Abstract
Aim
To study simultaneously the actions of maspin and CXCR4, which share several similar pathways in cancer, including apoptosis and angiogenesis.
Methods
Our material consisted of 151 invasive breast carcinomas arranged in a tissue microarray setting. Maspin and CXCR4 expression was evaluated by immunohistochemistry. Microvessel density was assessed by CD34 immunodetection and apoptosis by the Tdt‐mediated dUTP nick end labelling assay.
Results
Maspin expression was related to CXCR4 expression, apoptosis, patient age and the Nottingham prognostic index. The expression of both maspin and CXCR4 progressively increased in high‐grade tumours. In patients with lymph node negative breast cancer, maspin overexpression was associated with increased risk of death. High CXCR4 expression was associated with prolonged survival of patients with high maspin expression.
Conclusions
Our results show that maspin overexpression could prove to be a potentially useful marker, especially for the clinically important group of patients with lymph node negative breast cancer. The expression of CXCR4 is of less significance in our study, but may be informative for specific patient subsets or in a longer time frame.
Breast cancer is the commonest malignancy in the female population worldwide.1 Studies on putative prognostic factors are necessary to improve prediction and to determine appropriate therapeutic interventions.
Maspin is a unique serine protease inhibitor of the serpin superfamily with reported tumour‐suppressing activity in breast cancer.2 The anti‐tumour effects of maspin can be attributed to inhibition of pericellular proteolysis.3 Recent studies have shown the involvement of maspin in inhibition of angiogenesis4,5 and induction of apoptosis.6,7 Maspin has been detected in the myoepithelial cells of normal breast tissue, where it has been considered to act defensively against malignant growth.8 The clinical significance of maspin in breast cancer remains elusive, as contradictory results have been reported.2,9,10,11,12,13,14
The chemokine receptor CXCR4 (fusin) is a G‐protein‐coupled receptor that plays an important part in a number of cellular functions, including cell migration during morphogenesis15 and modulation of the immune response.16 CXCR4 was found to act as a cofactor for T cell infection by the HIV virus.17 Recently, CXCR4 was also shown to mediate chemotactic and invasive responses in breast cancer cells through its ligand CXCL‐12/stromal cell derived factor‐1 (SDF‐1).18 The CXCR4/SDF‐1 axis may have a putative prognostic role in breast cancer.19,20,21
Maspin and CXCR4 operate in closely connected pathways, but the possibility of their correlation has not been studied. Maspin expression has been associated with stromal lymphocyte infiltration,13 and SDF‐1, the CXCR4 ligand, is a highly efficacious lymphocyte chemoattractant.22 Furthermore, CXCR4 and maspin expression may affect both tumour angiogenesis and apoptosis4,5,6,7,23,24,25,26,27 through synergistic or competing pathways.
Materials and methods
Patients
The study included 151 women with invasive breast cancer who presented consecutively at the Queen Elizabeth Hospital in Birmingham, UK, from January 1999 to May 2001. All patients received appropriate surgical and adjuvant treatment according to stage and oestrogen receptor status. The median post‐surgical follow‐up was 47 months (interquartile range, 18 months) and 31 (21.4%) patients died of the disease during that time. Table 1 shows the clinical and pathological characteristics of the patients. Histological grading was estimated by the Nottingham Modification of the Bloom and Richardson system28,29 and oestrogen receptor status was evaluated according to the NHS guidelines.30,31
Table 1 Patient characteristics*.
| I | II | III | Missing | n | ||
|---|---|---|---|---|---|---|
| Stage | 46 (31) | 79 (52) | 11 (7) | 15 (10) | 146 | |
| 1 | 2 | 3 | ||||
| Grade | 20 (14) | 79 (52) | 50 (33) | 2 (1) | 149 | |
| Ductal | Lobular | |||||
| Tumour type | 119 (79) | 24 (16) | 8 (5) | 143 | ||
| Positive | Negative | |||||
| Lymph node status | 58 (38) | 78 (52) | 15 (10) | 146 | ||
| Oestrogen receptor status | 112 (74) | 39 (26) | 0 | 151 | ||
| Min | Median | Mean | Max | |||
| Age at diagnosis | 29 | 63 | 62.93 | 94 | 0 | 151 |
| NPI | 2.1 | 4.3 | 4.34 | 8 | 16 (11) | 135 |
NPI, Nottingham Prognostic Index.
*Numbers in parentheses are percentages.
Tissue microarray construction
Tissue cores for the construction of tissue microarrays (TMAs) were taken with a 3 mm skin punch biopsy needle (Stiefel Laboratories, Buckinghamshire, UK) from the paraffin wax blocks that were used for histological diagnosis. The tissue cores were inserted in the holes of the rectangular carrier made of liver tissue, punched out by a 4 mm punch biopsy needle (Stiefel Laboratories). The carrier facilitated the smooth cutting of sections with minimal artefacts in transition from paraffin wax to tissue. Each carrier had a grid of 4×5 holes and, in addition to the breast tumour tissue samples, contained one core of normal breast tissue as a control. Four cores per sample, from multiple areas of the same tumour, were included in the TMA and embedded in different blocks at different positions on the grid for redundancy. After core insertion, the tissue was re‐embedded in paraffin wax. A small number of cores were damaged during TMA construction or subsequent methods and were labelled as non‐informative. On average, 83.4% of the cores, or 3.3 cores per specimen, were informative and were used in the analysis.
Immunohistochemistry
Antibodies used
Maspin Ab‐1 (Clone EAW24, IgG2A mouse monoclonal antibody, corresponding to the N terminal region of human maspin; Lab Neomarkers, Athens, Greece)
MAB171 (IgG2A mouse monoclonal antibody, immunogen CXCR4 transfectants; R&D Systems, Anti‐Sel, Athens, Greece)
ab9002 (IgG sheep polyclonal antibody, immunogen LDH‐5, purified from human placenta; Abcam, Cambridge, UK)
QBend/10 (IgG3 mouse monoclonal antibody, immunogen CD34 isolated from human placenta; Lab Neomarkers, Athens, Greece)
Immunohistochemistry was performed according to the indirect biotin–streptavidin–hyperoxidase method, as previously described.32 The antibody dilutions were 1:75 for oestrogen receptors, 1:100 for maspin, 1:600 for CXCR4, 1:200 for LDH‐5 and 1:20 for CD34. Negative controls were created by omitting the primary antibody. Myoepithelial cells served as maspin internal positive controls.13
For all antibodies, tumour cells were counted in the entire core area at ×430 magnification. Subsequently, the percentage of cells with positive staining versus the total number of cells was calculated. Nuclear or cytoplasmic staining of tumour cells for maspin was considered positive and recorded separately. Similarly, nuclear or cytoplasmic staining for LDH‐5 and cytoplasmic staining for CXCR4 were considered positive. Microvessel density was assessed as described previously.33
Tdt‐mediated DUTP nick labelling assay
Double‐strand DNA breaks were detected by Tdt‐mediated dUTP nick end labelling assay, as described previously.34
Statistical analysis
The R language for statistics V.2.2 was used.35 All tests for difference and correlation were non‐parametric (Wilcoxon, Spearman). Both the log‐rank test for Kaplan–Meier estimates of survival and Cox proportional hazards models were used in the analysis of survival. When necessary, the expression of study parameters was stratified into “low” and “high” groups, below and above the median, respectively. The Bonferroni–Holm correction was applied for multiple comparisons where appropriate, and results were considered significant when p<0.05.
Results
TMA analysis and data validation
To validate our TMA analysis, the expression of every parameter was compared between the different cores of each specimen by intraclass correlation (one‐way analysis of variance). The values obtained were 0.72, 0.88, 0.91, 0.92 and 0.98 for maspin, CXCR4, LDH‐5, microvessel density and apoptotic index, respectively, reflecting high reliability for all parameters examined. This high reliability could be attributed to the large diameter of cores in our study, compared with 0.6 mm2 cores in other studies.36 The mean value of each specimen's cores was used in the analysis.
Immunohistochemical analysis
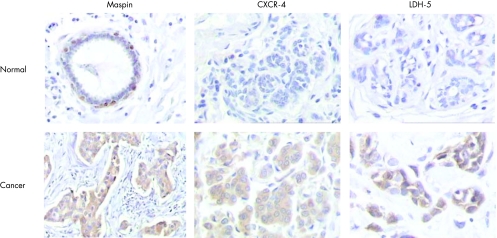
All samples displayed high cytoplasmic maspin expression with a median value of 92%. The standard deviation (SD 3.94) and other measures of variability (interquartile range 4.6) were very low and small differences in expression were reliably considered significant despite the abundance of immunohistochemical staining. Nuclear staining was also observed in 36 (26%) cases. Myoepithelial cells showed cytoplasmic and occasionally nuclear staining, whereas normal epithelial cells were not stained. Cytoplasmic and occasional membrane staining for CXCR4 was found in 133 (88%) tumours, whereas normal epithelium displayed insignificant focal staining. Cytoplasmic staining of LDH‐5, a marker of anaerobic metabolism, was observed in 146 (97%) specimens, whereas nuclear staining was limited to 32 (22%) specimens. Adjacent normal breast tissue showed weak LDH‐5 staining (fig 1 and table 2).
Figure 1 Immunohistochemistry for maspin, CXCR4 and LDH‐5. Sections of normal breast epithelium (original magnification ×100). Adjacent breast carcinoma sections (original magnification ×100 for maspin staining and ×200 for CXCR4 and LDH‐5 staining).
Table 2 Molecular characteristics of the tumours.
| Min | Median | Mean | Max | Missing* | n | |
|---|---|---|---|---|---|---|
| Maspin IHC (%)† | 70 | 92 | 91.1 | 97 | 3 (2) | 148 |
| CXCR4 IHC (%) | 0 | 53 | 47.9 | 91 | 0 (0) | 151 |
| LDH5 IHC (%)† | 0 | 63 | 58.4 | 100 | 0 (0) | 151 |
| AI | 0.28 | 0.85 | 1.26 | 4.2 | 6 (4) | 145 |
| MVD | 5.5 | 21.8 | 22.8 | 57.5 | 4 (3) | 147 |
| Present n* | Absent n* | Missing* | n | |||
| Nuclear maspin expression | 36 (24) | 102 (68) | 13 (8) | 138 | ||
| Nuclear LDH‐5 expression | 32 (21) | 116 (77) | 3 (2) | 148 |
AI, apoptotic index; MVD, microvessel density.
*Numbers in parentheses are percentages.
†Refers to cytoplasmic staining.
Correlation between maspin, CXCR‐4 and clinicopathological factors
The immunohistochemical expression of CXCR4 correlated with both maspin (p<0.001, r = 0.34) and LDH5 expression (p<0.001, r = 0.31). The expression of all three molecules varied significantly between ductal and lobular carcinomas (p = 0.001 for maspin, p = 0.03 for CXCR4 and p = 0.015 for LDH‐5). Maspin and CXCR4 expression progressively increased in higher grade tumours (p = 0.006 for maspin and p = 0.005 for CXCR4).
Analysis of survival
Simple univariate models were used as a starting point (table 4). As expected, most known prognostic factors (stage, lymph node status, patient age and oestrogen receptor status) were found to significantly affect survival. The prognostic value of the Nottingham prognostic index approached significance with p = 0.059. Histological type and grade were not significant predictors of survival.
Table 4 Univariate analysis of survival.
| Cox model | ||
|---|---|---|
| p Value | Hazard ratio | |
| Stage | 0.026 | 3.3, 3.6 |
| Grade | 0.26 | — |
| Lymph node status | 0.039 | 2.3 |
| Oestrogen receptor status* | 0.029 | 0.382 |
| Lobular or ductal | 0.140 | — |
| Age | 0.009 | 1.04 |
| NPI | 0.059 | 1.32 |
| Maspin | 0.048 | 1.7 |
| CXCR4 | 0.085 | 0.98 |
| LDH5 | 0.013 | 0.98 |
| AI | 0.2 | — |
| MVD | 0.61 | — |
| Nuclear maspin | 0.52 | — |
| Nuclear LDH5 | 0.27 | — |
AI, apoptotic index; MVD, microvessel density; NPI, Nottingham prognostic index.
*Result refers to patients who did receive hormonal treatment.
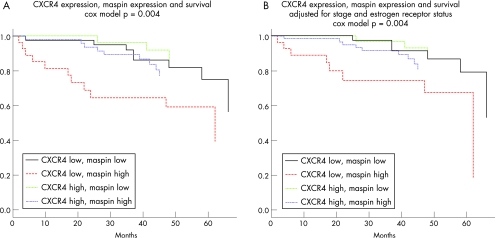
To further refine and explore our results, we proceeded with the construction of multivariate models of survival using the Cox proportional hazards method. After adjusting for disease stage or the Nottingham prognostic index, maspin expression is still associated with a worse prognosis (hazard ratio 1.2 or 1.17, p = 0.0098 or 0.02, respectively) and CXCR4 expression is associated with a small survival advantage that becomes significant (hazard ratio 0.98 or 0.98, p = 0.018 or 0.0094, respectively). Furthermore, the “high maspin, low CXCR4” group has worse survival than the “low maspin, high CXCR4” group, as is evident in fig 2A (model p = 0.004, maspin hazard ratio 1.17, p = 0.014, CXCR4 hazard ratio 0.98, p = 0.007) and fig 2B (model p = 0.004, maspin hazard ratio 1.19, p = 0.016, CXCR4 hazard ratio 0.98, p = 0.019). By contrast, LDH5 expression did not provide prognostic information when disease stage or lymph node status was included in the model.
Figure 2 Survival analysis. Maspin and CXCR4 coexpression patterns correlate to patient survival (A) even after adjustment for stage and oestrogen receptor status (B).
The division between lymph node negative and lymph node positive breast cancer is of profound clinical significance because lymph node positivity is assumed to indicate a high possibility of undetectable micrometastatic disease. In the group of patients with lymph node negative breast cancer (n = 75), maspin is associated with worse prognosis (hazard ratio 1.27, p = 0.038). In that group, 27% of patients with high maspin expression died during follow‐up, compared with only 14% of patients with low maspin expression. In the group of patients with lymph node positive breast cancer (n = 54), maspin has no prognostic value (p = 0.88). The exact opposite is true for CXCR4, whose expression is of no significance in patients with lymph node negative breast cancer (n = 75, p = 0.43) but does predict a small survival benefit in patients with lymph node positive breast cancer (n = 55, hazard ratio 0.978, p = 0.005).
The most powerful stepwise predictive model that we constructed included stage, oestrogen receptor status, maspin expression and CXCR4 expression. This model could not be improved with the inclusion of age, lymph node status, grade, histology, apoptosis, Nottingham prognostic index, LDH5 or maspin expression and should be considered conclusive for our dataset. In this model, maspin expression was again associated with a poorer prognosis (hazard ratio 1.19, p = 0.012) and CXCR4 expression was associated with a very small reduction in risk (hazard ratio 0.98, p = 0.016). Oestrogen receptor expression reduced risk by almost 60% (hazard ratio 0.44, p = 0.049).
Discussion
The abundant staining for maspin seems paradoxical at first, but has been reported elsewhere.12 One would normally expect the activity of a tumour suppressor protein like maspin to be down regulated in cancer. However, its staining does not have to diminish. The apparent overexpression of maspin can be attributed to the accumulation of dysfunctional isoforms, a phenomenon that occurs with mutant p53.37 Maspin can also form dimers and higher order polymers under certain conditions,38,39 but it is unclear whether this takes place in cancer cells and whether it explains the abundant staining.
Despite some studies to the contrary,9,14 our results associate maspin overexpression with adverse pathological characteristics. Similar results have also been published for colorectal cancer.40 The statistical trend between high maspin expression and lymph node positivity in our report cannot be considered conclusive, but it is consistent with the “aggressive” profile of maspin‐overexpressing tumours (table 3). Recent reports, in vitro and in mice,7,41,42 have focused on the proapoptotic abilities of maspin. Indeed, we observed higher apoptosis in tumours that overexpressed maspin, suggesting that it retains its proapoptotic ability in breast cancer. The increase of maspin expression in older patients has been reported in a recent study of normal skin samples and keratinocyte cell cultures,5 and it may reflect a senescent phenotype with protective paracrine antiangiogenic activity. Although maspin did not exert a significant effect on microvessel density in our study, possibly because of the antagonistic recruitment of other pathways by the tumour tissue microenvironment, we believe that the functional link between cellular senescence and maspin expression deserves further investigation.
Table 3 Correlation of molecular and clinicopathological variables with maspin and CXCR4 expression.
| Maspin | CXCR4 | |||||
|---|---|---|---|---|---|---|
| Median | Median | p Value | Median | Median | p Value | |
| Stage | 0.4 | 0.55 | ||||
| I vs II | 91.2 | 91.5 | 0.29 | 51.7 | 56.3 | 1 |
| II vs III | 91.5 | 92.1 | 0.96 | 56.3 | 25 | 1 |
| I vs III | 91.2 | 92.1 | 0.96 | 51.7 | 25 | 1 |
| Grade | 0.006 | 0.011 | ||||
| 1 vs 2 | 90.9 | 91.3 | 0.97 | 13.8 | 48.3 | 0.16 |
| 2 vs 3 | 91.3 | 93 | 0.01 | 48.3 | 65 | 0.08 |
| 1 vs 3 | 90.9 | 93 | 0.08 | 13.8 | 65 | 0.07 |
| Lobular | Ductal | Lobular | Ductal | |||
| Tumour type | 89.1 | 92.5 | 0.001 | 28.1 | 55 | 0.03 |
| Negative | Positive | Negative | Positive | |||
| Lymph node status | 91.5 | 93 | 0.06 | 50 | 67.7 | 0.15 |
| Oestrogen receptor status | 93 | 91.8 | 0.14 | 60 | 49.2 | 0.31 |
| Maspin low | Maspin high | CXCR4 low | CXCR4 high | |||
| Age (years) | 58 | 68 | 0.03 | 63.5 | 62 | 0.79 |
| NPI | 3.5 | 4.5 | 0.01 | 3.5 | 4.5 | 0.07 |
| Maspin IHC (%) | — | — | — | 90 | 93 | <0.001 |
| CXCR4 IHC (%) | 37.5 | 66.7 | <0.001 | — | — | — |
| LDH5 IHC (%) | 63.3 | 64.4 | 0.53 | 53.3 | 73.3 | <0.001 |
| AI | 0.8 | 1.2 | 0.049 | 0.9 | 0.9 | 0.56 |
| MVD | 22 | 21.4 | 0.81 | 22.8 | 21.4 | 0.55 |
AI, apoptotic index; IHC, immunohistochemistry; MVD, microvessel density; NPI, Nottingham prognostic index.
It is not surprising that maspin overexpression was associated with an increased hazard ratio and a shorter overall survival. Our results are in line with several studies in breast,10,11,43 lung,44 thyroid45 and other tumour types. The fact that this effect is prominent in patients with lymph node negative disease leads us to consider the possibility that maspin overexpression is associated with a high risk for clinically undetectable disease spread and subsequent recurrence. Maspin expression was not informative in patients with lymph node positive disease, but its levels were uniformly high in these cases, as discussed, and the division into “low” and “high” expression was far less pronounced within that group. The substantial increase of maspin expression in patients with lymph node positive disease (table 3) strengthens the assumption that maspin overexpression is associated with disease progression, but could weaken its discriminatory power in advanced stages. In that sense, the evaluation of maspin expression might prove to be primarily useful in the early stages of breast cancer.
The immunohistochemical staining of CXCR4 was detected in the cytoplasm and membrane of cancerous cells as expected for a membrane receptor. Others have reported the existence of nuclear CXCR4 staining19,21 that we could not verify. Like maspin expression, CXCR4 expression was associated with a higher tumour grade and a tendency towards higher NPI values and lymph node positivity. Similar findings have been reported previously.19,21,46
Generally, CXCR4 is believed to facilitate breast cancer metastasis18,20 by helping cells migrate to various target organs. However, we were unable to deduce any prognostic implications in patients with lymph node negativity who overexpressed CXCR4. Similarly, the survival of patients who had low levels of maspin and were therefore expected to be at a lower risk of death was not affected by the presence of high CXCR4. By contrast, high levels of CXCR4 were associated with a small but measurable survival advantage in patients who were lymph node positive or overexpressed maspin and were assumed to already harbour more advanced disease. The possibility that CXCR4 and maspin may have opposing functions should be considered, although such an antagonistic activity can only be reliably deduced from molecular studies. Alternatively, high CXCR4 expression in these patients could be a secondary effect of a protective reaction like lymphocyte infiltration and the corresponding up regulation of SDF‐1.22 Although we are reluctant to generalise this finding, others have reported a survival benefit in patients with familial chronic lymphocytic leukemia who express high CXCR447 and in patients with non‐small cell lung cancer48 who express nuclear CXCR4.
Take‐home messages
Both maspin and CXCR4 were overexpressed in breast cancer, in comparison with normal tissue.
Maspin and CXCR4 expression were related.
High maspin expression was associated with shorter survival, especially in the group of patients with lymph node negative disease.
The “high maspin, low CXCR4” patient group had worse prognosis than the “low maspin, high CXCR4” patient group.
The prognosis of patients with lymph node negative breast cancer varies considerably and the selection of an appropriate therapeutic regimen can be hard. We believe that there is now sufficient evidence to support a more extensive evaluation of maspin as a potentially sensitive prognostic marker in these patients. The prognostic value of CXCR4 in patients with lymph node negative disease was not significant in our study, but it may have to be assessed in a larger study. The unexpected finding of slightly longer survival in patients with lymph node positive disease with high levels of CXCR4 could be a special characteristic of their molecular profile and might fuel further investigations.
Abbreviations
SDF‐1 - stromal cell derived factor‐1
TMA - tissue microarray
Footnotes
Competing interests: None declared.
References
- 1.Ferlay J, Bray F, Pisane P.et al Globocan 2000: Cancer Incidence, Mortality and Prevalence Worldwide. IARC Press 2001
- 2.Zou Z, Anisowicz A, Hendrix M J.et al Maspin, a serpin with tumor‐suppressing activity in human mammary epithelial cells. Science 1994263526–529. [DOI] [PubMed] [Google Scholar]
- 3.Biliran H, Sheng S. Pleiotrophic inhibition of pericellular urokinase‐type plasminogen activator system by endogenous tumor suppressive maspin. Cancer Res 2001618676–8682. [PubMed] [Google Scholar]
- 4.Zhang M, Volpert O, Shi Y H.et al Maspin is an angiogenesis inhibitor. Nat Med 20006196–199. [DOI] [PubMed] [Google Scholar]
- 5.Nickoloff B J, Lingen M W, Chang B D.et al Tumor suppressor maspin is up‐regulated during keratinocyte senescence, exerting a paracrine antiangiogenic activity. Cancer Res 2004642956–2961. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, Yin S, Reddy N.et al Bax mediates the apoptosis‐sensitizing effect of maspin. Cancer Res 2004641703–1711. [DOI] [PubMed] [Google Scholar]
- 7.Zhang W, Shi H Y, Zhang M. Maspin overexpression modulates tumor cell apoptosis through the regulation of Bcl‐2 family proteins. BMC Cancer 2005550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sternlicht M D, Barsky S H. The myoepithelial defense: a host defense against cancer. Med Hypotheses 19974837–46. [DOI] [PubMed] [Google Scholar]
- 9.Maass N, Teffner M, Rosel F.et al Decline in the expression of the serine proteinase inhibitor maspin is associated with tumour progression in ductal carcinomas of the breast. J Pathol 2001195321–326. [DOI] [PubMed] [Google Scholar]
- 10.Umekita Y, Ohi Y, Sagara Y.et al Expression of maspin predicts poor prognosis in breast‐cancer patients. Int J Cancer 2002100452–455. [DOI] [PubMed] [Google Scholar]
- 11.Umekita Y, Yoshida H. Expression of maspin is up‐regulated during the progression of mammary ductal carcinoma. Histopathology 200342541–545. [DOI] [PubMed] [Google Scholar]
- 12.Mohsin S K, Zhang M, Clark G M.et al Maspin expression in invasive breast cancer: association with other prognostic factors. J Pathol 2003199432–435. [DOI] [PubMed] [Google Scholar]
- 13.Kim D H, Yoon D S, Dooley W C.et al Association of maspin expression with the high histological grade and lymphocyte‐rich stroma in early‐stage breast cancer. Histopathology 20034237–42. [DOI] [PubMed] [Google Scholar]
- 14.Hojo T, Akiyama Y, Nagasaki K.et al Association of maspin expression with the malignancy grade and tumor vascularization in breast cancer tissues. Cancer Lett 2001171103–110. [DOI] [PubMed] [Google Scholar]
- 15.Ma Q, Jones D, Borghesani P R.et al Impaired B‐lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in CXCR4‐ and SDF‐1‐deficient mice. Proc Natl Acad Sci USA 1998959448–9453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ameisen J C. HIV. Setting death in motion. Nature 1998395117–119. [DOI] [PubMed] [Google Scholar]
- 17.Berson J F, Long D, Doranz B J.et al A seven‐transmembrane domain receptor involved in fusion and entry of T‐cell‐tropic human immunodeficiency virus type 1 strains. J Virol 1996706288–6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muller A, Homey B, Soto H.et al Involvement of chemokine receptors in breast cancer metastasis. Nature 200141050–56. [DOI] [PubMed] [Google Scholar]
- 19.Kato M, Kitayama J, Kazama S.et al Expression pattern of CXC chemokine receptor‐4 is correlated with lymph node metastasis in human invasive ductal carcinoma. Breast Cancer Res 20035R144–R150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y M, Pan Y, Wei Y.et al Upregulation of CXCR4 is essential for HER2‐mediated tumor metastasis. Cancer Cell 20046459–469. [DOI] [PubMed] [Google Scholar]
- 21.Cabioglu N, Yazici M S, Arun B.et al CCR7 and CXCR4 as novel biomarkers predicting axillary lymph node metastasis in T1 breast cancer. Clin Cancer Res 2005115686–5693. [DOI] [PubMed] [Google Scholar]
- 22.Bleul C C, Fuhlbrigge R C, Casasnovas J M.et al A highly efficacious lymphocyte chemoattractant, stromal cell‐derived factor 1 (SDF‐1). J Exp Med 19961841101–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bachelder R E, Wendt M A, Mercurio A M. Vascular endothelial growth factor promotes breast carcinoma invasion in an autocrine manner by regulating the chemokine receptor CXCR4. Cancer Res 2002627203–7206. [PubMed] [Google Scholar]
- 24.Darash‐Yahana M, Pikarsky E, Abramovitch R.et al Role of high expression levels of CXCR4 in tumor growth, vascularization, and metastasis. FASEB J 2004181240–1242. [DOI] [PubMed] [Google Scholar]
- 25.Fernandis A Z, Prasad A, Band H.et al Regulation of CXCR4‐mediated chemotaxis and chemoinvasion of breast cancer cells. Oncogene 200423157–167. [DOI] [PubMed] [Google Scholar]
- 26.Datta S R, Brunet A, Greenberg M E. Cellular survival: a play in three Akts. Genes Dev 1999132905–2927. [DOI] [PubMed] [Google Scholar]
- 27.Helbig G, Christopherson K W, Bhat‐Nakshatri P.et al NF‐kappaB promotes breast cancer cell migration and metastasis by inducing the expression of the chemokine receptor CXCR4. J Biol Chem 200327821631–21638. [DOI] [PubMed] [Google Scholar]
- 28.Elston C W, Ellis I O. Pathological prognostic factors in breast cancer. The value of histological grades in breast cancer. Experience from a large study with long term follow‐up. Histopathology 199119403–410. [DOI] [PubMed] [Google Scholar]
- 29.Galea M H, Blamey R W, Elston C E.et al The Nottingham Prognostic Index in primary breast cancer. Breast Cancer Res Treat 199222207–219. [DOI] [PubMed] [Google Scholar]
- 30. NHSBSP Publication No 58: Pathology reporting of Breast Disease. Sheffield: NHS Breast Screening Programmes, 2005
- 31.Leake R, Barnes D, Pinder S.et al Immunohistochemical detection of steroid receptors in breast cancer: a working protocol. UK Receptor Group, UK NEQAS, The Scottish Breast Cancer Pathology Group, and The Receptor and Biomarker Study Group of the EORTC. J Clin Pathol 200053634–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mariatos G, Gorgoulis V G, Zacharatos P.et al Expression of p16(INK4A) and alterations of the 9p21–23 chromosome region in non‐small‐cell lung carcinomas: relationship with tumor growth parameters and ploidy status. Int J Cancer 200089133–141. [DOI] [PubMed] [Google Scholar]
- 33.Tsoli E, Zacharatos P, Dasiou‐Plakida D.et al Growth index is independent of microvessel density in non‐small‐cell lung carcinomas. Hum Pathol 200233812–818. [DOI] [PubMed] [Google Scholar]
- 34.Gavrieli Y, Sherman Y, Ben‐Sasson S A. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol 1992119493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Team R D C. R: A language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria; 2005. ISBN 3‐900051‐07‐0
- 36.Simon R, Sauter G. Tissue microarray (TMA) applications: implications for molecular medicine. Expert Rev Mol Med 200320031–12. [DOI] [PubMed] [Google Scholar]
- 37.Bartek J, Bartkova J, Vojtesek B.et al Aberrant expression of the p53 oncoprotein is a common feature of a wide spectrum of human malignancies. Oncogene 199161699–1703. [PubMed] [Google Scholar]
- 38.Pemberton P A, Wong D T, Gibson H L.et al The tumor suppressor maspin does not undergo the stressed to relaxed transition or inhibit trypsin‐like serine proteases. Evidence that maspin is not a protease inhibitory serpin. J Biol Chem 199527015832–15837. [DOI] [PubMed] [Google Scholar]
- 39.Silverman G A, Bird P I, Carrell R W.et al The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J Biol Chem 200127633293–33296. [DOI] [PubMed] [Google Scholar]
- 40.Bettstetter M, Woenckhaus M, Wild P J.et al Elevated nuclear maspin expression is associated with microsatellite instability and high tumour grade in colorectal cancer. J Pathol 2005205606–614. [DOI] [PubMed] [Google Scholar]
- 41.Jiang N, Meng Y, Zhang S.et al Maspin sensitizes breast carcinoma cells to induced apoptosis. Oncogene 2002214089–4098. [DOI] [PubMed] [Google Scholar]
- 42.Latha K, Zhang W, Cella N.et al Maspin mediates increased tumor cell apoptosis upon induction of the mitochondrial permeability transition. Mol Cell Biol 2005251737–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bieche I, Girault I, Sabourin J C.et al Prognostic value of maspin mRNA expression in ER alpha‐positive postmenopausal breast carcinomas. Br J Cancer 200388863–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hirai K, Koizumi K, Haraguchi S.et al Prognostic significance of the tumor suppressor gene maspin in non‐small cell lung cancer. Ann Thorac Surg 200579248–253. [DOI] [PubMed] [Google Scholar]
- 45.Ito Y, Yoshida H, Tomoda C.et al Maspin expression is directly associated with biological aggressiveness of thyroid carcinoma. Thyroid 20041413–18. [DOI] [PubMed] [Google Scholar]
- 46.Su Y C, Wu M T, Huang C J.et al Expression of CXCR4 is associated with axillary lymph node status in patients with early breast cancer. Breast 2005 [DOI] [PubMed]
- 47.Ishibe N, Albitar M, Jilani I B.et al CXCR4 expression is associated with survival in familial chronic lymphocytic leukemia, but CD38 expression is not. Blood 20021001100–1101. [DOI] [PubMed] [Google Scholar]
- 48.Spano J P, Andre F, Morat L.et al Chemokine receptor CXCR4 and early‐stage non‐small cell lung cancer: pattern of expression and correlation with outcome. Ann Oncol 200415613–617. [DOI] [PubMed] [Google Scholar]