Abstract
Background
The use of tissue microarrays (TMAs) is now a generally accepted method for the investigation of solid tumours. However, little is known about the applicability of the TMA technique for analysis of patients with acute leukaemia. A bone marrow (BM)‐TMA analysis with 15 different immunohistochemical markers was performed. The TMA was validated by comparison with the corresponding full tissue sections.
Materials and methods
A BM‐TMA comprising 148 cases of acute leukaemia, including 115 acute myeloid leukaemia (AML) and 33 acute lymphoblastic leukaemia (ALL) cases, was constructed. Expression of CD3, CD10, CD15, CD20, CD34, CD61, CD68, CD79a, CD99, CD117, CD138, myeloperoxidase, haemoglobin A1, glycophorin and terminal deoxynucleotidyl transferase was immunohistochemically analysed. 50 cases of the TMA were directly compared with the corresponding full tissue section to validate the results.
Results
Morphologically and immunohistochemically, 6 (4%) of 148 cases and 765 (11%) cores of 6912 individual analyses were not evaluable. A direct comparison of TMA cases with conventional full sections showed a concordance of the results of 100%.
Conclusions
The small size of bone‐marrow biopsies and the presence of bony trabeculae do not preclude construction and analysis of acute leukaemia TMAs. Acute leukaemia cases on TMA displayed the characteristic phenotypic profiles expected in different AML and ALL subtypes. Therefore, the TMA technique is also a promising method for high‐throughput analysis of combined marker expression and clinicopathological correlations in patients with leukaemia.
The World Health Organization classification of acute leukaemias includes morphological, immunohistochemical, cytogenetic and molecular features.1 The spectrum of classical molecular diagnostic methods has recently been extended by the use of gene expression profiling (GEP). GEP of patients with acute myeloid leukaemia (AML) has shown differentially expressed genes in various leukaemia cases, thereby allowing delineation of new subtypes with particular biological features and clinical outcome.2,3 Similarly, in patients with acute lymphoblastic leukaemia (ALL), it has been possible to identify clinically relevant subtypes by the use of GEP.2,4 On the one hand, these data corroborate the existing acute leukaemia classification system; on the other hand, they prompt to search for new markers at the protein level that might be of diagnostic and/or prognostic relevance for the individual patient.
The tissue microarray (TMA) technology allows the simultaneous analysis of a large number of tumours to investigate the genetic changes and protein expression in a single experiment under highly standardised conditions.5,6 Advances in genomics and proteomics are increasing the need to evaluate large numbers of molecular targets with regard to their prognostic and predictive value in clinical oncology.7 Originally, TMAs were used in the research of solid tumours. For these tumours, especially carcinomas, this technique is now a recognised and established method. Recently, the TMA approach was also applied successfully to fluorescent in‐situ hybridisation and immunohistochemical studies of lymphomas.8,9,10 The combination of GEP using cDNA arrays with TMA technique in lymphoma research is promising because new possible surrogate markers or combinations thereof can rapidly be evaluated for their use in daily routine diagnostics. This has been shown in various studies for the immunohistochemical evaluation of diffuse large B cell lymphomas confirming the molecular classification.11,12,13,14
Up to now, very little information is available about the applicability of the TMA technique for the analysis of bone‐marrow biopsies, because of the specific difficulties in constructing arrays generated from small tissue cylinders, including bone structures. Only two very recent studies describe the use of bone marrow (BM)‐TMA, albeit with small case numbers.15,16 Therefore, the aim of the present study was to assess the feasibility of BM‐TMA construction. A TMA consisting of 148 acute leukaemia cases was constructed, and the quality of morphology and immunohistochemistry of 15 markers was investigated. Additionally, we validated the methodology by directly comparing a panel of immunohistochemical marker expression on the tissue cores of the TMA with the corresponding conventional bone‐marrow biopsies.
Materials and methods
Patients and specimens
Bone‐marrow biopsies of 148 patients were used in this study. Patient characteristics showed a male:female ratio of 1.69:1 and a mean (range) age of 49.7 (1–93) years. Of the 107 patients for whom further data were available, a de novo acute leukaemia was diagnosed in 68 patients and secondary acute leukaemia in 38 patients. One additional AML case was a patient in relapse. The bone‐marrow specimens comprised 115 AML cases subclassified according to the French–American–British (FAB) classification (10 M0, 12 M1, 30 M2, 5 M3, 29 M4, 15 M5, 1 M6, 2 M7 and 11 not further specified) and 33 ALL cases (23 B‐ALL and 5 T‐ALL, 5 not classified; table 1). The FAB classification was based on a combination of cytological, cytochemical, histological, immunohistochemical and other relevant factors.
Table 1 Detailed summary of the results: immunophenotyping on an acute leukaemia tissue microarray.
| Diagnosis | Total | CD3 | CD10 | CD15 | CD20 | CD34 | CD61 | CD79a | CD99 | CD117 | CD138 | MPO | HbA1 | Glyc | TdT | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | ||
| AML M0 | 10 | 0 | 0 | 2 | 20 | 0 | 5 | 50 | 0 | 0 | 4 | 40 | 5 | 50 | 0 | 5 | 50 | 0 | 0 | 3 | 30 | |||||||||
| AML M1 | 12 | 0 | 0 | 5 | 42 | 0 | 7 | 58 | 0 | 1 | 8 | 2 | 17 | 7 | 58 | 0 | 11 | 92 | 0 | 0 | 0 | |||||||||
| AML M2 | 30 | 0 | 0 | 10 | 33 | 0 | 12 | 40 | 0 | 0 | 9 | 30 | 13 | 43 | 0 | 28 | 93 | 0 | 0 | 2 | 7 | |||||||||
| AML M3 | 5 | 0 | 0 | 1 | 20 | 0 | 1 | 20 | 0 | 0 | 4 | 80 | 4 | 80 | 0 | 4 | 100 | 0 | 0 | 0 | ||||||||||
| AML M4 | 25 | 0 | 1 | 4 | 9 | 36 | 0 | 9 | 36 | 0 | 0 | 2 | 8 | 7 | 28 | 0 | 16 | 64 | 0 | 0 | 3 | 12 | ||||||||
| AML M4eo | 4 | 0 | 0 | 2 | 50 | 0 | 1 | 25 | 0 | 0 | 0 | 2 | 50 | 0 | 4 | 100 | 0 | 0 | 0 | |||||||||||
| AML M5 | 15 | 0 | 0 | 4 | 27 | 0 | 1 | 7 | 0 | 0 | 2 | 13 | 2 | 13 | 0 | 5 | 33 | 0 | 0 | 0 | ||||||||||
| AML M6 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||||||||||||
| AML M7 | 2 | 0 | 0 | 1 | 50 | 0 | 0 | 2 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| AML nc | 11 | 0 | 1 | 17 | 2 | 33 | 0 | 5 | 45 | 0 | 0 | 4 | 40 | 3 | 50 | 0 | 4 | 57 | 0 | 0 | 1 | 17 | ||||||||
| B‐ALL | 23 | 1 | 4 | 16 | 70 | 2 | 9 | 7 | 30 | 12 | 52 | 0 | 9 | 41 | 4 | 19 | 1 | 4 | 0 | 4 | 17 | 0 | 0 | 18 | 78 | |||||
| T‐ALL | 5 | 2 | 40 | 1 | 20 | 0 | 0 | 2 | 40 | 0 | 1 | 20 | 4 | 80 | 0 | 0 | 0 | 0 | 0 | 3 | 60 | |||||||||
| ALL nc | 5 | 0 | 2 | 40 | 1 | 20 | 0 | 3 | 60 | 0 | 1 | 20 | 1 | 20 | 1 | 20 | 0 | 1 | 20 | 0 | 0 | 3 | 60 | |||||||
ALL, acute lymphoblastic leukaemia; AML, acute myeloid leukaemia; glyc, glycophorin; HbA1, haemoglobin A1; MPO, myeloperoxidase; nc, not classified; TdT, terminal deoxynucleotidyl transferase.
These cases were obtained from the surgical pathology files of the Institute of Pathology, University Hospital of Basel, Basel, Switzerland (n = 76), and the Institute of Haematology and Clinical Oncology “L. & A. Seràgnoli”, University of Bologna, Bologna, Italy (n = 72), between 1990 and 2002. The material was either Susa‐fixed or “B5‐fixed” and paraffin‐embedded. Susa‐fixative solution is made of Hg2Cl2, sodium chloride, distilled water, trichloroacetic acid, glacial acetic acid and formaldehyde (37%). B5 solution consists of mercuric chloride, sodium acetate and formalin. Other fixatives have not been tested.
For routine diagnostic purposes, these cases had been extensively analysed by immunohistochemistry and each case has been reviewed by at least two experienced haematopathologists (SD, SAP, PW) in combination with the involved haematologist.
Construction of tissue microarrays
For TMA construction, a haematoxylin and eosin‐stained (H&E) slide from each block was used to define the representative regions displaying leukaemic infiltrates. Tissue cylinders (cores) of 0.6 mm were punched from the previously defined areas and brought into a recipient paraffin block using a precision instrument (Beecher Instruments, Silver Spring, Maryland, USA), as reported previously.6 To increase the number of evaluable cases, all recipient blocks contained at least two donor tissue cylinders. Individually, 60 cases were punched twice and 88 cases were punched three‐times. Altogether, 384 cores of 148 different acute leukaemias were brought into an array format (table 2).
Table 2 Technical data of the bone marrow tissue microarray of acute leukaemias.
| n | % | |
|---|---|---|
| Number of acute leukaemia cases | 148 | |
| Number of punches (cores) | 348 | |
| Morphological analysis (H&E): | ||
| Punches missing tissue | 28/348 | 7 |
| Punches ⩽20% tumour cells | 14/384 | 4 |
| Number of cases not evaluable | 6/148 | 4 |
| Immunohistochemical analysis: | ||
| Number of punches | 5760 | 5 |
| Punches missing tissue | 286/5760 | 8 |
| Punches ⩽20% tumour cells | 437/5760 | 13 |
| Punches not evaluable | 723 | |
| Summary (H&E and IHC): | ||
| Number of punches | 6912 | 11 |
| Punches not evaluable | 765/6912 |
H&E, haematoxylin and eosin; IHC, immunohistochemistry.
Morphological analysis
Serial sections, 4‐μm thick, of the TMA blocks were transferred to an adhesive‐coated glass slide system (Instrumedics Inc. Hackensack, New Jersey, USA), dried overnight and stained with H&E, Giemsa and periodic acid Schiff (PAS). Only cores containing a minimum of 20% tumour tissue were included in the study.
Immunohistochemistry
Table 3 lists the primary antibodies against CD3, CD10, CD15, CD20, CD34, CD61, CD68, CD79a, CD99, CD117, CD138, myeloperoxidase (MPO), haemoglobin A1, glycophorin and terminal deoxynucleotidyl transferase (TdT), their dilutions and pretreatment conditions.
Table 3 Antibodies and antigen‐retrieval techniques applied.
| Antibody | Source | Dilution | Retrieval |
|---|---|---|---|
| CD3 | DAKO | 1:100 | MW, 800 W, 100°C/30 min, EDTA |
| CD10 | Novocastra | 1:80 | MW, 800 W, 100°C/30 min, EDTA |
| CD15 | DAKO | 1:500 | MW, 800 W, 100°C/15 min, citrate |
| CD20 | DAKO | 1:2000 | MW, 800 W, 100°C/30 min, EDTA |
| CD34 | DAKO | 1:20 | MW, 800 W, 100°C/30 min, EDTA |
| CD61 | NeoMarkers | 1:100 | MW, 800 W, 100°C/15 min, EDTA |
| CD68 (PG‐M1) | DAKO | 1:2000 | None |
| CD79a | NeoMarkers | 1:200 | Pressure cooker, 121°C, 3 min, EDTA |
| CD99 | NeoMarkers | 1:50 | None |
| CD117 | DAKO | 1:250 | MW, 800 W, 100°C/30 min, EDTA |
| CD138 | DAKO | 1:50 | MW, 800 W, 100°C/15 min, EDTA |
| Glycophorin A | DAKO | 1:500 | None |
| HbA1 | DAKO | 1:10000 | None |
| MPO | DAKO | 1:20000 | None |
| TdT | Novocastra | 1:250 | Pressure cooker, 121°C, 3 min, EDTA |
HbA1, haemoglobin A1; MPO, myeloperoxidase; MW, microwave; TdT, terminal deoxynucleotidyl transferase.
Bound secondary antibodies were visualised by standard avidin‐biotin‐peroxidase and alkaline phosphatase/anti‐alkaline phosphatase techniques using diaminobenzidine and fast red TR as chromogens, respectively. A case was considered positive if ⩾20% of tumour cells (blasts) displayed a specific staining pattern. For positive controls, appropriate tissues (normal bone marrow and tonsils) were used. In the negative control experiments, the primary antibodies were omitted.
For direct comparison of TMA cases with corresponding large sections, 50 cases were randomly chosen from the surgical pathology files of the Institute of Pathology. Of these, 42 cases could be re‐evaluated. They consisted of two AML M0, six AML M2, one AML M3, 12 AML M4, two AML M5, two AML not classified, eight B‐ALL, five T‐ALL and four ALL not classified.
Results
Technical aspects and morphology
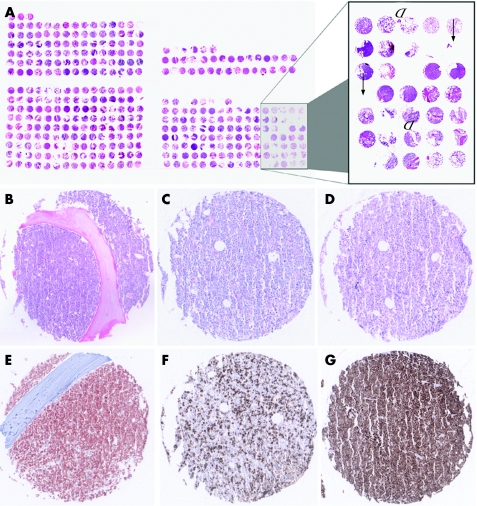
The construction of the BM‐TMA was hampered as expected by the presence of the bony trabeculae in the bone‐marrow biopsies. This problem was tackled, firstly, by carefully marking representative areas (ie, rich in leukaemic blasts) in the bone‐marrow biopsies selected for TMA construction and, secondly, by punching these selected areas exactly. Thus, the punched bone‐marrow tissue only very rarely turned out to be non‐representative due to necrotic or normal non‐neoplastic bone‐marrow tissue (<5%). Also, punches containing no tissue at all (“empty spots”) lead to a loss of evaluable cases (<5%) in morphological analysis (fig 1A) similar to that seen in TMAs without bone structures.
Figure 1 (A) Technical problems of bone marrow tissue microarray (TMA) procedure. Arrows point to punched tissue cores with reduced tissue area due to a loss of tissue during construction of the acute leukaemia TMA. Arrow heads point to diminished tissue area due to bony trabecula. The trabecula was partially or totally torn away by manufacturing the TMA sections highlighting arising problems in TMA cutting procedure. (B–G) Morphology and examples of immunohistochemistry on acute leukaemia TMA. (B) B‐acute lymphoblastic leukaemia (ALL), haematoxylin and eosin staining, ×100. (C) Acute myeloid leukaemia (AML), M4, periodic acid Shiff base staining, ×100. (D) AML M4, Giemsa staining, ×100. (E) B‐ALL, CD34 immunohistochemistry, ×100. (F) AML, M4, myeloperoxidase immunohistochemistry, ×100. (G) B‐ALL, terminal deoxynucleotidyl transferase immunohistochemistry, ×100. Overall, excellent staining quality was achieved in all investigated cases.
In 148 cases of acute leukaemia, a total of 384 punches were performed. Taken together all TMA slides analysed in this study, a total number of 6912 single‐tissue cores were morphologically and immunohistochemically investigated. Among the total number of 384 punches in H&E morphology, analysis failure of 42 (11%) punches was linked to the array technology; among 14 (4%) punches, tissue in at least one core was missing (“no tissue”); among 28 (7%) punches, the cores contained no tumour cells (table 2, fig 1A). Overall, morphology seen by H&E, PAS and Giemsa stainings displayed an excellent quality (fig 1B–D).
Of 148 acute leukaemia cases included, 142 (96%) were representative by H&E morphology (table 2). Only 6 (4%) cases were non‐informative because of missing tissue or a lack of tumour cells in all arrayed tissue cores per punched case. They consisted of five AML and one B‐ALL.
Immunohistochemistry
In all, 5760 tissue cores were immunohistochemically analysed. Of them, 723 (13%) cores were not evaluable (table 2). A case was diagnosed to be positive if ⩾20% of tumour cells (blasts) displayed the specific staining pattern. Table 1 lists the immunophenotypic markers of the acute leukaemia cases. Immunohistochemical staining displayed a moderate or even strong colour pattern in all tissue cylinders. All CD markers (ie, CD3, CD10, CD15, CD20, CD34, CD61, CD68, CD79a, CD99, CD117 and CD138) showed the expected membranous immunoreactivity (fig 1E–G). TdT immunoreactivity showed a strong and uniform nuclear staining. MPO positivity in AML cases displayed the specific intracytoplasmatic location.
For direct validation with the conventional bone‐marrow biopsies, CD3, CD10, CD20, CD34, CD68, CD117 and TdT immunostains were compared. On TMA, 3 (6%) cases were positive for CD3, 18 (36%) cases for CD10, 5 (10%) cases for CD20, 27 (54%) cases for CD34, 9 (18%) cases for CD68, 5 (10%) cases for CD117 and 24 (48%) cases for TdT. The direct comparison with the corresponding large tissue section displayed a 100% concordance in each of the seven evaluated markers.
Discussion
In this study, we report the construction and validation of a BM‐TMA used for combined immunophenotypic profiling of a large series of acute leukaemia cases. The arrays contained 384 cores of 148 acute leukaemia samples encompassing 115 AML and 33 ALL cases of all subtypes according to the FAB classification.
Technical problems in constructing the BM‐TMA were caused by the heterogeneity of the samples used. First of all, punched bony trabeculae, necrotic and normal bone‐marrow tissue reduced the area of evaluable tumour tissue. Also, these fragments of bony material within punches increased the difficulties in manufacturing the TMA sections for further analysis. Finally, punches containing no tissue at all (empty spots) led to a further loss of evaluable cases in morphological analysis. The empty spots are mostly due to the different core length in the recipient block leading to non‐informative spots in deeper sections. Interestingly, in that context, very recently Obermann et al15 reported the construction of a small BM‐TMA (75 cases) using the “stack method”. Thereby, the different core lengths of individual biopsies can be avoided. Nevertheless, only 6 (4%) cases were lost for morphological evaluation in our series of 148 cases, and these results are even better than our observations in a large series of Hodgkin's lymphoma and non‐Hodgkin's lymphoma TMAs.9,10,12,17,18,19 These results are also in line with another BM‐TMA comprising 64 ALL cases.16
The results of our immunohistochemical expression analysis of the various markers are in agreement with the frequency and distribution as reported in the literature and as expected specifically in these cases, extrapolating the results from the diagnostic reports. This further shows the feasibility of the technique used. About 100–150 good‐quality sections of a bone‐marrow trephine can be cut, allowing the analysis of a large number of different markers under highly standardised conditions. The tumour mass analysed is extremely diminished due to the square area of 0.6 mm for each punched tissue core. However, in large studies of various and often heterogeneous neoplasms, these small tissue areas have already proved to be highly representative in direct comparison with the corresponding conventional large tissue sections.19,20,21,22 Previously, Hedvat et al20 stated that by using four core samples from each case the agreement between lymphoma TMA and large tissue sections is 97–100%. For comparison, in a classical Hodgkin's lymphoma (cHL) TMA study, we could find a correlation of 94–100% between large tissue section and the TMA by using only two core samples per case.9,10 We observed a concordance of 100% between the TMA and conventional large sections for the expression of LMP‐1––a marker that is homogenously and consistently expressed in all Hodgkin/Reed–Sternberg cells of EBV‐associated cHL.9 Moreover, a direct validation in cHL showed that the inconsistently expressed CD20 on the TMA matched in 94% (63/67) of cases with the corresponding conventional full tissue sections.10 In this acute leukaemia series, immunohistochemical information obtained from the minute arrayed samples was 100% concordant with the corresponding full section. These results show that intratumour heterogeneity does not affect the ability of the TMA technology to be representative and reliable in acute leukaemia. Thus, TMA technology provides an ideal tool for high‐throughput analysis even for acute leukaemia in bone‐marrow biopsies. For this pilot study of the suitability of a BM‐TMA in acute leukaemia, however, we selected morphologically clear‐cut cases with an extensive and diffuse bone‐marrow involvement by leukaemic blasts. Cases with a variable percentage of tumour cells or a less homogeneous pathology might cause greater technical problems and might potentially reduce the high degree of congruity between TMA and full tissue immunohistochemistry. Nonetheless, similar to other heterogeneous tumours, careful selection of the area to be punched and construction of a TMA with duplicate/triplicate course will in all likelihood reduce this problem.
Normally, acute leukaemia is diagnosed by integrating clinical data with morphological, immunophenotypic and cytogenetic data using a bone‐marrow aspirate and/or biopsy. Fresh material of bone‐marrow aspirates and/or blood samples can be used for further molecular studies (eg, GEP). Using blotting techniques, the translation of GEP results at the protein level is time and material consuming. The use of the TMA technique accelerates combined protein profiling and at the same time spares the material available. Moreover, even years after the bone‐marrow biopsy has been performed, new markers can be examined successfully on TMA sections containing large series of tumour tissues, which makes the TMA technique a powerful tool in the evaluation of archive material. Unlike the situation with bone‐marrow aspirates, simultaneous in‐situ analysis of many cases is possible. Similar to lymphomas and solid tumours, this tool may serve to identify rapidly the biological or clinical significance of molecular changes in acute leukaemia. One such example is the evaluation of CD117 expression and mutational status in a large series of solid tumours and lymphomas to clarify the question of possible new therapeutic targets for imatinib.18,23
Nonetheless, it is evident that the range of demonstrable genetic and phenotypic targets in fixed tissues is limited. Only a prospective collection of cases in a TMA format along with concurrent accrual of cytogenetic and molecular genetic data from fresh blast cells is the ideal way to draw meaningful genetic/phenotypic conclusions and to identify potential prognostic and/or predictive markers.
Another limitation is the fact that immunophenotyping of trephine sections is restricted compared with the immunofluorescent phenotyping by flow cytometry using blood or aspirated marrow cells. But both techniques have their specific advantages and disadvantages. Although flow cytometry offers the possibility of a multiple colour analysis and a virtual selection and gating, the assessment of the topographical distribution and a correlation of the immunophenotype with the morphology of the cells is possible only in paraffin‐embedded specimens. Moreover, in the event of a dry tap, the diagnosis might rely entirely on histopathological and immunohistochemical examination.
In summary, in this study we were able to show that retrospective protein profiling of acute leukaemia bone‐marrow biopsies is possible. The TMA method is able to simplify immunophenotypic and cytogenetic assessment in a larger cohort of tumours, and thus serve as a powerful tool to assess protein expression profiles in combination with gene expression profiles.24
Acknowledgements
We thank H Weisskopf, T Schürch and J Schwegler for their excellent technical assistance and A Tzankov for editorial help.
Abbreviations
ALL - acute lymphoblastic leukaemia
AML - acute myeloid leukaemia
BM‐TMA - bone marrow tissue microarray
cHL - classical Hodgkin's lymphoma
FAB - French–American–British
GEP - gene expression profiling
H&E - haematoxylin and eosin
MPO - myeloperoxidase
TdT - terminal deoxynucleotidyl transferase
TMA - tissue microarray
Footnotes
Funding: This study was partly supported by a grant from AIRC (Milan).
Competing interests: None.
References
- 1.Brunning R D, Matutes E, Harris N L.et al Acute myeloid leukemias. Precursor B‐cell and T‐cell neoplasms. In: Jaffe ES, Harris NL, Stein H, Vardiman J, eds, World Health Organization classification of tumours, pathology and genetics of tumours of heamatopoietic and lymphoid tissues. Lyon: IARC Press 200175–118.
- 2.Kohlmann A, Schoch C, Schnittger S.et al Molecular characterization of acute leukemias by use of microarray technology. Genes Chromosomes Cancer 200337396–405. [DOI] [PubMed] [Google Scholar]
- 3.Valk P J, Verhaak R G, Beijen M A.et al Prognostically useful gene‐expression profiles in acute myeloid leukemia. N Engl J Med 20043501617–1628. [DOI] [PubMed] [Google Scholar]
- 4.Yeoh E J, Ross M E, Shurtleff S A.et al Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell 20021133–143. [DOI] [PubMed] [Google Scholar]
- 5.Bubendorf L, Kononen J, Koivisto P.et al Survey of gene amplifications during prostate cancer progression by high‐throughout fluorescence in situ hybridization on tissue microarrays. Cancer Res 199959803–806. [PubMed] [Google Scholar]
- 6.Kononen J, Bubendorf L, Kallioniemi A.et al Tissue microarrays for high‐throughput molecular profiling of tumor specimens. Nat Med 19984844–847. [DOI] [PubMed] [Google Scholar]
- 7.Torhorst J, Bucher C, Kononen J.et al Tissue microarrays for rapid linking of molecular changes to clinical endpoints. Am J Pathol 20011592249–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia J F, Camacho F I, Morente M.et al Hodgkin and Reed‐Sternberg cells harbor alterations in the major tumor suppressor pathways and cell‐cycle checkpoints: analyses using tissue microarrays. Blood 2003101681–689. [DOI] [PubMed] [Google Scholar]
- 9.Tzankov A, Zimpfer A, Lugli A.et al High‐throughput tissue microarray analysis of G1‐cyclin alterations in classical Hodgkin's lymphoma indicates overexpression of cyclin E1. J Pathol 2003199201–207. [DOI] [PubMed] [Google Scholar]
- 10.Tzankov A, Zimpfer A, Pehrs A C.et al Expression of B‐cell markers in classical Hodgkin lymphoma: a tissue microarray analysis of 330 cases. Mod Pathol 2003161141–1147. [DOI] [PubMed] [Google Scholar]
- 11.Hans C P, Weisenburger D D, Greiner T C.et al Confirmation of the molecular classification of diffuse large B‐cell lymphoma by immunohistochemistry using a tissue microarray. Blood 2004103275–282. [DOI] [PubMed] [Google Scholar]
- 12.Tzankov A, Pehrs A C, Zimpfer A.et al Prognostic significance of CD44 expression in diffuse large B cell lymphoma of activated and germinal centre B cell‐like types: a tissue microarray analysis of 90 cases. J Clin Pathol 200356747–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tzankov A, Went P, Zimpfer A.et al Tissue microarray technology: principles, pitfalls and perspectives—lessons learned from hematological malignancies. Exp Gerontol 200540737–744. [DOI] [PubMed] [Google Scholar]
- 14.Zinzani P L, Dirnhofer S, Sabattini E.et al Identification of outcome predictors in diffuse large B‐cell lymphoma. Immunohistochemical profiling of homogeneously treated de novo tumors with nodal presentation on tissue micro‐arrays. Haematologica 200590341–347. [PubMed] [Google Scholar]
- 15.Obermann E C, Marienhagen J, Stoehr R.et al Tissue microarray construction from bone marrow biopsies. Biotechniques 200539822–826. [DOI] [PubMed] [Google Scholar]
- 16.Bueso‐Ramos C, Xu Y, McDonnell T J.et al Protein expression of a triad of frequently methylated genes, p73, p57Kip2, and p15, has prognostic value in adult acute lymphocytic leukemia independently of its methylation status. J Clin Oncol 2005103932–3939. [DOI] [PubMed] [Google Scholar]
- 17.Went P T, Zimpfer A, Pehrs A C.et al High specificity of combined TRAP and DBA.44 expression for hairy cell leukemia. Am J Surg Pathol 200529474–478. [DOI] [PubMed] [Google Scholar]
- 18.Zimpfer A, Went P, Tzankov A.et al Rare expression of KIT (CD117) in lymphomas: a tissue microarray study of 1166 cases. Histopathology 200445398–404. [DOI] [PubMed] [Google Scholar]
- 19.Camp R, Charette L, Rimm D. Validation of tissue microarray technology in breast carcinoma. Lab Invest 2000801943–1949. [DOI] [PubMed] [Google Scholar]
- 20.Hedvat C V, Hegde A, Chaganti R S. Application of tissue microarray technology to the study of non‐Hodgkiǹs and Hodgkiǹs lymphoma. Hum Pathol 200233968–974. [DOI] [PubMed] [Google Scholar]
- 21.Hoos A, Urist M, Stojadinovic A.et al Validation of tissue microarrays for immunohistochemical profiling of cancer specimens using the example of human fibroblastic tumors. Am J Pathol 20011581245–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nocito A, Bubendorf L, Tinner E M.et al Microarrays of bladder cancer tissue are highly representative of proliferation index and histological grade. J Pathol 2001194349–357. [DOI] [PubMed] [Google Scholar]
- 23.Went P T, Dirnhofer S, Bundi M.et al Prevalence of KIT expression in human tumors. J Clin Oncol 2004224514–4522. [DOI] [PubMed] [Google Scholar]
- 24.Warford A, Howat W, McCafferty J. Expression profiling by high‐throughput immunohistochemistry. J Immunol Methods 200429081–92. [DOI] [PubMed] [Google Scholar]