Abstract
The Nova paraneoplastic antigens are neuron-specific RNA binding proteins that participate in the control of alternative splicing. We have used the yeast two-hybrid system to isolate Nova interacting proteins and identify an RNA binding protein that is closely related to the polypyrimidine tract-binding protein (PTB). The expression of this protein, brPTB, is enriched in the brain, where it is expressed in glia and neurons. brPTB interacts with Nova proteins in cell lines and colocalizes with Nova within neuronal nuclei. We previously found that Nova binds to a pyrimidine-rich RNA element present upstream of an alternatively spliced exon, E3A, in glycine receptor α2 (GlyRα2) pre-mRNA, and this binding is implicated in Nova-dependent regulation of splicing. Cotransfection assays with a GlyRα2 minigene demonstrate that brPTB antagonizes the action of Nova to increase utilization of GlyRα2 E3A. brPTB binds to a 90-nt GlyRα2 RNA adjacent to the Nova binding site, but with an affinity that is more than 10-fold lower than Nova. When a putative binding site for brPTB on the GlyRα2 RNA is mutated, binding is abolished and the inhibitory effect on Nova-dependent exon selection disappears. These results suggest that brPTB is a tissue-restricted RNA binding protein that interacts with and inhibits the ability of Nova to activate exon selection in neurons.
Neurons make extensive use of alternative splicing to regulate functional differences in proteins. A wide variety of neurotransmitter receptor activities are regulated by alternative splicing, including NR1 N-methyl-d-aspartate (NMDA) receptor subcellular localization (1) and interaction with neurofilaments (2), the physiology of the glutamate (3) and NMDA (4) receptors, and the ability of agrin to induce clustering of acetylcholine receptors (5). Moreover, several neurologic diseases such as spinal muscular atrophy, amyotrophic lateral sclerosis, and paraneoplastic opsoclonus-myoclonus ataxia (POMA) have been associated with defects in proteins involved in generating the splicing machinery or in the accurate splicing of target RNAs (6–8).
Since the discovery of tissue-specific splicing of the calcitonin/calcitonin gene-related peptide (CGRP) transcript in neurons, there has been an extensive search for cis-acting RNA elements and trans-acting RNA binding proteins that mediate neuron-specific splicing. The first example of cis-acting regulatory elements in neuronal pre-mRNAs identified was in calcitonin/CGRP pre-mRNA (9), and a number of specific sequences have been identified that are responsible for calcitonin/CGRP tissue-specific processing (10, 11). Subsequent work identified regulatory sequences near other neuron-specific exons such as the N1 exon of src (12) and a 24-nt exon of the γ-aminobutyric acid type A receptor γ2 subunit (13).
The identification of trans-acting factors that regulate neuronal splicing has been a greater challenge. Two general mechanisms might account for the way such factors could mediate regulation of neuronal splicing. Brain-specific variants in splicing patterns could be achieved by differing levels or modifications of ubiquitously expressed splicing factors. For example, antagonism between the heterogeneons nuclear ribonudeoprotein (hnRNP) A1 and ASF/SF2 proteins determines the selection of 5′ splice site (14), and varying ratios of these proteins have been shown to affect neuron-specific splicing of clathrin exon EN (15). Moreover, posttranslational modifications such as phosphorylation influence the activity of general splicing factors including ASF/SF2 (16, 17). Such variations might be difficult to detect if they are transient or localized to specific cells especially in the brain, where there is great complexity in cell type and number.
A second and perhaps more direct way to regulate pre-mRNA splicing in neurons would be to use brain-restricted regulatory proteins. Evidence for such trans-acting factors has been limited. The polypyrimidine tract binding protein (PTB) helps regulate neuron-specific splicing of calcitonin-CGRP pre-mRNA (11, 18), and the existence of a brain-enriched protein related to PTB has been suggested (19–21). Neuron-specific splicing of n-src involves several generally expressed RNA binding proteins (20, 22, 23), and KH-type splicing regulatory protein, which is enriched in brain (23). Finally, the RNA binding protein elav may regulate alternative splicing in Drosophila neurons (24).
Recently, we provided evidence that the neuron-specific RNA binding protein Nova-1 regulates alternative splicing in neurons (7, 25). In vitro RNA selection experiments identified RNAs harboring UCAU elements to which Nova proteins bound with sequence specificity and high affinity (26, 27). In addition, the cocrystal structure of Nova protein bound to a UCAY RNA element has been solved and confirms the specific nature of the Nova–RNA interaction (28, 29). The Nova-1 RNA selection consensus sequence matched an intronic UCAU repeat element in glycine receptor α2 (GlyRα2) pre-mRNA and identified it as a candidate Nova target. Coprecipitation and cross-linking experiments demonstrated that Nova in brain extracts specifically interacted with the GlyRα2 UCAU repeat element, and cotransfection assays demonstrated that Nova-1 enhanced utilization of an adjacent 3′ splice site of an alternatively spliced exon, E3A (7, 26). Conversely, Nova-1 null mice have a specific defect in utilization of GlyRα2 exon E3A (7), demonstrating that Nova-1 is necessary for accurate GlyRα2 exon selection in vivo.
Several findings support the idea that Nova proteins may undergo protein–protein interactions in addition to RNA binding. The cocrystal structure of a Nova–RNA complex reveals that most of the hnRNPK homology (KH) domain is accessible for protein–protein interactions, even when bound to RNA (29). Second, Nova contains several proline- and alanine-rich stretches in the spacer region linking the second and third KH domains that may represent protein interaction motifs (27, 30).
To examine whether Nova interacts specifically with other proteins, and to assess whether these interactions might modulate the ability of Nova to regulate alternative splicing, we have performed yeast two-hybrid screens using Nova proteins as bait. We identified a PTB-like protein, termed brPTB, that is specifically enriched in the brain and it interacts with Nova in vivo. In transfected cells, brPTB antagonizes Nova-1's ability to increase the inclusion of GlyRα2 exon E3A. Filter binding assays demonstrate that brPTB is able to bind to the GlyRα2 intronic element upstream of E3A, but with significantly lower affinity than Nova-1, and that this binding depends on a specific sequence. When this sequence is mutated, brPTB no longer inhibits Nova-1 function in splicing assays. These results demonstrate that brPTB functions to block the action of the neuron-specific splicing factor Nova, most likely by forming an inhibitory complex that binds to an intronic enhancer element.
Materials and Methods
Yeast Two-Hybrid Screening.
Nova bait constructs were used to screen adult mouse brain or E11 Matchmaker libraries (1.52 × 107 transformants; CLONTECH). Four of six clones identified are shown in Table 1. A 1.3-kb EcoRI fragment from the brPTB yeast two-hybrid clone was used to screen an adult mouse brain cDNA library (5.4 × 105 phage clones; Uni-ZAP XR, Stratagene) and identified two overlapping clones spanning the 1.6-kb predicted ORF and a 1.4-kb 3′ untranslated region.
Table 1.
Yeast two-hybrid interactions of Nova proteins
| Prey (GAD) | Bait (LexA)
|
||||
|---|---|---|---|---|---|
| Lac Z
|
His−
|
||||
| TRF1 | Nova-1 | Nova-2 | Nova-1 | Nova-2 | |
| TD1 | — | — | — | — | — |
| brPTB | — | +++ | +++ | +++ | +++ |
| hnRNP I/PTB | — | ND | +++ | ND | +++ |
| hnRNP L | — | ND | +++ | ND | +++ |
| Matrin3 | — | +++ | +++ | +++ | +++ |
Yeast host strain LD40 was transformed with the indicated bait (LexA fusion) and prey (GAD fusion) constructs and their interaction was scored by staining for LacZ expression and growth on His− media. Negative controls included an unrelated protein fused to GAD (TD1) and LexA (TRF1). +++, Robust staining and growth; —, No staining or growth. GAD: Gal4 activation domain; ND: not determined.
Fusion Proteins and Pull-Down Assays.
In vitro glutathione S-transferase (GST) pull-down assays were performed (27) by using T7/His tagged brPTB and PTBs made in pET21 (Novagen). The indicated amounts of recombinant proteins were incubated in 100 μl of low stringency binding buffer at room temperature for 45 min followed by precipitation with glutathione-Sepharose 4B beads (Amersham Pharmacia). In vivo GST pull-down assays were performed by transfecting (Fugene6, Boehringer Mannheim) N2A cells with Nova-1 and Nova-2 GST fusion protein constructs (in pEBG vector). Cells were harvested 2 days later and lysed on ice for 45 min (1% Triton X-100/10 mM Tris⋅Cl, pH 7.6/50 mM NaCl/30 mM sodium pyrophosphate/50 mM NaF/1 mM EDTA) before GST precipitation.
Northern and Western Blot Analysis.
Total RNA was obtained from wild-type mice by using a modified guanidine-acid phenol protocol (27). The membranes were UV cross-linked and hybridized (QuikHyb, Stratagene) with 32P-labeled DNA probes (Prime-It Kit, Stratagene). Proteins from fresh tissue specimens (adult wild-type mice) were isolated, separated by 10% SDS/PAGE, and transferred to poly(vinylidene difluoride) membranes (Millipore). Membranes were blocked and incubated with primary antibodies including anti-T7 (Novagen), M5 anti-flag (Sigma), and anti-GST (Sigma). Polyclonal anti-brPTB (used at a 1:5,000 dilution) antibody was developed by injection of rabbits with full-length recombinant fusion protein and Freund's adjuvant (Pocono Rabbit Farm, Canadensis, PA).
Immunohistochemistry and Immunofluorescence.
Adult rat (male Sprague–Dawley), embryonic day (E) 16 rat embryo, and P5 mouse (male CD1) brain sections were prepared (31) and probed with anti-brPTB polyclonal antiserum (1:5,000 dilution), anti-MAP2 mAb (Sigma, 1:500), anti-glial fibrillary acidic protein mAb (Dako, 1:25) or POMA patient sera (1:500) and incubated at 4°C overnight. Proteins were visualized by using Cy2 anti-mouse IgG and Cy3 anti-rabbit IgG (see Fig. 4 H and I) or Cy5 anti-human IgG and Cy2 anti-rabbit IgG (see Fig. 5) (Jackson ImmunoResearch) by confocal microscopy. Sections stained with brPTB/Cy2 gave no signal when detected at the wavelength of Cy5 in the absence of POMA/Cy5.
Figure 4.
brPTB colocalizes with Nova protein in spinal cord motor neurons. (A and D) Confocal laser image of brPTB expression in a single 1.2-μm optical section, visualized with anti-brPTB antibody and Cy-2-conjugated anti-rabbit antibody. (B and E) Same 1.2-μm section as A and D visualized with POMA patient serum and Cy-5-conjugated anti-human antibody. (C and F) Fusion of images in A and D and B and E, respectively, showing colocalization of both brPTB and Nova proteins within motor neuronal nuclei; some cells (presumably glia; see Fig. 3H) stain with anti-brPTB antibody but not with POMA serum. (A–C) P5 mouse spinal cord horizontal section; (D–F) adult rat spinal cord horizontal section.
Figure 5.
brPTB antagonizes the action of Nova by binding to the GlyRα2 intronic RNA. (A) Schematic and nucleotide sequence of the 90-nt fragment of GlyRα2 intronic RNA that contains Nova binding sites (UCAU; solid underline) and a candidate binding site for brPTB (UCUU in a pyrimidine context; dashed underline, see text and ref. 33). The C → T mutation in the brPTB binding site is indicated. (B) Filter binding assays of purified fusion proteins Nova-1 and brPTB with the wild-type and C → T GlyRα2 RNA and an unrelated fragment of rRNA. Mutating the brPTB binding site as shown in A abolished its binding (open squares), whereas it had no effect on Nova-1 binding (data not shown). (C) In vivo splicing assays. 293T cells were cotransfected with the indicated amount of Nova-1 and brPTB expression constructs plus a wild-type (WT) or mutant (C → T) GlyRα2 minigene containing exons 2–4 (7). Ratios of included exon 3A over 3B were determined by reverse transcription–PCR, followed by SspI digestion (which only cuts within exon 3A; see ref. 7). Each point represents the average of duplicate transfections, and error bars represent the SD. The same results were obtained in three independent experiments, each performed in duplicate. (D) Western blot analysis of cell extracts. One-twentieth the amount of protein in each transfection was separated on SDS/PAGE, followed by immunoblotting with α-flag mAb (brPTB) or POMA patient serum (Nova-1) to confirm that increasing amounts of transfected DNA correlated with proportionally increasing amount of protein synthesized by the cells.
In Vivo Splicing and Protein–RNA Filter Binding Assays.
Assays were performed as described (7, 26, 27). The brPTB binding site on GlyRα2 was mutated by using the QuikChange Site-Directed Mutagenesis Kit (Stratagene).
Results
Identification of a PTB-Like Protein That Interacts with Nova.
To identify Nova-interacting proteins we performed yeast two-hybrid screens of adult brain or E11 mouse cDNA libraries with full-length Nova-1 or Nova-2 bait constructs. As shown in Table 1, a number of different RNA binding proteins were isolated, among which was a novel sequence with striking similarity to the hnRNP I/PTB splicing regulator, which we named brain-enriched PTB (brPTB; see below).
To confirm the results of the yeast two-hybrid assay and to demonstrate that the Nova–brPTB interaction does not depend on additional yeast proteins or RNA, we assayed the interaction between brPTB and the Nova proteins in vitro and in vivo, by using a GST pull-down assay. In vitro, purified brPTB and Nova fusion proteins specifically interact, and this interaction was not affected by pretreating each protein sample with RNase A (Fig. 1A). In N2A cells, a neuroblastoma cell line that expresses endogenous brPTB (see below), brPTB coprecipitates equally well with transfected Nova-1 and Nova-2 GST fusion proteins, but not with GST protein alone (Fig. 1B). Taken together, these data demonstrate that brPTB and Nova proteins are capable of interacting directly and independently of an RNA intermediate.
Figure 1.
Nova and brPTB proteins interact in vitro and in vivo. (A) In vitro GST pull-down assay. Five micrograms of purified GST, GST-Nova-1, and GST-Nova-2 fusion proteins as indicated was incubated with 2 μg of T7-tagged brPTB fusion protein (brPTB). The amount of T7-brPTB that was pulled down was assayed by Western blot (WB) using an anti-T7-tag mAb (Top, α-T7). The presence of equal amounts of GST fusion proteins in each reaction was confirmed by probing blots with an anti-GST polyclonal antibody (Middle, α-GST). GST runs at 27 kDa, GST-Nova-1 at 82 kDa, and GST-Nova-2 at 80 kDa. To examine whether RNA was needed for the Nova-brPTB interaction, pull-down assays were repeated in the presence (Left, +RNase) or absence (Right, −RNase) of 1 μg/ml RNaseA. To confirm the activity of the RNase, reactions were spiked with 32P-labeled RNA and aliquots were run on denaturing urea/polyacrylamide gels and exposed by autoradiography (Bottom). (B) In vivo GST pull-down assay. Equal amounts of GST alone, GST-Nova-1, and GST-Nova-2 expression vectors were transfected into N2A cells as indicated. Cell lysates were incubated with glutathione Sepharose beads and the amount of endogenous brPTB pulled down was assayed by Western blot (WB) using a brPTB-specific antibody (Upper, α-brPTB; see Fig. 2B). A lysate from untransfected N2A cells (cell lysate, first lane) identified the endogenous brPTB band. The blot also was probed with a polyclonal GST antibody (Lower, α-GST) to confirm expression of the transfected GST proteins. (C) Yeast two-hybrid interactions of Nova-2 and brPTB truncation constructs. A number of bait constructs encoding the indicated Nova-1 or Nova-2 amino acids (rows) were cotransformed with different Nova-1, Nova-2 or brPTB activation constructs as indicated (columns). Interaction was measured by β-galactosidase activity (++, very strong staining; +, strong staining; +/−, weak staining or not all colonies positive; −, no staining). Control bait (control, pVA3) and prey (control, pTD1) transformations showed no β-galactosidase activity.
We further dissected the interaction between brPTB and the Nova proteins by using truncation constructs in the yeast-two hybrid assay (Fig. 1C). The spacer domain of Nova-2 (amino acids 230–407) is sufficient in producing as robust an interaction with brPTB as were full-length Nova-1 or Nova-2. In contrast, no single brPTB deletion construct was adequate in mediating a strong interaction with Nova. Nova-1 truncation constructs were found to activate LacZ transcription in the absence of a cotransformed prey (Gal4 activation domain) construct (data not shown).
We cloned a 3.0-kb brPTB cDNA encoding the predicted full-length ORF from an adult mouse brain cDNA library. The 1.6-kb ORF encodes for 532 aa and is followed by a 1.4-kb 3′ untranslated region (GenBank accession no. AF095718). Sequence analysis shows that brPTB protein shares 73% identity and 84% similarity with PTB at the amino acid level. These numbers increase to 80% and 91%, respectively, over the four RNA recognition motif (RRM) domains. Interestingly, the RRM-containing proteins that were pulled down from the yeast two-hybrid screen with Nova (hnRNP I/PTB, hnRNP L, and matrin3) form a distinct subgroup of the RRM-type family of RNA binding proteins based on amino acid homologies over the RRM domains.
brPTB Expression in the Nervous System.
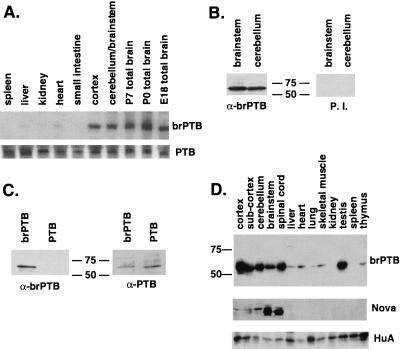
To examine the expression pattern of brPTB, we performed Northern blot analysis on several mouse tissues (Fig. 2A). brPTB mRNA was detected preferentially in brain, including the cortex and hindbrain, with very reduced levels evident in some organs outside the nervous system, most notably in heart. brPTB is expressed at similar levels across different developmental stages, ranging from E18 to adult. In contrast, a PTB probe detected mRNA in all tissues, although we cannot rule out the possibility that this probe might cross-react with brPTB (Fig. 2A).
Figure 2.
brPTB mRNA and protein levels are enriched in the brain. (A) A Northern blot of total RNA from the indicated mouse tissues and developmental stages (20 μg/lane) was probed with a 1.3-kb DNA probe from the coding sequence of brPTB. The blot was stripped and reprobed with a 1.2-kb probe from the coding sequence of PTB. The two messages were approximately the same size (3.6 kb). (B) Production of anti-brPTB antibody. Western blot of rat cerebellum and brainstem probed with polyclonal anti-brPTB or preimmune serum (P.I.) shows that a band of approximately 57 kDa is recognized only after the immunization with full-length brPTB fusion protein. (C) Polyclonal antibody is brPTB specific. PTB and brPTB purified fusion proteins (50 ng/lane) were probed with anti-brPTB serum (dilution 1:5,000) or PTB polyclonal antibody (1:500). (D) Western blot of the indicated mouse tissues (50 μg total protein/lane) probed with various primary antibodies. brPTB antibody recognizes a 57-kDa band most prominently in brain tissues and the testis. POMA patients' serum shows that Nova proteins are restricted in the brain. HuA polyclonal antibody recognizes a ubiquitously expressed Hu isoform (44) and served as a loading control.
To evaluate the tissue specificity of brPTB, we generated a brPTB-specific antibody (Fig. 2B). This polyclonal antibody reacts with purified brPTB but not PTB fusion proteins (Fig. 2C). Western blot analysis using this brPTB antibody revealed that brPTB, like its mRNA, is enriched in neural tissues (Fig. 2D). brPTB also is expressed at high levels in the testis and at considerably lower levels in the liver, heart, lung, skeletal muscle, and thymus. This pattern contrasts with the strict brain-specific expression of Nova proteins (Fig. 2D; refs. 30 and 32).
We performed immunohistochemical studies to further define the expression pattern of brPTB in rat and mouse tissues (Fig. 3). Strong reactivity to the brPTB-specific antibody was present in most brain tissues, including cerebellum, brainstem, spinal cord, and hypothalamus (Fig. 3 A, B, and D). brPTB also was expressed in the peripheral nervous system and neural crest derivatives, including the dorsal root and trigeminal ganglia (Fig. 3 C and D), the cochlear spiral and intestinal ganglion cells (Fig. 3 E and F), and the adrenal medulla (Fig. 3C). Non-neural tissues that expressed brPTB included the heart and, at low levels, the liver (Fig. 3C and data not shown).
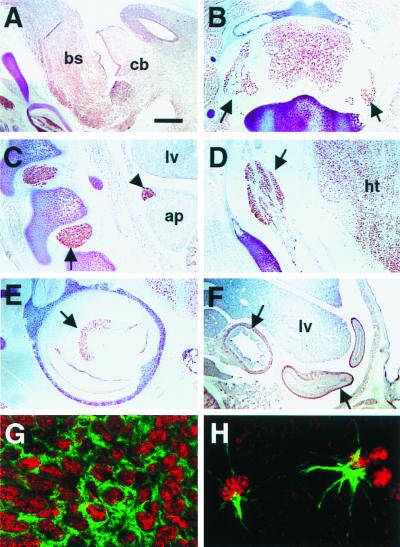
Figure 3.
Distribution of brPTB immunoreactivity in rat and mouse tissues. (A) Sagittal section from E16 rat brain stained with anti-brPTB polyclonal antibody shows strong reactivity with most cells in brain including cerebellum (cb) and brainstem (bs). (B) brPTB immunoreactivity in adult rat spinal cord and dorsal root ganglia (arrows). (C and D) brPTB expression in the developing peripheral nervous system. Immunostaining of E16 rat sagittal sections indicate that brPTB expression is strong in embryonic dorsal root ganglia (C, arrow), trigeminal ganglia (D, arrow), developing adrenal medulla (C, arrowhead) and hypothalamus (D, ht); however, it is very low in liver (C, lv) and adrenal primordium (C, ap). (E) brPTB staining in P5 mouse cochlea. Note the intensive staining in cochlear spiral ganglion cells (arrow). (F) brPTB expression in the ganglion cells of the small intestine (E16 rat embryo). (G) Confocal image of immunofluorescence double exposure of MAP2 (green) and brPTB (red) in a sagittal section of E16 rat cortex. (H) Confocal image of immunofluorescence double exposure of glial fibrillary acidic protein (green) and brPTB (red) in a sagittal section of adult rat cerebellum. Control sections stained with preimmune serum or with brPTB antibody preadsorbed with brPTB fusion protein were negative (data not shown). (Scale bar: 80 μm in A–E; 160 μm in F; 3.4 μm in G; 2.3 μm in H.)
To examine the subcellular distribution of brPTB and Nova, we used immunofluorescence microscopy on mouse and rat sections (Figs. 3 G and H and 4). brPTB is predominantly nuclear and stains both neurons and astrocytes in confocal microscopy images of cells doubly stained for brPTB and the neuronal marker MAP2 (Fig. 3G) or the astrocyte marker glial fibrillary acidic protein (Fig. 3H). Furthermore, in rat or mouse spinal cord sections, Nova and brPTB proteins colocalize in the nuclei of motor neurons, but not in glia where Nova is absent (Fig. 4).
brPTB Down-Regulates Nova-Dependent Splicing Activation of GlyRα2.
In earlier work we have shown that Nova-1 specifically binds to a GlyRα2 UCAU-repeat RNA element. The Nova-1 binding site is located just upstream of the first of the two mutually exclusive exons (Fig. 5A). Interestingly, a consensus sequence binding site for PTB exists proximal to the Nova binding element (Fig. 5A; ref. 33). Given the high degree of sequence identity between brPTB and PTB, we tested whether brPTB also might bind to this GlyRα2 intronic element. We performed filter binding assays with brPTB and a radiolabeled 90-nt RNA that included the Nova-1 and PTB binding sites (Fig. 5B). brPTB bound to this RNA, but with approximately a 12-fold reduced affinity (Kd = 559 nM) relative to Nova (Kd = 46 nM). Nonetheless, this binding is specific because no detectable binding was observed to an unrelated RNA. Furthermore, when the putative brPTB binding site on this 90-nt RNA was mutated (cytidine to thymidine) its binding was reduced to background levels (Fig. 5B).
We previously have demonstrated that Nova protein regulates alternative splicing of the GlyRα2 pre-mRNA by specifically increasing the inclusion of E3A at the expense of E3B. To determine whether brPTB is able to affect the ability of Nova to activate GlyRα2 E3A splicing, we cotransfected 293T cells with the GlyRα2 minigene together with varying concentrations of brPTB and Nova-1. Nova-1 transfection alone stimulated E3A exon inclusion by approximately 2-fold, as previously reported (7). In cotransfections, brPTB suppressed the effect of Nova-1 on exon inclusion in a dose-dependent fashion (Fig. 5C). In control experiments, increasing amounts of transfected brPTB DNA in the absence of Nova-1 did not alter the splicing pattern of GlyRα2 pre-mRNA. These results suggest that the interaction between brPTB and Nova-1 is functional in vivo.
When these experiments were repeated by using a mutant GlyRα2 minigene harboring the C → T mutation that abrogated brPTB binding (Fig. 5B), brPTB was no longer able to block the ability of Nova to stimulate E3A inclusion (Fig. 5C). We confirmed the presence of transfected brPTB and Nova-1 in these experiments by Western blot analysis (Fig. 5D). Taken together, these results suggest that brPTB inhibits Nova-1-dependent activation of exon inclusion and that this action is mediated at least in part by brPTB binding to GlyRα2 pre-mRNA upstream of the Nova-1 binding site.
Discussion
Nova Interacts with a PTB-Like Protein.
We have identified a PTB-like protein that specifically interacts with the neurologic disease antigen Nova. The present work extends previous findings, made through a convergence of biochemical and genetic experiments, that Nova-1 acts to promote exon inclusion in neurons. We previously have identified GlyRα2 pre-mRNA as a candidate Nova ligand in vivo (26) and demonstrated in cell transfection studies that Nova-1 activates inclusion of GlyRα2 exon E3A by a direct action on the intronic GlyRα2 UCAU-repeat element (7). Moreover, utilization of GlyRα2 E3A is decreased in the hindbrain of Nova-1 null mice (7). The data presented here suggest that brPTB may antagonize Nova-dependent enhancement of splicing. This observation is consistent with numerous studies implicating PTB in the negative regulation of alternative splicing in neurons and other tissues (20, 21, 34–36).
Several studies predicted the existence of brPTB. Three distinct isoforms of PTB are known, each with different sizes of a spacer region between the first and second RNA binding domains (37). A fourth PTB isoform was predicted from studies of cell extracts that were able to replicate aspects of neuron-specific alternative splicing in vitro (reviewed in ref. 21). These studies documented a PTB-like protein species enriched in nuclear extracts of brain tissue (38) and retinoblastoma cell lines (20) that migrated more slowly on SDS/PAGE than PTB. The brPTB cDNA clone identified here predicts a protein that is slightly larger than PTB (57.6 vs. 57.2 kDa; ref. 37) and is most likely the previously identified p59/PTB brain-enriched species.
Yeast two-hybrid screens using Nova proteins as bait identified several PTB-related RNA binding proteins. It is remarkable that four of the most robust interacting proteins in these screens were PTB, hnRNP L, matrin3, and brPTB. These proteins all constitute a subfamily of RRM-type RNA binding proteins, as first suggested by comparisons of PTB and hnRNP L (39), because their RRM motifs are strongly homologous with each other but only weakly homologous to the canonical RRM consensus sequence. The observation of interactions between Nova proteins and the various members of this subfamily of RRM proteins suggests that they may be mediated by a domain shared among the RRM proteins, which seems likely to include the atypical RRM motif. However, mapping of the interaction between Nova and brPTB failed to reveal a deletion mutant that interacted strongly with Nova, suggesting either that the full protein is necessary for the interaction or that several different domains are important for a robust interaction.
brPTB Functions to Inhibit Nova-Dependent Activation of Splicing.
The action of brPTB to inhibit Nova-dependent activation of exon inclusion is consistent with the general role described for PTB in suppressing exon inclusion of pre-mRNAs. This action of PTB is believed to be mediated by PTB binding to polypyrimidine-rich elements and blocking the assembly of a splicing competent complex in a manner analogous to the action of Sxl to block U2AF65 polypyrimidine tract binding (40). For example, PTB acts to repress smooth muscle-specific inclusion of alternatively spliced exons in the α-tropomyosin and α-actinin pre-mRNAs, most likely by binding to regulatory elements upstream of the 3′ splice site (33, 34, 41). In neuronal cells, PTB inhibits inclusion of the n-src exon, in part by an action on 3′ splice-site selection (20). Similarly, previous studies with p59/PTB as well as current studies on brPTB all point to an inhibitory action on splice site selection. Even in the single instance where PTB enhances exon inclusion in the alternative splicing of the calcitonin-CGRP pre-mRNA, it is believed to act by disrupting U2AF recognition of an enhancer pseudoexon, thereby indirectly promoting correct exon utilization (11).
Given several instances in which PTB acts on alternatively spliced neuronal pre-mRNAs to inhibit the inclusion of neuronal exons, it has been suggested that a general action of PTB might be to inhibit neuron-specific exon utilization (21, 36). In previous work, the presence of brPTB correlated with the presence of neuron-specific splicing in vitro, whereas the addition of the general form of PTB inhibited neuron-specific exon utilization (19, 38). These results had suggested the possibility that the inhibitory action of PTB is replaced in brain by a permissive or stimulatory action of a brain-PTB isoform (21). The proposal that brPTB promotes neuron-specific splicing is not supported by our experiments, where brPTB antagonizes Nova's action in promoting neuron-specific exon inclusion.
A Model for the Effect of brPTB on Nova Function.
There are several possible mechanisms by which brPTB could regulate Nova activity. One model is that brPTB competes with the ability of Nova to bind the GlyRα2 UCAU element, in a manner reminiscent of the action of PTB to block U2AF binding. However, we find that brPTB binds to the isolated GlyR 90-nt RNA with a 12-fold weaker affinity than Nova-1. Moreover, brPTB appears to supershift a Nova-RNA complex, suggesting that it does not interfere with the ability of Nova to bind the RNA (unpublished observation). Therefore, it is unlikely that brPTB and Nova compete for the same site on the RNA and that this competition inhibits the ability of Nova to enhance exon inclusion.
Notably, immediately upstream of the GlyRα2 UCAU repeat element exists a potential PTB consensus binding element, as defined by Patton and colleagues (33): a UCUU motif in the context of a pyrimidine-rich element. Although this sequence does not clearly define a PTB binding site (for example see ref. 35), and brPTB and PTB binding specificity may differ, our results suggest that Nova and brPTB may bind adjacent intronic sequence elements. We cannot rule out the possibility that brPTB binds exclusively to the RNA target or to the Nova protein. We find that brPTB and Nova proteins interact and that brPTB binds the GlyRα2 RNA weakly but with sequence specificity. Furthermore, binding of the RNA is necessary for the inhibitory action of brPTB on Nova-1 function. The simplest model consistent with this data is that brPTB binds the GlyRα2 pre-mRNA at a site adjacent to Nova and mediates an inhibitory effect on Nova-dependent exon inclusion through protein–protein interactions, perhaps by preventing assembly of a multiprotein complex necessary to activate splicing.
Our model bears some similarities to studies of factors that regulate n-src splicing in neurons. A core sequence termed the downstream control sequence (DCS) is necessary for enhancement of n-src splicing in vitro (42, 43). Repression of n-src splicing in non-neuronal cells requires both the upstream element and sequences within and adjacent to the DCS. The DCS RNA element has been used to clone trans-acting factors that may bind to brPTB in a manner analogous to the proposed Nova–brPTB interaction to regulate neuronal splicing. For example, a complex of at least six proteins cross links to DCS RNA, including a positive acting factor related to Nova by the presence of multiple hnRNPK homology-type RNA binding domains, termed KSRP (23). In addition, the same brPTB identified here recently was shown to bind DCS RNA as part of a larger complex including KSRP and hnRNP H and to exhibit weak repression of n-src splicing (V. Markovtsov and D. Black, personal communication). These observations and our observations on Nova and brPTB suggest that protein–RNA complexes acting to regulate splice site selection may include brPTB, and that brPTB acts as an antagonist of neuron-specific splicing mediated by positive acting factors.
In conclusion, this work extends our understanding of the mechanism by which neurons differentially regulate alternative splicing. We suggest that the function of the neuron-specific protein Nova to promote exon inclusion can be modulated by interaction with additional proteins. brPTB is not strictly neuron specific as is the Nova protein: brPTB also is expressed in glial cells, and to a lesser but significant extent, in cells outside of the nervous system, particularly the heart and testis. Nonetheless, the existence of a brain-enriched protein interacting with a strictly neuron-specific protein suggests a layer of neuronal regulation different from a model in which tissue-specific alternative splicing is regulated by varying levels or modifications of general splicing factors.
Acknowledgments
We are grateful to Dr. Titia de Lange for generously providing yeast two-hybrid reagents, TRF2 fusion protein, and pcDNAflag vector; Dr. Doug Black for anti-PTB antibody and for communicating results before publication; Dr. Joan Steitz for anti-HuA antibody; Dr. T. Shishido for pEBG vector; and to members of our laboratory, particularly Kirk Jensen and Jennifer Darnell, for helpful discussions. We thank Nathaniel Heintz for critical reading of the manuscript. This work was supported by the National Institutes of Health (RO1 NS34389), an Irma T. Hirschl Career Scientist Award (R.B.D.), National Research Service Award Training Grant CA 09673–18 (Y.Y.L.Y.), the Ataxia Telangiectasia Children's Project (H.J.O.), and the Cornell/Rockefeller/Sloan-Kettering Tri-Institutional MD/Ph.D. program (A.D.P.).
Abbreviations
- PTB
polypyrimidine tract-binding protein
- brPTB
brain-enriched PTB
- GlyRα2
glycine receptor α2
- POMA
paraneoplastic opsoclonus-myoclonus ataxia
- CGRP
calcitonin gene-related peptide
- hnRNP
heterogeneous nuclear ribonucleoprotein
- GST
glutathione S-transferase
- RRM
RNA recognition motif
- E(n)
embryonic day
- DCS
downstream control sequence
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF095718).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.110128397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.110128397
References
- 1.Ehlers M D, Tingley W G, Huganir R L. Science. 1995;269:1734–1737. doi: 10.1126/science.7569904. [DOI] [PubMed] [Google Scholar]
- 2.Ehlers M D, Fung E T, O'Brien R J, Huganir R L. J Neurosci. 1998;18:720–730. doi: 10.1523/JNEUROSCI.18-02-00720.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sommer B, Keinanen K, Verdoorn T A, Wisden W, Burnashev N, Herb A, Kohler M, Takagi T, Sakman B, Seeburg P H. Science. 1990;249:1580–1585. doi: 10.1126/science.1699275. [DOI] [PubMed] [Google Scholar]
- 4.Hollmann M, Boulter J, Maron C, Beasley L, Sullivan J, Pecht G, Heinemann S. Neuron. 1993;10:943–954. doi: 10.1016/0896-6273(93)90209-a. [DOI] [PubMed] [Google Scholar]
- 5.Tsim K W, Ruegg M A, Escher G, Kroger S, McMahan U J. Neuron. 1992;8:677–689. doi: 10.1016/0896-6273(92)90089-v. [DOI] [PubMed] [Google Scholar]
- 6.Lin C L G, Bristol L A, Jin L, Dykes-Hoberg M, Crawford T, Clawson L, Rothstein J D. Neuron. 1998;20:589–602. doi: 10.1016/s0896-6273(00)80997-6. [DOI] [PubMed] [Google Scholar]
- 7.Jensen K B, Dredge B K, Stefani G, Zhong R, Buckanovich R J, Okano H J, Yang Y Y, Darnell R B. Neuron. 2000;25:359–371. doi: 10.1016/s0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 8.Fischer U, Liu Q, Dreyfuss G. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- 9.Amara S G, Jonas V, Rosenfeld M G, Ong E S, Evans R. Nature (London) 1982;298:240–244. doi: 10.1038/298240a0. [DOI] [PubMed] [Google Scholar]
- 10.Emeson R B, Hedjran F, Yeakley J M, Guise J W, Rosenfeld M G. Nature (London) 1989;341:76–80. doi: 10.1038/341076a0. [DOI] [PubMed] [Google Scholar]
- 11.Lou H, Helfman D M, Gagel R F, Berget S M. Mol Cell Biol. 1999;19:78–85. doi: 10.1128/mcb.19.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Black D L. Cell. 1992;69:795–807. doi: 10.1016/0092-8674(92)90291-j. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L, Ashiya M, Sherman T G, Grabowski P J. RNA. 1996;2:682–698. [PMC free article] [PubMed] [Google Scholar]
- 14.Horowitz D S, Krainer A R. Trends Genet. 1994;10:100–106. doi: 10.1016/0168-9525(94)90233-x. [DOI] [PubMed] [Google Scholar]
- 15.Caceres J F, Stamm S, Helfan D M, Krainer A R. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 16.Colwill K, Pawson T, Andrews B, Prasad J, Manley J L, Bell J C, Duncan P I. EMBO J. 1996;15:265–275. [PMC free article] [PubMed] [Google Scholar]
- 17.Petersen-Mahrt S K, Estmer C, Ohrmalm C, Matthews D A, Russell W C, Akusjarvi G. EMBO J. 1999;18:1014–1024. doi: 10.1093/emboj/18.4.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lou H, Gagel R F, Berget S M. Genes Dev. 1996;10:208–219. doi: 10.1101/gad.10.2.208. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L, Liu W, Grabowski P J. RNA. 1999;5:117–130. doi: 10.1017/s1355838299981530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan R C, Black D L. Mol Cell Biol. 1997;17:4667–4676. doi: 10.1128/mcb.17.8.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grabowski P J. Cell. 1998;92:709–712. doi: 10.1016/s0092-8674(00)81399-9. [DOI] [PubMed] [Google Scholar]
- 22.Chou M Y, Rooke N, Turck C W, Black D L. Mol Cell Biol. 1999;19:69–77. doi: 10.1128/mcb.19.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Min H, Turck C W, Nikolic J M, Black D L. Genes Dev. 1997;11:1023–1036. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- 24.Koushika S P, Soller M, White K. Mol Cell Biol. 2000;20:1836–1845. doi: 10.1128/mcb.20.5.1836-1845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grabowski P J. Neuron. 2000;25:254–256. doi: 10.1016/s0896-6273(00)80888-0. [DOI] [PubMed] [Google Scholar]
- 26.Buckanovich R J, Darnell R B. Mol Cell Biol. 1997;17:3194–3201. doi: 10.1128/mcb.17.6.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Y Y L, Yin G L, Darnell R B. Proc Natl Acad Sci USA. 1998;95:13254–13259. doi: 10.1073/pnas.95.22.13254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jensen, K. B., Musunuru, K., Lewis, H. A., Burley, S. K. & Darnell, R. B. (2000) Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 29.Lewis H A, Musunuru K, Jensen K B, Edo C, Chen H, Darnell R B, Burley S K. Cell. 2000;100:323–332. doi: 10.1016/s0092-8674(00)80668-6. [DOI] [PubMed] [Google Scholar]
- 30.Buckanovich R J, Posner J B, Darnell R B. Neuron. 1993;11:657–672. doi: 10.1016/0896-6273(93)90077-5. [DOI] [PubMed] [Google Scholar]
- 31.Okano H J, Park W Y, Corradi J P, Darnell R B. Genes Dev. 1999;13:2087–2098. doi: 10.1101/gad.13.16.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buckanovich R J, Yang Y Y, Darnell R B. J Neurosci. 1996;16:1114–1122. doi: 10.1523/JNEUROSCI.16-03-01114.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perez I, Lin C H, McAfee J G, Patton J G. RNA. 1997;3:764–778. [PMC free article] [PubMed] [Google Scholar]
- 34.Lin C-H, Patton J. RNA. 1995;1:234–245. [PMC free article] [PubMed] [Google Scholar]
- 35.Singh R, Valcarcel J, Green M R. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 36.Valcarcel J, Gebauer F. Curr Biol. 1997;7:R705–R708. doi: 10.1016/s0960-9822(06)00361-7. [DOI] [PubMed] [Google Scholar]
- 37.Gil A, Sharp P A, Jamison S F, Garcia-Blanco M A. Genes Dev. 1991;5:1224–1236. doi: 10.1101/gad.5.7.1224. [DOI] [PubMed] [Google Scholar]
- 38.Ashiya M, Grabowski P J. RNA. 1997;3:996–1015. [PMC free article] [PubMed] [Google Scholar]
- 39.Ghetti A, Pinol-Roma S, Michael W M, Morandi C, Dreyfuss G. Nucleic Acids Res. 1992;20:3671–3678. doi: 10.1093/nar/20.14.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valcarcel J, Singh R, Zamore P D, Green M R. Nature (London) 1993;362:171–175. doi: 10.1038/362171a0. [DOI] [PubMed] [Google Scholar]
- 41.Southby J, Gooding C, Smith C W. Mol Cell Biol. 1999;19:2699–2711. doi: 10.1128/mcb.19.4.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Modafferi E F, Black D L. RNA. 1999;5:687–706. doi: 10.1017/s1355838299990155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Modafferi E F, Black D L. Mol Cell Biol. 1997;17:6537–6545. doi: 10.1128/mcb.17.11.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fan X C, Steitz J A. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]