Abstract
The extrinsic fluorescence dye 8-anilino-1-naphthalene sulfonate (ANS) is widely used for probing conformational changes in proteins, yet no detailed structure of ANS bound to any protein has been reported so far. ANS has been successfully used to monitor the induced-fit mechanism of MurA [UDPGlcNAc enolpyruvyltransferase (EC 2.5.1.7)], an essential enzyme for bacterial cell wall biosynthesis. We have solved the crystal structure of the ANS⋅MurA complex at 1.7-Å resolution. ANS binds at an originally solvent-exposed region near Pro-112 and induces a major restructuring of the loop Pro-112–Pro-121, such that a specific binding site emerges. The fluorescence probe is sandwiched between the strictly conserved residues Arg-91, Pro-112, and Gly-113. Substrate binding to MurA is accompanied by large movements especially of the loop and Arg-91, which explains why ANS is an excellent sensor of conformational changes during catalysis of this pharmaceutically important enzyme.
The fluorescence dye 8-anilino-1-naphthalene sulfonate (ANS) is a valuable probe for the detection and analysis of conformational changes in proteins and in the studies of biological membranes (1). It has a low fluorescence yield in polar environments, which is greatly enhanced on interaction with many proteins. ANS binds noncovalently to proteins and its fluorescence varies with changes in the probe environment. ANS also is used for the evaluation of folding/unfolding processes in proteins (2). Although the interaction of ANS with proteins was recognized almost 50 years ago (3), no detailed structural data for a protein liganded with ANS is available. In 1979, a crystal structure of chymotrypsin liganded with ANS at 2.8-Å resolution was reported, but the data were of limited quality, allowing only the naphthalene moiety to be identified (4). This ANS binding site appeared to be solvent-exposed, and the authors suggested that ANS might well bind to proteins through polar interactions, a conclusion that contrasts with the common opinion that ANS binds preferentially to hydrophobic cavities (5).
MurA is the first enzyme of the biosynthetic pathway of the bacterial cell wall. It catalyzes a two-substrate reaction, the transfer of the enolpyruvyl moiety of phosphoenolpyruvate (PEP) to UDP-N-acetylglucosamine (UDPGLcNAc), via an induced-fit mechanism (6, 7). On ligand binding, the two domains of MurA approach each other and the loop Pro-112–Pro-121 containing the catalytically essential Cys-115 moves toward the interdomain cleft. The only other enzyme known to catalyze the transfer of the intact enolpyruvyl moiety of PEP to a substrate is 5-enolpyruvylshikimate-3-phosphate (EPSP) synthase (EC 2.5.1.19), an essential enzyme in the biosynthesis of aromatic amino acids in plants and microbes. The dynamic properties of MurA upon ligand binding have been characterized by small-angle x-ray scattering and fluorescence spectroscopy using ANS (8). A change in the overall structure was accompanied by a quenching of the fluorescence signal. Recently, the inactivation of Enterobacter cloacae MurA by the naturally occurring antibiotic fosfomycin (9) has been re-evaluated by using inhibition kinetics and ANS-fluorescence decay, revealing that inactivation and structural change are parallel processes (10). Fosfomycin covalently attaches to the thiol group of Cys-115, and the substitution of Ser for Cys-115 resulted in altered fluorescence kinetics.
We thus were motivated to crystallize MurA in the presence of ANS to confirm our interpretations of the fluorescence experiments by elucidating the structural basis for the interaction of ANS with MurA. Initial attempts failed because of the poor solubility of ANS in the crystallization medium used for obtaining the first unliganded MurA structure (6). Instead, we have grown excellent crystals from E. cloacae MurA in complex with ANS based on the alternative crystallization conditions found recently for unliganded MurA (11). We report here the crystal structure of the open state of MurA liganded with ANS at 1.7-Å resolution, which reveals details of the interaction of this important fluorescence probe with a protein.
Materials and Methods
Cloning, overexpression, and purification of wild-type E. cloacae MurA and Escherichia coli EPSP synthase have been described (12, 13). Purified enzymes were transferred to 50 mM sodium/potassium phosphate buffer (pH. 6.9) containing 1 mM DTT by using a PD-10 column (Amersham Pharmacia), and concentrated to 80 mg/ml in Centricon-30 devices (Amicon). Aliquots were frozen in liquid nitrogen and stored at −80°C. Protein concentration was determined by using the Coomassie reagent from Pierce with BSA as a standard. ANS ammonium salt was from Sigma.
Fluorescence experiments were performed with an Aviv Associates (Lakewood, NJ) model ATF-105 spectrofluorometer, essentially as described (8, 10). The buffer used for all measurements was 50 mM sodium/potassium phosphate (pH 6.9) with 2 mM DTT. The concentration of protein (MurA or EPSP synthase) in the assay was 3 μM. Fluorescence of ANS was excited at 360 nm, and emission spectra were recorded between 400 and 600 nm. For data evaluation, ANS emission spectra in buffer were subtracted from the corresponding ANS/protein spectra, thereby giving the fluorescence intensities (y in Eq. 1) as a function of the ANS concentration as indicated in Fig. 1. The dissociation constant of ANS binding to MurA was determined by fitting data to
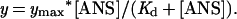 |
1 |
Crystallization was carried out by the procedure described in ref. 11, in which MurA crystallizes from PEG 20000 as a monomeric species. For cocrystallization of MurA with ANS the precipitant consisted of 25 mM Mes/NaOH (pH = pK), 5% PEG 20000, including 2.5 mM ANS. The molar ratio of MurA and ANS in the crystallization droplet was ca. 1:2. Crystals were fully grown after 2 days at 19°C and were harvested in 25 mM Mes/10% PEG 20000/2.5 mM ANS. For crystallographic analysis, one crystal was flash-frozen in harvesting buffer containing 25% (vol/vol) glycerol (120 K, cooling device: Xstream, MSC, Houston). Complete diffraction data to 1.7 Å were recorded on a RaxisIV image plate (MSC) using CuKα X-rays generated by a Rigaku rotating anode (RU300), and focused by Osmic mirror optics (MSC). Data reduction was performed with the hkl program package (14). A summary of data collection statistics is given in Table 1. Molecular replacement (MR) and refinement were performed with the cns program package (15). The search model for MR was the 1.8-Å resolution structure of unliganded type 2 MurA stripped of all solvent molecules (11). The initial model of ANS was that of Cody and Hazel (16) as deposited in the Cambridge Structural Database. Several rounds of minimization, simulated annealing (2,500 K starting temperature) and restrained individual B factor refinement were carried out. The resulting electron density maps (Fo−Fc, 2Fo−Fc) were evaluated with o (17). Throughout the refinement reflections with σ(I)>0 were used, leaving 3% of the data aside for calculation of the free R value. Solvent molecules were added to the model at chemically reasonable positions where the difference density of the Fo−Fc synthesis exceeded 3 σ, and the 2Fo−Fc synthesis exceeded 1 σ. Refinement statistics are summarized in Table 2. Model validation with procheck (18) and what_check (19) showed 93.6% of all 419 residues of MurA to have dihedral angles within the most favored regions of the Ramachandran plot, 5.8% within additionally allowed regions, one residue (Ser-349) in a generously allowed region; residue Asn-67, which is an l-isoaspartate (11), is not recognized by common validation programs.
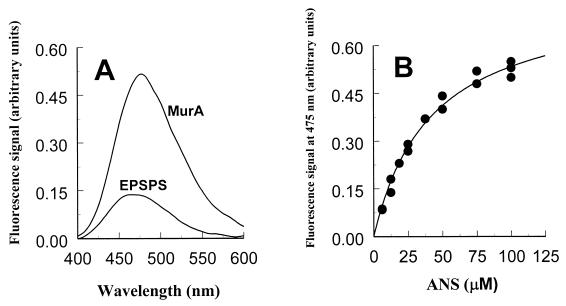
Figure 1.
ANS-dependent fluorescence response in MurA and EPSP synthase. (A) Comparison of the fluorescence spectra of MurA and EPSP synthase at 75 μM ANS. The protein concentration was 3 μM. (B) Fluorescence signals of MurA as a function of the ANS concentration. The dissociation constant of ANS in its reaction with free MurA was determined by fitting the data to y = ymax* [ANS]/(Kd + [ANS]).
Table 1.
Summary of data collection
| Data collection statistics | |
|---|---|
| Space group | P212121 |
| Unit cell dimensions, Å | a = 62.1, b = 66.6, c = 107.8 |
| Molecules/asym. unit | 1 |
| Wavelength, Å | 1.542 |
| Temperature, K | 120 |
| Resolution range, Å | 30.0–1.7 |
| Measured reflections | 1,222,364 |
| Unique reflections | 49,890 |
| Completeness, % | 99.5 (98.3) |
| 〈I/σ(I)〉 | 20.0 (2.6) |
| Rsym,* % | 6.2 (28.9) |
Values for the highest resolution shell (1.73–1.70 Å) are given in parentheses.
Rsym = ∑h∑i|Ihi − Ih|/∑hiIhi, where h indicates unique reflection indices.
Table 2.
Summary of refinement
| Refinement statistics | |
|---|---|
| Resolution range, Å | 30–1.7 |
| No. of reflections used in refinement | 49,890 |
| No. of reflections used for Rfree | 1,541 |
| Rcryst,* % | 18.0 |
| Rfree,† % | 21.1 |
| Protein atoms (nonhydrogen) | 3,143 |
| ANS atoms (nonhydrogen) | 21 |
| Solvent molecules | 512 |
| Mean B factor (protein), Å2 | 20.1 |
| Mean B factor (water), Å2 | 33.3 |
| Mean B factor (ANS), Å2 | 22.9 |
| rmsd‡ bond lengths, Å | 0.017 |
| rmsd‡ bond angles, ° | 1.78 |
Rcryst = ∑|Fobs − Fmodel|/∑Fobs, where Fobs and Fmodel are observed and calculated structure factor amplitudes, respectively.
R factor calculated for 3% randomly chosen reflections that were excluded from the refinement.
rmsd: rms deviations from ideal values.
Results and Discussion
ANS Binding to MurA and EPSP Synthase Assayed by Fluorescence Spectroscopy.
ANS binding to substrate-free MurA can be detected through the appearance of a characteristic fluorescence spectrum with the maximum of the emission curve at 475 nm (Fig. 1A). The fluorescence signal upon interaction of MurA with ANS is established rapidly (i.e., in less than 10 sec), and increasing the molar ratio of ANS to MurA results in a saturation curve. Up to ANS concentrations around 100 μM the data can be fit to Eq. 1 (Fig. 1B), yielding a dissociation constant of 40.8 ± 3.3 μM for the ANS interaction with MurA. ANS/MurA molar ratios of 40 and higher result in a loss of signal, also observed for BSA interaction with ANS (data not shown). Therefore, the dissociation constant is an approximate value.
EPSP synthase, which is structurally and mechanistically highly homologous to MurA (20), interacts only weakly with ANS (Fig. 1A). The fluorescence quantum yield is low compared with that of the ANS/MurA interaction, and the maximum of the emission curve is shifted toward shorter wavelengths, indicating a different mode of action of EPSP synthase with ANS. Neither substrates nor the inhibitor glyphosate quench the fluorescence of EPSP synthase/ANS solutions, whereas substrate binding to MurA is accompanied by a significant quenching of the fluorescence intensity (8, 10). This implies that only in the case of MurA is the ANS binding site affected by the enzyme's transition from the open to the closed state.
The ANS Binding Site in MurA.
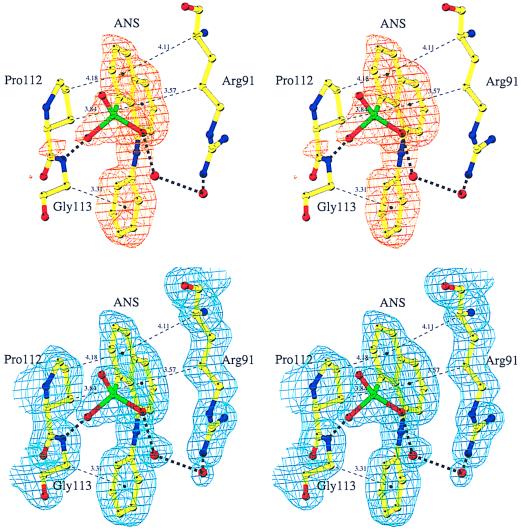
ANS binds to MurA in a solvent-exposed region of the unliganded enzyme (Fig. 2). Difference Fourier analysis shows unambiguous density for ANS (Fig. 3 Upper). The refined structure returns rather low temperature factors for the ANS molecule and the neighboring protein atoms, reflecting specific binding of the fluorophore to MurA. The naphthalene ring of ANS is sandwiched between Pro-112 and the hydrophobic side-chain part of Arg-91, whereas the anilino ring opposes the α-carbon atom of Gly-113 (Fig. 3). The sulfonate group is hydrogen-bonded to the main-chain amide of Gly-113, and, via two solvent molecules, to the guanidinium group of Arg-91. It is remarkable that none of the 19 aromatic residues, including three tryptophans, in MurA are involved in coordinating the fluorophore, because the aromatic structure of ANS suggested that binding to proteins occurs primarily through aromatic-aromatic interactions (2, 5). Apparently the aliphatic nature of Gly, Pro, and Arg is sufficient to bind the hydrophobic moieties of ANS. The hydrophobic interaction of aliphatic portions of extended charged residues like arginine with aromatic groups is unusual, but also occurs in, for example, the cytokine receptor superfamily, where indole rings of tryptophans stack with arginine or lysine side chains (23). Arg-91, Pro-112, and Gly-113 are strictly conserved in MurA. The importance and the specificity of the ANS interactions, especially with Arg-91 and Pro-112, are emphasized by the different characteristics of ANS binding to EPSP synthase. The equivalent residues in the family of EPSP synthases are a glycine instead of the arginine and a predominantly hydrophilic residue at the position of the proline.
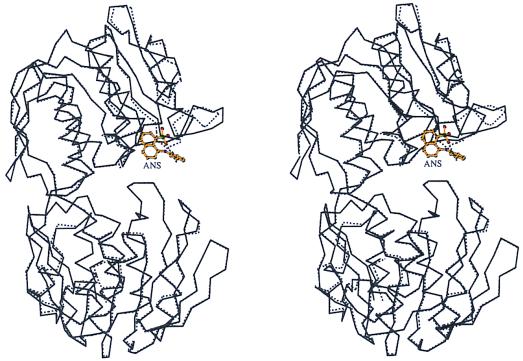
Figure 2.
Comparison of the overall structures of free and ANS liganded MurA. Stereo representation of the optimally superimposed α-carbon chains of unliganded MurAtype2 (dotted line) (11) and ANS-liganded enzyme (solid line). Figs. 2 and 4 were drawn by using molscript (21).
Figure 3.
The ANS binding site in MurA. (Upper) Representation of the Fo−Fc difference density contoured at 3 σ in the region of the ANS binding site. ANS was omitted in the refinement and the calculation of the Fourier synthesis. (Lower) The final 2Fo−Fc electron density contoured at 1 σ. ANS was included in the refinement and the calculation of the Fourier synthesis. Carbon atoms are colored yellow, nitrogen atoms blue, oxygen atoms red, sulfur atoms green. Solvent molecules are shown as red spheres. Thick dashed lines represent hydrogen bonds. To designate hydrophobic interactions, distances between ring centers and closest carbon atoms are given along thin dashed lines. The figure was drawn with bobscript (22).
Comparison of the ANS structure bound to MurA with the two small-molecule structures deposited in the Cambridge Structural Database (16, 24) reveals considerable differences in the orientation of the anilino ring with respect to the naphthalene moiety. Despite the rotational freedom around the C-N-C bond, ANS adopts a rigid conformation in its MurA-bound state, with lowest temperature factors for the anilino ring.
Structural Changes in MurA Induced by ANS Binding.
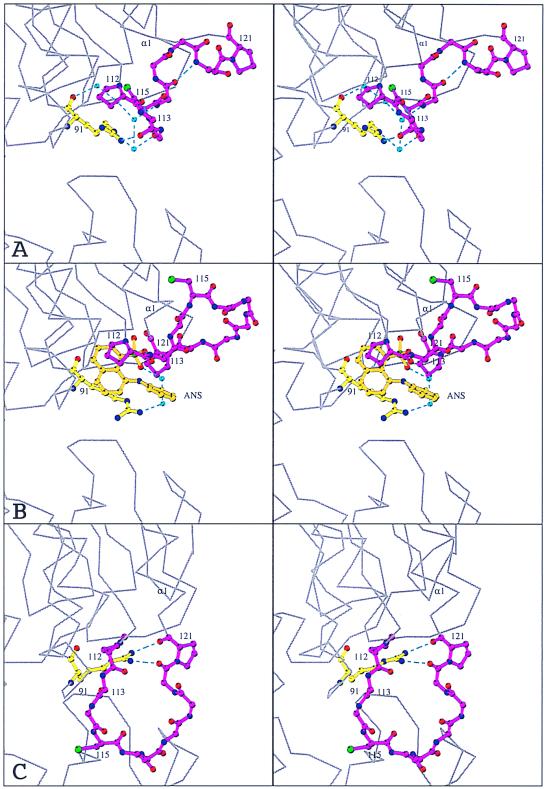
The complex MurA⋅ANS has been crystallized under conditions identical to those used with the unliganded enzyme (11). Both crystals belong to space group P212121, but differ by 30 Å in the length of their c axes, resulting in completely different crystal contacts. Yet, most of the enzyme structure is well conserved (Fig. 2). The structural differences are confined to residues 112–123 in the upper domain of MurA, which suggests that the conformation of this region is responsible for the crystal packing and not vice versa. ANS induces a conformational change in the catalytically important loop Pro-112-Gly-Gly-Cys-Ala-Ile-Gly-Ala-Arg-Pro-121 (Fig. 4). In unliganded MurA this loop exists in at least two distinct states, an extended (6) and a coiled conformation (11). Under the crystallization conditions used here, the loop adopts the coiled conformation in the absence of any ligand (Fig. 4A), but unwinds upon the interaction with ANS (Fig. 4B). The N-terminal part of the coiled loop is stabilized through a hydrogen-bonded water chain linking the carbonyl oxygen and the guanidinium group of Arg-91 with the main chain of Gly-114 and Ala-116, as well as through a direct hydrogen bond from the guanidinium group of Arg-91 to the carbonyl oxygen of Gly-113. Binding of ANS results in a disruption of these hydrogen bonds and a displacement of the water molecules, thus inducing a major reorientation of the loop (Fig. 4B). The two β-turns 112–115 and 116–119 are lost while the loop stretches into a U-like shape. The most striking feature is a shift of almost 180° in the Ψ angle of Asp-123, completing the first turn of helix α1 (subunit IIc) and bringing Pro-121 more than 9 Å closer to Pro-112. The orientation of the two loop-anchoring prolines and the resulting U-like shape of the loop are approximately as in the closed state of MurA (Fig. 4C). In the closed state, the loop has moved toward the cleft, and Arg-91 is shifted to stabilize the new loop conformation through hydrogen bonds to the main-chain oxygens of Pro-121 and Leu-122 (Fig. 4C). Assuming that the side chain of Arg-91 is essential to assure ANS binding to unliganded MurA, conformational alterations of this residue will result in dissociation of ANS. Thus, it is likely that the shift of Arg-91 precedes the loop movement during catalysis.
Figure 4.
Conformational changes in the loop around Cys-115. Stereo representations of the loop in unliganded MurAtype2 (11) (A), ANS-liganded MurA (B), and the closed state of MurA (7) (C). Arg-91 (yellow), the ANS molecule (orange), and the main chain of the loop Pro-112-Gly-Gly-Cys-Ala-Ile-Gly-Ala-Arg-Pro-121 together with the side chains of Pro-112, Cys-115, and Pro-121 (magenta) are represented as ball and stick. Noncarbon atoms are color-coded as in Fig. 3. Turquoise spheres and dashed lines designate water molecules and hydrogen bonds, respectively. The surrounding protein is shown as a gray α-carbon trace. The view corresponds to a zoom of Fig. 2. The structures were aligned through residues Asp-231, Asp-305, Arg-371, and Arg-331 of the bottom domain of MurA.
Conclusion
ANS binds specifically to a single site of MurA, which is fully occupied even at the low excess of ANS used in the crystallization setup. Apparently, the closely related EPSP synthase does not possess such a binding site, as reflected in the different fluorescence characteristics of MurA and EPSP synthase. In MurA, ANS binds in a solvent exposed region and induces the formation of a hydrophobic environment for its aromatic rings, thus merging the opposing views of Stryer (5) and Weber et al. (4). The lack of hydrophobic interactions reported for ANS binding to α-chymotrypsin (4) and cytochrome c (25) may be attributed to the low pH (2.0 and 3.6) used in these studies. It now appears, that at neutral pH both types of interactions are required to specifically bind ANS and that the resulting structural reorganization of the protein into a more nonpolar environment appears to be a common feature (26).
The nature of the ANS binding site in MurA corroborates the previous interpretation of the fluorescence data with respect to the underlying induced-fit mechanism (8, 10). ANS is not located in the interdomain cleft where catalysis takes place and therefore does not compete with substrates for binding. Instead, the fluorescence quench observed upon substrate/inhibitor interaction with MurA reflects the gross conformational changes of the entire loop and neighboring residues, in turn affecting the ANS binding site.
The MurA⋅ANS structure is evidence for the existence of an MurA⋅ligand complex in the open state. The ability of the loop Pro-112–Pro-121 to serve as a regular binding site even in the absence of sugar nucleotide might well be of significance for an alternative approach to the development of novel antibiotics. Therefore, the known loop structures should aid the design of compounds, other than ground-state substrate analogs, to inhibit the conformational changes required for the reaction of MurA.
Acknowledgments
We thank Drs. A. Young-Woody and R. Woody (Colorado State University, Fort Collins) as well as Dr. J. Wray (Max-Planck Institute for Medical Research, Heidelberg, Germany) for critical reading of the manuscript.
Abbreviations
- ANS
8-anilino-1-naphthalene sulfonate
- EPSP
5-enolpyruvylshikimate-3-phosphate
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1EYN).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.120120397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.120120397
References
- 1.Slavik J. Biochim Biophys Acta. 1982;694:1–25. doi: 10.1016/0304-4157(82)90012-0. [DOI] [PubMed] [Google Scholar]
- 2.Semisotnov G V, Rodionova N A, Razgulyaev O I, Uversky V N, Gripas A F, Gilmanshin R I. Biopolymers. 1991;31:119–128. doi: 10.1002/bip.360310111. [DOI] [PubMed] [Google Scholar]
- 3.Weber G. Biochem J. 1952;51:155–167. doi: 10.1042/bj0510155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weber L D, Tulinsky A, Johnson J D, El-Bayoumi M A. Biochemistry. 1979;18:1297–1303. doi: 10.1021/bi00574a028. [DOI] [PubMed] [Google Scholar]
- 5.Stryer L. J Mol Biol. 1965;13:482–495. doi: 10.1016/s0022-2836(65)80111-5. [DOI] [PubMed] [Google Scholar]
- 6.Schönbrunn E, Sack S, Eschenburg S, Perrakis A, Krekel F, Amrhein N, Mandelkow E. Structure (London) 1996;4:1065–1075. doi: 10.1016/s0969-2126(96)00113-x. [DOI] [PubMed] [Google Scholar]
- 7.Skarzynski T, Mistry A, Wonacott A, Hutchinson S E, Kelly V A, Duncan K. Structure (London) 1996;4:1465–1474. doi: 10.1016/s0969-2126(96)00153-0. [DOI] [PubMed] [Google Scholar]
- 8.Schönbrunn E, Svergun D I, Amrhein N, Koch M H J. Eur J Biochem. 1998;253:406–412. doi: 10.1046/j.1432-1327.1998.2530406.x. [DOI] [PubMed] [Google Scholar]
- 9.Kahan F M, Kahan J S, Cassidy P J, Kropp H. Ann NY Acad Sci. 1974;235:364–385. doi: 10.1111/j.1749-6632.1974.tb43277.x. [DOI] [PubMed] [Google Scholar]
- 10.Schönbrunn E, Eschenburg S, Krekel F, Luger K, Amrhein N. Biochemistry. 2000;39:2164–2173. doi: 10.1021/bi991091j. [DOI] [PubMed] [Google Scholar]
- 11.Eschenburg, S. & Schönbrunn, E. (2000) Proteins Struct. Funct. Genet., in press. [DOI] [PubMed]
- 12.Wanke C, Falchetto R, Amrhein N. FEBS Lett. 1992;301:271–276. doi: 10.1016/0014-5793(92)80255-f. [DOI] [PubMed] [Google Scholar]
- 13.Krekel F. Zurich, Switzerland: Eidgenössiche Technische Hochschule; 1998. [Google Scholar]
- 14.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 15.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve W, Jiang J-S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 16.Cody V, Hazel J. Acta Crystallogr B. 1977;33:3180–3183. [Google Scholar]
- 17.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 18.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 19.Hooft R W W, Vriend G, Sander C, Abola E E. Nature (London) 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 20.Stallings W C, Abdel-Meguid S S, Lim L W, Shie H-S, Dayringer H E, Leimgruber N K, Stegemann R A, Anderson K S, Sikorski J A, Padgette S R, Kishore G M. Proc Natl Acad Sci USA. 1991;88:5046–5050. doi: 10.1073/pnas.88.11.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 22.Esnouf R M. Mol Graph. 1997;15:132–134. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 23.Livnah O, Stura E A, Johnson D L, Middleton S A, Mulcahy L S, Wrighton N C, Dower W J, Jolliffe L K, Wilson I A. Science. 1996;273:464–471. doi: 10.1126/science.273.5274.464. [DOI] [PubMed] [Google Scholar]
- 24.Weber L D, Tulinsky A. Acta Crystallogr B. 1980;36:611–614. [Google Scholar]
- 25.Ali V, Prakash K, Kukarni S, Ahmad A, Madhusudan K P, Bhakuni V. Biochemistry. 1999;38:13635–13642. doi: 10.1021/bi9907835. [DOI] [PubMed] [Google Scholar]
- 26.Shi L, Palleros D R, Fink A L. Biochemistry. 1994;33:7536–7546. doi: 10.1021/bi00190a006. [DOI] [PubMed] [Google Scholar]