Abstract
Malignant transformation is a multistep process that may involve dysregulation of oncogenes and tumour suppressor genes, and monoclonal gammopathy of undetermined significance (MGUS) is believed to be a precursor of multiple myeloma. To investigate whether aberrant promoter methylation might be involved in the evolution of MGUS to multiple myeloma, we examined the p16, protein tyrosine phosphatase, non‐receptor type 6 (SHP1), death‐associated protein (DAP) kinase, E‐cadherin and oestrogen receptor genes, most being tumour suppressor genes, by methylation‐specific polymerase chain reaction. In 32 cases of multiple myeloma and 19 cases of MGUS, significantly more frequent methylation of p16 (p = 0.001), SHP1 (p⩽0.001) and E‐cadherin (p⩽0.001) genes was found in multiple myeloma than in MGUS. Methylation of DAP kinase and oestrogen receptor genes was comparable in multiple myeloma and MGUS. In conclusion, methylation of p16, SHP1 and E‐cadherin genes might be involved in the progression of MGUS to multiple myeloma.
Oncogenesis is a multistep process involving dysregulation of oncogenes and tumour suppressor genes. Multiple myeloma is characterised by neoplastic proliferation of monoclonal plasma cells. Multiple myeloma cells arise from postgerminal centre B cells, which migrate to the bone marrow, adhere to the stroma, proliferate, and trigger subsequent bone resorption and a paracrine cytokine loop.1,2 The disease may progress from monoclonal gammopathy of undetermined significance (MGUS) to multiple myeloma. MGUS occurs in approximately 3% of people >70 years old.2 Multiple myeloma develops in about one quarter of MGUS patients after a median of 10 years.3 Therefore, MGUS is regarded as a precursor of multiple myeloma, with the rate of transformation to multiple myeloma estimated to be 1% per year.3
DNA methylation involves the addition of a methyl group to the carbon 5 position of the cytosine ring in CpG dinucleotide. At gene promoters, DNA methylation leads to recruitment of a transcriptional repressor complex, resulting in suppression of transcription.4 In cancers, the CpG islands of selected genes are aberrantly methylated, resulting in silencing of these genes. Therefore, methylation is an alternative mechanism of gene inactivation, in addition to deletion and mutation.
Frequent methylation of the oestrogen receptor, E‐cadherin, protein tyrosine phosphatase, non‐receptor type 6 (SHP1), death‐associated protein (DAP) kinase and p16 genes has been shown in multiple myeloma.5,6,7 During the evolution of MGUS to multiple myeloma, multiple genetic aberrations are sequentially acquired. In this study, we tested the hypothesis that aberrant promoter methylation might be one of the steps involved in the progression of MGUS to multiple myeloma.
Materials and methods
Patients, diagnosis and treatment
Diagnosis of MGUS was based on an asymptomatic monoclonal gammopathy (immunoglobulin G (IgG)⩽3.5 g/dl, IgA⩽2 g/dl, and urinary Bence Jones protein⩽1 g/24 h), <10% plasma cells in the bone marrow and absence of lytic bone lesions. Diagnosis of multiple myeloma was based on standard criteria.3 Monoclonal paraproteins were identified by serum electrophoresis, immunoelectrophoresis or immunofixation. The study was approved by the review board of Queen Mary Hospital, Hong Kong.
Methylation‐specific polymerase chain reaction
High‐molecular‐weight DNA was isolated by standard protocols from bone marrow aspirates at diagnosis, and from eight marrow aspirates of healthy donors. Methylation‐specific polymerase chain reaction (MSP) was performed as described.6 Briefly, treatment of DNA with bisulphite for conversion of unmethylated, but not methylated, cytosine to uracil was performed with the CpGenome DNA modification kit (Intergen, New York, New York, USA). Primers for the methylated MSP and unmethylated MSP gene promoter regions were as described previously (fig 1). In all experiments, DNA from normal donors was used as negative control, and methylated control DNA (CpGenome Universal Methylated DNA, Intergen) as positive control. MSP was performed in a thermal cycler (9700, Perkin‐Elmer Biosystems, Foster City, California, USA). The polymerase chain reaction mixture contained 50 ng of bisulphite‐treated DNA, 0.2 mM deoxynucleoside triphosphates, 2 mM MgCl2, 10 pmol of each primer, 1× PCR buffer II and 2.5 U AmpliTaq Gold (Perkin‐Elmer Biosystems) in a final volume of 25 μl. The identity of the MSP products was confirmed by DNA sequencing.
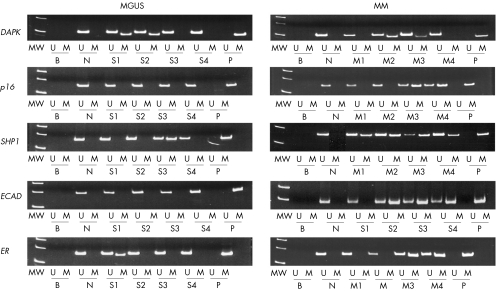
Figure 1 Methylation‐specific polymerase chain reaction of death‐associated protein kinase (DAPK) , p16, suppressor of high‐copy PP1 (SHP1), E‐cadherin (ECAD) and oestrogen receptor (ER) genes in monoclonal gammopathy of undetermined significance (MGUS) and multiple myeloma (MM). Samples tested were marrow aspirates obtained at diagnosis. The sequences for primers were DAPK (methylated, forward: 5′‐GGA TAG TCG GAT CGA GTT AAC GTC‐3′, reverse: 5′‐CCC TCC CAA ACG CCG A‐3′; unmethylated, forward: 5′‐GGA GGA TAG TTG GAT TGA GTT AAT GTT‐3′, reverse: 5′‐CAA ATC CCT CCC AAA CAC CAA‐3′), p16 (methylated, forward: 5′‐TTA TTA GAG GGT GGG GCG GAT CGC‐3′, reverse: 5′‐GAC CCC GAA CCG CGA CCG TAA‐3′; unmethylated, forward: 5′‐TTA TTA GAG GGT GGG GTG GAT TGT‐3′, reverse: 5′‐TTA TTA GAG GGT GGG GTG GAT TGT‐3′), SHP1 (methylated, forward: 5′‐GAA CGT TAT TAT AGT ATA GCG TTC‐3′, reverse: 5′‐TCA CGC ATA CGA ACC CAA ACG‐3′; unmethylated, forward: 5′‐GTG AAT GTT ATT ATA GTA TAG TGT TTG G‐3′, reverse: 5′‐TTC ACA CAT ACA AAC CCA AAC AAT‐3′), E‐cadherin (methylated, forward: 5′‐GTG GGC GGG TCG TTA GTT TC‐3′, reverse: 5′‐CTC ACA AAT ACT TTA CAA TTC CGA CG‐3′; unmethylated, forward: 5′‐GGT GGG TGG GTT GTT AGT TTT GT‐3′, reverse: 5′‐AAC TCA CAA ATC TTT ACA ATT CCA ACA‐3′) and oestrogen receptor (methylated, forward: 5′‐CGA GTT GGA GTT TTT GAA TCG TTC‐3′, reverse: 5′‐CTA CGC GTT AAC GAC GAC CG‐3′; unmethylated: forward: 5′‐ATG AGT TGG AGT TTT TGA ATT GTT T‐3′, reverse: 5′‐ATA AAC CTA CAC ATT AAC AAC AAC CA‐3′). MW, molecular weight markers; U, unmethylated MSP; M, methylated MSP; B, reagent blank; N, normal negative control; P, methylated positive control; S1–S4, four MGUS marrow samples; M1–M4, four primary myeloma marrow samples.
Statistical analysis
Frequencies of gene methylation in multiple myeloma and MGUS were compared by χ2 or Fisher's exact test (SPSS V.12). p Values are two sided.
Results
Methylation‐specific polymerase chain reaction
The positive control, but none of the eight normal marrow DNA samples, showed complete methylation of DAP kinase, p16, SHP1, E‐cadherin and oestrogen receptor genes (fig 1). In samples showing negative methylated‐MSP amplification, integrity of DNA was confirmed by positive unmethylated‐MSP amplification.
MSP of multiple myeloma and MGUS
There were 32 patients with multiple myeloma and 19 patients with MGUS (table 1). Frequencies of methylation of DAP kinase and oestrogen receptor genes were comparable in multiple myeloma and MGUS. E‐cadherin gene methylation was found only in multiple myeloma (p<0.001). p16 and SHP1 methylation was significantly more frequent in multiple myeloma than in MGUS (p16 53.1% v 5.3%, p<0.001; SHP1 84.4% v 31.6%, p<0.001; fig 1, table 2).
Table 1 Demographic data of patients with monoclonal gammopathy of undetermined significance and multiple myeloma.
| MGUS | MM | |
|---|---|---|
| n | 19 | 32 |
| Sex | ||
| Male | 10 (52.6%) | 17 (53.1%) |
| Female | 9 (47.4%) | 15 (46.9%) |
| Age (years) | ||
| Median | 76 | 63 |
| Range | 55–85 | 25–87 |
| Paraprotein subtype | ||
| IgG | 12 (85.7%) | 18 (56.3%) |
| IgA | 2 (14.3%) | 8 (25.0%) |
| IgD | 1 (3.1%) | |
| Light chain | 5 (15.6%) | |
| Bone marrow plasma cells | 3–7% | 38–93% |
| Paraprotein level | ||
| Median | – | 37.5 g/l |
| Range | – | 12–78 g/l |
| Durie–Salmon stage | ||
| I | – | 4 (12.5%) |
| II | – | 9 (28.1%) |
| III | – | 19 (59.4%) |
–, not applicable.
MGUS, monoclonal gammopathy of undetermined significance; MM, multiple myeloma.
Table 2 Frequencies of aberrant methylation of death‐associated protein kinase, p16, SHP1, E‐cadherin and oestrogen receptor genes.
| MGUS | MM (%) | p Value | |
|---|---|---|---|
| n | 19 | 32 | |
| DAP kinase | |||
| M | 6 (31.6) | 18 (56.3) | 0.146 |
| U | 13 (68.4) | 14 (43.7) | |
| p16 | |||
| M | 1 (5.3) | 17 (53.1) | 0.001 |
| U | 18 (94.7) | 15 (46.9) | |
| SHP1 | |||
| M | 6 (31.6) | 27 (84.4) | <0.001 |
| U | 13 (68.4) | 5 (15.6) | |
| ECAD | |||
| M | 0 (0) | 18 (56.3) | <0.001 |
| U | 19 (100) | 14 (43.7) | |
| ER | |||
| M | 3 (15.8) | 13 (40.6) | 0.117 |
| U | 16 (84.2) | 19 (59.3) |
DAP, death‐associated protein gene; ECAD, E‐cadherin gene; ER, oestrogen receptor gene; M, methylated gene status; MGUS, monoclonal gammopathy of undetermined significance; MM, multiple myeloma; SHP1, suppressor of high‐copy PP1 gene; U, unmethylated gene status.
M and U values are n (%).
Discussion
Our study is based on the hypothesis that if methylation of these genes were involved in the progression of MGUS to multiple myeloma, the frequency of gene hypermethylation would be significantly higher than that of MGUS. On the other hand, if hypermethylation of these genes were part of the initiating event that leads to immortalisation of the plasma cell, the frequency of hypermethylation of these genes in multiple myeloma would be expected to be comparable to that of MGUS. We showed that p16, SHP1 and E‐cadherin gene methylation was significantly more frequent in multiple myeloma than in MGUS, suggesting that methylation of these genes might be involved in clonal evolution of MGUS to multiple myeloma.
p16 is a cyclin‐dependent kinase inhibitor targeting cyclin‐dependent kinases 4 and 6. We showed preferential methylation of p16 in multiple myeloma. Previous studies have also shown more frequent p16 methylation in multiple myeloma than in MGUS.8,9 Furthermore, p16 methylation was also more frequent in plasma‐cell leukaemia.10 However, in CD138‐purified plasma cells, p16 methylation has been found to be equally frequent in MGUS and multiple myeloma.10
E‐cadherin, a homotypic cell adhesion molecule, is considered a suppressor of metastasis. Our finding of preferential E‐cadherin gene methylation in multiple myeloma instead of MGUS was consistent with previous reported results.10 As the progression of MGUS involves metastasis of the myeloma plasma cells to other parts of the bone marrow, E‐cadherin gene methylation might be expected to play a role in the evolution of MGUS to multiple myeloma.
SHP1, a cytoplasmic protein tyrosine phosphatase, is a putative tumour suppressor negatively regulating the interleukin 6/Jak/STAT signalling in plasma cells.6 In myeloma cells with complete SHP1 methylation, STAT3 was constitutively activated. Demethylation with re‐expression of SHP1 led to down regulation of phosphorylated STAT3, showing that SHP1 methylation is biologically relevant in myeloma pathogenesis.6
Taken together, p16 and SHP1 gene methylation might confer a proliferative and survival advantage on neoplastic plasma cells, and E‐cadherin gene methylation might enhance the metastatic potential and thus progression of MGUS.
Similar to another study,11 we showed a comparable frequency of DAP kinase methylation in patients with MGUS and multiple myeloma, implying that DAP kinase is probably not involved in the progression of MGUS. However, there was a relatively high frequency of DAP kinase methylation (35.7%) in MGUS, suggesting that this might be involved in the development of MGUS.
Take-home messages
Tumour suppressor genes are implicated in the pathogenesis of multiple myeloma.
Hypermethylation of ECAD, SHP1 and p16 is implicated in the progression from MGUS to mutiple myeloma.
Ideally, methylation of p16, E‐cadherin and SHP1 genes should be investigated serially in MGUS and multiple myeloma samples in the same patient. However, as the annual rate of progression of MGUS to multiple myeloma is only 1%, this would require a long period of observation.
In conclusion, there was preferential methylation of some tumour suppressor genes in MGUS. Therefore, inactivation of tumour suppressor genes by gene methylation might contribute to disease progression from MGUS to multiple myeloma.
Acknowledgements
We express our sincere thanks to the Department of Pathology for helping with the diagnosis, and YY Chan and her team in K20N, Queen Mary Hospital, for excellent nursing care of the patients.
Abbreviations
DAP - death‐associated protein
MGUS - monoclonal gammopathy of undetermined significance
MSP - methylation‐specific polymerase chain reaction
SHP1 - protein tyrosine phosphatase, non‐receptor type 6.
Footnotes
Competing interests: None declared.
References
- 1.Kuehl W M, Bergsagel P L. Multiple myeloma: evolving genetic events and host interactions. Nat Rev Cancer 2002275–87. [DOI] [PubMed] [Google Scholar]
- 2.Hideshima T, Bergsagel P L, Kuehl W M.et al Advances in biology of multiple myeloma: clinical applications. Blood 2004104607–618. [DOI] [PubMed] [Google Scholar]
- 3.Kyle R A, Therneau T M, Rajkumar S V.et al A long‐term study of prognosis in monoclonal gammopathy of undetermined significance. N Engl J Med 2002346564–569. [DOI] [PubMed] [Google Scholar]
- 4.Chim C S, Liang R, Kwong Y L. Gene promoter hypermethylation in hematologic malignancies. Hematol Oncol 200220167–176. [DOI] [PubMed] [Google Scholar]
- 5.Herman J G, Baylin S B. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 20033492042–2054. [DOI] [PubMed] [Google Scholar]
- 6.Chim C S, Fung T K, Cheung J.et alSOCS1 and SHP1 hypermethylation in multiple myeloma: implications for epigenetic activation of the Jak/STAT pathway. Blood 20041034630–4635. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi T, Shivapurkar N, Reddy J.et al DNA methylation profiles of lymphoid and hematopoietic malignancies. Clin Cancer Res 2004102928–2935. [DOI] [PubMed] [Google Scholar]
- 8.Mateos M V, Garcia‐Sanz R, Lopez‐Perez R.et alP16/INK4a gene inactivation by hypermethylation is associated with aggressive variants of monoclonal gammopathies. Hematol J 20012146–149. [DOI] [PubMed] [Google Scholar]
- 9.Uchida T, Kinoshita T, Ohno T.et al Hypermethylation of p16INK4A gene promoter during the progression of plasma cell dyscrasia. Leukemia 200115157–165. [DOI] [PubMed] [Google Scholar]
- 10.Galm O, Wilop S, Reichelt J.et al DNA methylation changes in multiple myeloma. Leukemia 2004181687–1692. [DOI] [PubMed] [Google Scholar]
- 11.Guillerm G, Gyan E, Wolowiec D.et al p16(INK4a) and p15(INK4b) gene methylations in plasma cells from monoclonal gammopathy of undetermined significance. Blood 200198244–246. [DOI] [PubMed] [Google Scholar]