS1 and SHP1 hypermethylation in multiple myeloma: implications for epigenetic activation of the Jak/STAT pathway. Blood 20041034630–4635. [DOI] [PubMed] [Google Scholar]
- 16.Wong K F, Chan J K C. Cytogenetic abnormalities in chronic B‐cell lymphoproliferative disorders in Chinese. Cancer Genet Cytogenet 199911155–60. [DOI] [PubMed] [Google Scholar]
- 17.Juliusson G, Merup M. Cytogenetics in chronic lymphocytic leukemia. Semin Oncol 19982519–26. [PubMed] [Google Scholar]
- 18.Nakamura M, Sakaki T, Hashimoto H.et al Frequent alterations of the p14 and p16(INK4a) genes in primary central nervous system lymphomas. Cancer Res 2001616335–6339. [PubMed] [Google Scholar]
- 19.Wahlfors J, Hiltunen H, Heinonen K.et al Genomic hypomethylation in human chronic lymphocytic leukemia. Blood 1992802074–2080. [PubMed] [Google Scholar]
- 20.Rush L J, Raval A, Funchain P.et al Epigenetic profiling in chronic lymphocytic leukemia reveals novel methylation targets. Cancer Res 2004642424–2433. [DOI] [PubMed] [Google Scholar]
- 21.Decker T, Schneller F, Hipp S.et al Cell cycle progression of chronic lymphocytic leukemia cells is controlled by cyclin D2, cyclin D3, cyclin dependent kinase 4 and the cdk inhibitor p27. Leukemia . 2002;16627–634. [DOI] [PubMed]
- 22.Corcoran M, Parker A, Orchard J.et al ZAP‐70 methylation status is associated with ZAP‐70 expression status in chronic lymphocytic leukemia. Haematologica 2005901078–1088. [PubMed] [Google Scholar]
- 23.Valganon M, Giraldo P, Agirre X.et al p53 aberrations do not predict individual response to fludarabine in patients with B‐cell chronic lymphocytic leukaemia in advanced stages Rai III/IV. Br J Haematol 200512953–59. [DOI] [PubMed] [Google Scholar]
- 24.Martel V, Guerci A, Humbert J C.et al De novo methylation of tumour suppressor genes CDKN2A and CDKN2B is a rare finding in B‐cell chronic lymphocytic leukaemia. Br J Haematol 199799320–324. [DOI] [PubMed] [Google Scholar]
- 25.Melki J R, Clark S J. DNA methylation changes in leukaemia. Semin Cancer Biol 200212347–357. [DOI] [PubMed] [Google Scholar]
- 26.Drexler H G. Review of alterations of the cyclin‐dependent kinase inhibitor INK4 family genes p15, p16, p18 and p19 in human leukemia‐lymphoma cells. Leukemia 199812845–859. [DOI] [PubMed] [Google Scholar]
- 27.Latres E, Malumbres M, Sotillo R.et al Limited overlapping roles of P15(INK4b) and P18(INK4c) cell cycle inhibitors in proliferation and tumorigenesis. EMBO J 2000193496–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez‐Aguilera A, Delgado J, Camacho F I.et al Silencing of the p18INK4c gene by promoter hypermethylation in Reed‐Sternberg cells in Hodgkin lymphomas. Blood 20041032351–2357. [DOI] [PubMed] [Google Scholar]
- 29.Otsuki T, Jaffe E S, Wellmann A.et al Absence of p18 mutations or deletions in lymphoid malignancies. Leukemia 199610356–360. [PubMed] [Google Scholar]
- 30.Mertens D, Wolf S, Schroeter P.et al Down‐regulation of candidate tumor suppressor genes within chromosome band 13q14.3 is independent of the DNA methylation pattern in B‐cell chronic lymphocytic leukemia. Blood 2002994116–4121. [DOI] [PubMed] [Google Scholar]
- 31.Shen L, Kondo Y, Issa J P.et al Lack of p21(CIP1) DNA methylation in acute lymphocytic leukemia. Blood 20021003432–3433. [DOI] [PubMed] [Google Scholar]
- 32.Ying J, Srivastava G, Gao Z.et al Promoter hypermethylation of the cyclin‐dependent kinase inhibitor (CDKI) gene p21WAF1/CIP1/SDI1 is rare in various lymphomas and carcinomas. Blood 2004103(2)743–746. [DOI] [PubMed] [Google Scholar]
- 33.Liang J, Zubovitz J, Petrocelli T.et al PKB/Akt phosphorylates p27, impairs nuclear import of p27 and opposes p27‐mediated G1 arrest. Nat Med 200281153–1160. [DOI] [PubMed] [Google Scholar]
- 34.Tetsu O, McCormick F. Proliferation of cancer cells despite CDK2 inhibition. Cancer Cell 20033233–245. [DOI] [PubMed] [Google Scholar]