Abstract
Chromosomal imbalances represent an important mechanism in cancer progression. A clear association between DNA copy‐number aberrations and prognosis has been found in a variety of tumours. Comparative genomic hybridisation studies have detected copy‐number increases affecting chromosome 6p in several types of cancer. A systematic analysis of large tumour cohorts is required to identify genomic imbalances of 6p that correlate with a distinct clinical feature of disease progression. Recent findings suggest that a central part of the short arm of chromosome 6p harbours one or more oncogenes directly involved in tumour progression. Gains at 6p have been associated with advanced or metastatic disease, poor prognosis, venous invasion in bladder, colorectal, ovarian and hepatocellular carcinomas. Copy number gains of 6p DNA have been described in a series of patients who presented initially with follicle centre lymphoma, which subsequently transformed to diffuse large B cell lymphoma. Melanoma cytogenetics has consistently identified aberrations of chromosome 6, and a correlation with lower overall survival has been described. Most of the changes observed in tumours to date map to the 6p21–p23 region, which encompasses approximately half of the genes on all of chromosome 6 and one third of the number of CpG islands in this chromosome. Analyses of the genes that cluster to the commonly amplified regions of chromosome 6p have helped to identify a small number of molecular pathways that become deregulated during tumour progression in diverse tumour types. Such pathways offer promise for new treatments in the future.
Abnormalities of chromosome number leading to aneuploidy are the most common cytogenetic aberrations in cancer. Both genomic and cytogenetic analyses of different types of tumours have shown recurrent DNA copy‐number increases associated with progression. A clear association between DNA copy‐number aberrations and prognosis has been found in a variety of tumours.1
Partial or whole gain of 6p is much more common than loss. Molecular cytogenetic analysis of chromosome 6p has shown three modes of 6p gain:
1. The most frequent mechanism for generating 6p gain is the formation of an isochromosome, in which centromeric misdivision leads to triplication of 6p and monosomy of 6q;2
2. Acquisition of an unbalanced chromosome translocation can also lead to partial or complete 6p gain; and
3. Focal amplification of a discrete region of 6p typically involves a much larger copy number increase (>5‐fold).
The last mechanism of gain may involve the formation of double minute chromosomes or homogeneous staining regions.
Genomic imbalance can be detected and mapped by comparative genomic hybridisation (CGH). This method has been used to detect copy‐number increases on chromosome 6p in several human neoplasias,3,4,5,6,7 suggesting that this region may contain genes involved directly or indirectly in the pathogenesis and pathways of cancers. This review focuses on gains and amplifications that have been established to be recurrent for chromosome 6p in different classes of solid tumours (fig 1 and table 1), with emphasis on the most commonly amplified genomic interval of 6p21–p23. Such observations are strongly suggestive that this central part of the short arm of chromosome 6p harbours one or more oncogenes directly involved in tumour progression.
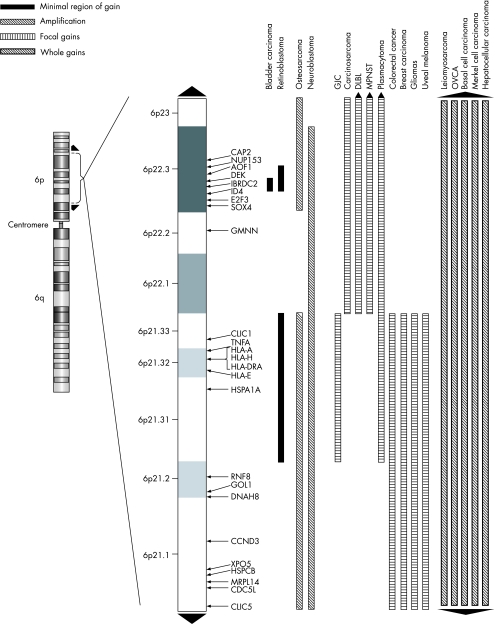
Figure 1 A map of the 6p21–p23 cytobands showing regions of gains and amplifications for specific tumours with genes that were previously published. The black up arrow indicates gain/amplification to the pter and the black down arrow indicates gain/amplification to the centromere. The solid black bar indicates the minimal region of gain; the white bar with black hashed lines indicates a region of amplification; the white bar with black vertical lines indicates a focal gain; and the back bar with white hashed lines indicates a region of gain for the whole 6p arm.
Table 1 Tumours with 6p21–p23 gain or amplification.
| Type of tumour | Number of tumours | Tumours with gain in 6p21–p23 (%) | Tumours with amplification in 6p21–p23 (%) |
|---|---|---|---|
| Carcinomas* | |||
| Hepatocellular carcinoma | 409 | 22.61 | 0.20 |
| Merkel cell carcinoma | 48 | 27.10 | 2.10 |
| Basal cell carcinoma | 16 | 40.30 | 0.00 |
| Ovarian serous carcinoma | 56 | 28.60 | 0.00 |
| Transitional cell carcinoma | 133 | 9.68 | 0.51 |
| Lymphoid tumours | |||
| Large B cell lymphoma | 360 | 8.36 | 0.07 |
| Plasmacytoma | 21 | 22.76 | 0.00 |
| Sarcomas | |||
| Osteosarcoma | 137 | 33.19 | 3.39 |
| Malignant peripheral nerve sheath | 70 | 23.93 | 0.00 |
| Leiomyosarcoma | 136 | 9.98 | 1.64 |
| Melanomas | 91 | 31.78 | 0.00 |
| Retinoblastoma | 133 | 39.36 | 10.50 |
| Glioblastoma | 108 | 4.60 | 0.90 |
| Neuroblastoma | 303 | 22.10 | 0.17 |
| Carcinosarcoma | 23 | 21.70 | 6.31 |
*This site does not currently allow the determination of the level of imbalance in a subset of breast and colon carcinomas.
Summary of genomic imbalance datasets from Progenetix (http://www.progenetix.com).
Carcinomas
In carcinomas, there is a general association between gains of 6p and tumour progression. In bladder carcinomas, gains of 6p have been reported in invasive bladder tumours, present in 7–55% of patients.8,9,10,11,12,13,14,15 Significant associations have been found between 6p22 gain and high histological grade,9,16 high tumour cell proliferative activity14 and metastases at initial presentation,16 suggesting that acquisition of 6p gain may confer a growth advantage and lead to disease progression.
As in advanced stages of transitional cell carcinomas in vivo, gains and amplifications of 6p were often found in transitional cell carcinoma lines with a common region of amplification at the 6p21.3–p23 locus.17 Over‐representation of 6p22 was the only individual change that was significantly linked to high tumour grade in a series of 54 pT1 urinary bladder carcinomas. The risk of progression was significantly associated with the number of deletions, but not with the number of gains.18 Moreover, gain at chromosome 6p22–p23 was noted in two patients with metastasis at diagnosis among pTa, pT1 and pT2–T4G3 tumours and gains of 6p and 10p were more frequent in pT1G3 in comparison to pT1G2 tumours.16 In high‐stage lesions 6p22 gains occurred as late events,19 and this gain was the only CGH change strongly associated to a high proliferative activity in the tumour cells and independent of grade and stage of the tumour.14
In small‐cell carcinomas, which represent a rare histological subtype of urinary bladder cancer, gains of DNA sequences were most prevalent at 8q, 5p, 6p and 20q.20 High‐level amplifications were detected most often at 6p22.3 (E2F3) and at four other genomic locations in 41 primary bladder tumours. Interestingly, there was a significant complementary association between gain of cyclin D1 at 11q13 and gain of E2F3, although there was no significant relationship between copy number changes and tumour stage or grade.21 Using quantitative multiplex polymerase chain reaction to study DNA from 59 bladder tumours, the focal region of genomic gain on 6p22 was mapped to a minimal region, spanning a genomic distance of 0.5 Mb.15 The E2F3 gene has been recently implicated as the target of 6p22 genomic gain in bladder cancer.22,23 In a study using bladder tumour‐derived cell lines, NM_017774 showed an expression related to the 6p22.3 amplicon.24 A recent paper reported overexpression of ID4 gene, which maps between E2F3 and DEK, in bladder cancer.25
Increased copy number of chromosome arm 6p has been associated with advanced stages of colorectal cancer (Dukes' stage D) and metastasis.7 In a recent CGH study on liver metastasis of colorectal carcinoma (CRC), gain in 6p21 was found in 41% of the patients. Taken together, both these observations suggest that acquisition of 6p gain may be contributing to progression in colorectal cancer, and that part of the short arm of this chromosome might harbour one or more oncogenes.26 A subtractive CGH analysis using paired samples from 20 patients with CRC with primary tumours and synchronous or metachronous liver metastases detected frequent gains in DNA copy number at 6p. Analysis of 11 CRC cell lines using array‐based CGH showed one 6p candidate gene, cyclin D3, which was significantly up regulated by quantitative reverse transcriptase‐polymerase chain reaction in liver‐metastatic lesions compared with primary lesions.
DNA amplification data from CGH and serial analyses of gene expression showed several chromosomal arms—for example, that chromosome 6 had frequent DNA amplifications that showed frequent changes in gene expression in gastroesophageal junction carcinomas. Despite the relatively large DNA amplification regions, overexpressed genes often mapped and clustered to small chromosomal regions at early‐replicating bands such as 1q21.3 (nine genes), 6p21.3 (five genes) and 17q21 (eight genes).27
CGH studies have shown that the most frequent gains in hepatocellular carcinoma (HCC) occurred at 8q, 1q, 3q, 6p and 17q.28,29,30 Chromosomal aberrations investigated in HCC cell lines that had hepatitis B virus integration showed chromosomal gains at 6p.31 Amplifications at 1q and 6p appeared to be independent factors for venous invasion in HCC.32 Significantly, a recent study described that physical clusters of two or more genes within predefined distance thresholds were detected non‐randomly on chromosomal regions 1q, 6p, 8q, 20q and Xq, indicating that HCC‐related genes are physically clustered at specific chromosomal locations.33 A previous study that mapped genes overexpressed in HCC to chromosomal locations found 13 regions of frequent cytogenetic change, including 6p gain.34 A recent explorative CGH meta‐analysis of HCC showed that the most marked amplifications were present at 1q (57.1%), 8q (46.6%), 6p (22.3%) and 17q (22.2%).35
In breast carcinomas, genomic imbalances with statistical significance were observed in poorly differentiated (G3) and oestrogen receptor negative tumours, including one region of 6p gain.36 One of the most frequent chromosomal aberration found in patients with high‐risk stage II/III breast cancer was 6p gain.37 Gain on 6p21 was identified in 14 of 31 (45%) formalin‐fixed and paraffin‐wax‐embedded primary advanced breast tumours analysed by microarray‐based CGH.
Recurrent gains of chromosome 6 have been detected in Merkel cell carcinomas.6,38 Although no significant correlation between genomic aberrations and clinicopathological factors was shown, primary tumours expressing changes in DNA were predominantly distinguished in large Merkel cell carcinomas, and risk of metastatic dissemination was threefold compared with tumours without chromosomal changes.38
In basal cell carcinoma (BCC) of the skin, recurrent changes were observed in 47% of the patients at 6p21‐pter. Interestingly, it was found that regional gain of 9p was strongly associated with 6p gain; however, no correlation between this association and clinical or histological appearance was observed in these patients.5 These results correlate with a previous study that identified clonal trisomy of chromosome 6 in BCC direct preparations by conventional cytogenetics and fluorescent in situ hybridisation analysis. No correlations between the BCC cytogenetic results and clinical parameters such as site, age, sex and recurrence rate have been shown.39
Gains of 6p were more common in stage IIIc than in stage IIIa+b ovarian serous papillary adenocarcinomas.40 A study of ovarian epithelial tumours using CGH found that high‐grade serous carcinomas had more than twice as much chromosomal imbalance as low‐grade serous carcinomas and also had pronounced changes. Overlapping changes occurring in serous and non‐serous carcinomas were gains on 3q and 6p, as well as losses on 4q. Among other chromosomal imbalances, 6p gain was associated with poor prognosis of ovarian carcinomas,41 and 6p22.1–p21.2 was one of several regions associated with acquisition to drug resistance in serous carcinomas.42
Lymphoid tumours
Amplification of the 6p21 locus has been documented in non‐Hodgkin's lymphoma. Among 27 specimens of patients with plasmacytoma, 25% of the specimens showed gain of 6p, with a minimal overlapping region at 6p21.3‐pter.43 Acquisition of 6p has a significantly higher frequency in large B cell tumours, and it was an independent prognostic factor with losses of 11q21–q23.1 and 17p.44 In another study of diffuse large B cell lymphoma with primary tumours and recurrent tumours excised after chemotherapy or radiotherapy, 21% showed 6p gain with minimal common regions at 6p22‐pter. High‐level amplifications were observed at 6p23‐pter and occurred only in different recurrent tumours.45 In a series of 23 patients, 21 showed genomic changes, with a mean value of 11 aberrations/sample (range 2–29). Among other regions, the minimal common region of 6p22‐pter was most frequently gained.46 In non‐Hodgkin's lymphomas, it has been shown and suggested that gains on 6p among other changes may be important in the transformation from low‐grade to high‐grade disease.47 Recently, over‐representation of 6p12.3–p21 among other genomic aberrations was reported in samples from patients who presented initially with follicle centre lymphoma, which subsequently transformed to diffuse large B cell lymphoma as measured by array CGH.48 A subset of genes that mapped to locations of gain or amplification exhibited coordinated increase or decrease of expression.49
Melanomas
Melanoma cytogenetics has consistently identified aberrations of chromosome 6 as the most common change. CGH analyses have identified recurrent chromosomal abnormalities at several locations including 6p gain in choroid and ciliary body melanomas (30–85%),50,51,52,53,54,55,56 in acral and superficial,57,58 and in sinonasal mucosal melanomas (93%). Gain of 6p occurred frequently in combination with a loss of copy number or a normal copy number of the opposite chromosome arm, suggesting isochromosome formation.59
Changes on 6p, detected by microsatellite analysis in 15 of 30 (50%) patients with uveal melanoma, were preferentially observed in tumours without chromosome 3 changes.53 Another study of 12 patients with uveal melanomas showed under‐representation of chromosome 3 exclusively in tumours with ciliary body involvement, whereas anomalies of chromosome 6 were described only in pure choroidal melanomas.60 Recently, an inverse relationship between monosomy 3 and gain of 6p has been detected in uveal and in cutaneous melanomas.56,61 A CGH study using 16 primary and 12 metastatic melanoma specimens showed that the pattern of chromosomal aberrations was similar in the two subgroups. The most frequent change was gain of 6p in 63% of primary and in 50% of metastatic tumours.62 Another CGH study has shown that tumours with 6p or 1q gain had lower overall survival in comparison to those without gain, thus implying that acquisition of 6p or 1q gain may confer prognostic differences.63
Sarcomas
Gain of 6p is one of the most frequent chromosomal change in osteosarcomas.64,65,66,67,68 A CGH study with 31 primary high‐grade osteosarcomas found gain of 6p12–p21.3 in 10 (32%) patients. Other frequent DNA sequence copy‐number changes were described, without prognostic significance, affecting chromosomal regions not previously associated with tumorigenesis of osteosarcomas.69 The same authors described an association between copy‐number increase at 6p and reduced overall and distant disease‐free survival.70 In another study chromosome 6 was involved in 37 rearrangements, of which nine were mapped to 6p12–p21.67 The same panel of 25 tumours showed amplification at 6p12–p21 in seven tumours. Three non‐overlapping BAC clones were identified within a 6p amplicon and several genes present in these clones. In this study, amplification of 6p12–p21 appeared to be significant and early event in the pathogenesis of osteosarcomas as it was observed in all specimen types: biopsy, definitive surgery and metastatic lesion. These chromosomal bands were also involved in the chromosomal rearrangements identified by SKY. These findings suggested the presence of oncogenes at 6p12–p21 whose overexpression may lead to the malignant phenotype in osteosarcomas. On the basis of the combined array CGH and fluorescent in situ hybridisation analysis, the same group suggested CDC5L, HSPCB, NFKBIE, HGNC and MRPL14 as the target genes from the 6p12–p21 amplicon (fig 1).68 A microarray CGH study showed that 6 of 9 osteosarcomas with a 6p gain contained the common region of gain of band 6p12.66 Focal amplifications occurred at 6p22.3–p25.1, 6p21 and 6p12; the broader regions of gain seen at 6p22.3–p24.2 and 6p21.2–p21.31 were consistent with a potential role for the breakage fusion bridge cycle in generation of amplification of 6p in one osteosarcoma cell line.71 In all, 10 of 23 patients showed gain but not high‐level amplifications of 6p22‐pter in a CGH study of malignant peripheral nerve sheath tumours, although this change did not have any prognostic relevance.72 High‐level gain of DNA copy number was detected in 6p and 17p in a study of 17 leiomyosarcomas.73
Retinoblastoma
Gains of the entire chromosome arm 6p have been reported very frequently in retinoblastoma.2,4,74,75,76,77 Interestingly, by far the most frequent mechanism of gain is by acquisition of an isochromosome of 6p.2,74,78,79,80 Karyotype studies showed small additional fragments of 6p, often including the distal segment 6p25–p22. This led to the assumption that increased copy number of genes localised in this region of 6p may confer a selective growth advantage to retinoblastoma cells.81 One of the most frequent imbalance observed in a series of primary retinoblastoma (13/24) cases was 6p gain, and no evident correlation was found between any of the imbalances identified and either the differentiation or the histoprognostic risk.74 Gains at 6p were also identified in another CGH study in 11 (42.3%) tumours, suggesting that 6p22–p25 might be a candidate chromosome region for containing presumptive retinoblastoma growth‐promoting genes.75 Subsequent CGH results have shown that the common region of gain maps to chromosome band 6p22.4 Using quantitative multiplex polymerase chain reaction, the same group narrowed down the region to a 0.6‐Mb region spanning the UniSTS markers X64229 and WI‐19208.82 Recently, it was found that only three of seven genes, DEK, NUP153 and E2F3, in this region analysed using real‐time reverse transcriptase‐polymerase chain reaction are overexpressed in retinoblastoma tumours and cell lines with documented genomic gain of 6p22. E2F3 and DEK mRNA overexpression was always associated with protein overexpression of primary tumours relative to the adjacent normal retina. E2F3 was strongly expressed in actively proliferating cells, whereas DEK was overexpressed in all tumour cells. It was concluded that both E2F3 and DEK were promising targets of the 6p22 genomic gain in retinoblastoma.83 Another study narrowed the minimal region of gain on 6p to the band 6p22.3 using a panel of retinoblastomas and retinoblastoma cell lines. The data indicated that the minimal region of chromosomal imbalance in 6p22.3 includes the NUP 153, KIF 13A, TPMT, DEK, ID4, E2F3 and TTRAP genes. Copy number gains were most consistently identified in the region containing the DEK and E2F3, thus suggesting these genes as potential targets of 6p gains.84 A high‐resolution matrix‐CGH analysis performed on a series of 17 primary retinoblastomas and 4 retinoblastoma cell lines showed on 6p a minimally overlapping gained region at 6p21.31–p21.33 that was 2.31 Mb in size and suggested the tumour necrosis factor α as a potential candidate gene.77
Other tumours
One frequent over‐representation was seen at 6p (minimal region of overlap in six patients, 6p22–p23) in a CGH analysis of 19 uterine and extrauterine carcinosarcomas. Amplifications of 6p were detected in two patients.85 Amplification and overexpression of cyclin D3 gene located in the 6p21 region has been detected in glioblastoma cell lines.86 It was suggested that the increased amounts of cyclin D3 caused by gene amplification could be involved in the development or progression of that glioblastoma.46 Common gains observed in 29 neuroblastomas were on chromosomes 7, 6 and 18. High‐level amplifications were detected at 2p23–p25 (MYCN region), at 4q33–q35 and at 6p11–p22.87 A CGH analysis of clinically non‐progressing stage 4s neuroblastomas showed a high rate of whole‐chromosome aberrations (73.4%), with an over‐representation of chromosomes 2, 6, 7, 12, 13, 17 and 18 and an under‐representation of chromosomes 3, 4, 11 and 14.88
Conclusions and future prospects
CGH studies have identified frequent amplifications and rearrangements involving 6p in a wide variety of different types of tumours. These observations suggest that genes more generally associated with tumour progression may map to this genomic location. In all, 624 genes are seen in the 33‐Mb region of 6p21–23, which represents about half of the genes on whole chromosome 6 (table 2). Several lines of evidence presented in this review implicate that one or more putative oncogenes may underlie the association between gain of 6p and tumour progression. The genomic architecture of the 6p region is noteworthy. The number of CpG islands in the 6p21–p23 region corresponds to one third of the number of CpG islands in chromosome 6. Recently, it has been shown that gene density, guanine‐cytosine content and replication timing all co‐correlate strongly with transcriptional activity, thus confirming that expressed genes tend to be replicated early in the S phase.89
Table 2 Genomic features of 6p21–p23.
| Genomic features of:* | ||
|---|---|---|
| 6p21–p23 region | Chromosome 6 | |
| Size of the region (Mb) | 33 | 171 |
| Number of genes | 624 | 1394 |
| Repeats | 59 854 | 265 300 |
| CpG island | 5508 | 17 622 |
| Variations (SNPs) | 141 464 | 601 204 |
| OMIM morbid | 62 | 121 |
| Mitelman breakpoint elements | 78 | 665 |
SNP, single nucleotide polymorphism.
*Data acquired from http://www.ncbi.nlm.nih.gov/mapview/.
In solid tumours, significantly high levels of chromosome abnormalities have been detected, but distinction between critical and consequential events has been a major challenge. It has been proposed that correlations between imbalances would show possible cytogenetic pathways. Some tumours described in this review present concordant genomic changes associated with 6p gain. In the future, the application of systematic higher resolution array CGH studies will help to show correlations and linked behaviour among genes that cluster to chromosome 6p and other specific regions, thus unravelling different pathways for an improved understanding of tumour progression. Analysis of the genes that cluster to the commonly amplified regions of chromosome 6p has helped to identify a small number of molecular pathways that become deregulated during tumour progression in diverse tumour types.21,61 Identification of such pathways will offer promise for new approaches to treatment in the future.
Take‐home messages
A clear association between DNA copy‐number aberrations and prognosis has been found in a variety of tumours. CGH studies have identified frequent amplifications and rearrangements involving 6p in different types of tumours.
Several lines of evidence presented in this review suggest that a part of the short arm of chromosome 6p harbours one or more oncogenes directly involved in tumour progression.
Some tumours described in this review present concordant genomic changes associated with 6p gain showing possible cytogenetic pathways. The application of systematic array CGH studies can help to show correlations and linked behaviour among genes that cluster to chromosome 6p and other specific regions. Such an approach will unravel different pathways for an improved understanding of tumour progression.
Abbreviations
BCC - basal cell carcinoma
CGH - comparative genomic hybridisation
CRC - colorectal carcinoma
HCC - hepatocellular carcinoma
Footnotes
Competing interests: None declared.
Funding: GdCS was supported by CAPES—Ministry of Education, Brazil. Financial support for this project was provided by the National Cancer Institute of Canada (NCIC), with funds from the Canadian Cancer Society.
References
- 1.Pinkel D, Albertson D G. Array comparative genomic hybridization and its applications in cancer. Nat Genet 200537(Suppl)S11–S17. [DOI] [PubMed] [Google Scholar]
- 2.Squire J, Phillips R A, Boyce S.et al Isochromosome 6p, a unique chromosomal abnormality in retinoblastoma: verification by standard staining techniques, new densitometric methods, and somatic cell hybridization. Hum Genet 19846646–53. [DOI] [PubMed] [Google Scholar]
- 3.Knuutila S, Bjorkqvist A M, Autio K.et al DNA copy number amplifications in human neoplasms: review of comparative genomic hybridization studies. Am J Pathol 19981521107–1123. [PMC free article] [PubMed] [Google Scholar]
- 4.Chen D, Gallie B L, Squire J A. Minimal regions of chromosomal imbalance in retinoblastoma detected by comparative genomic hybridization. Cancer Genet Cytogenet 200112957–63. [DOI] [PubMed] [Google Scholar]
- 5.Ashton K J, Weinstein S R, Maguire D J.et al Molecular cytogenetic analysis of basal cell carcinoma DNA using comparative genomic hybridization. J Invest Dermatol 2001117683–686. [DOI] [PubMed] [Google Scholar]
- 6.Larramendy M L, Koljonen V, Bohling T.et al Recurrent DNA copy number changes revealed by comparative genomic hybridization in primary Merkel cell carcinomas. Mod Pathol 200417561–567. [DOI] [PubMed] [Google Scholar]
- 7.Al‐Mulla F, Keith W N, Pickford I R.et al Comparative genomic hybridization analysis of primary colorectal carcinomas and their synchronous metastases. Genes Chromosomes Cancer 199924306–314. [DOI] [PubMed] [Google Scholar]
- 8.Bruch J, Wohr G, Hautmann R.et al Chromosomal changes during progression of transitional cell carcinoma of the bladder and delineation of the amplified interval on chromosome arm 8q. Genes Chromosomes Cancer 199823167–174. [DOI] [PubMed] [Google Scholar]
- 9.Richter J, Beffa L, Wagner U.et al Patterns of chromosomal imbalances in advanced urinary bladder cancer detected by comparative genomic hybridization. Am J Pathol 19981531615–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hovey R M, Chu L, Balazs M.et al Genetic alterations in primary bladder cancers and their metastases. Cancer Res 1998583555–3560. [PubMed] [Google Scholar]
- 11.Koo S H, Kwon K C, Ihm C H.et al Detection of genetic alterations in bladder tumors by comparative genomic hybridization and cytogenetic analysis. Cancer Genet Cytogenet 199911087–93. [DOI] [PubMed] [Google Scholar]
- 12.Czerniak B, Li L, Chaturvedi V.et al Genetic modeling of human urinary bladder carcinogenesis. Genes Chromosomes Cancer 200027392–402. [PubMed] [Google Scholar]
- 13.Simon R, Burger H, Semjonow A.et al Patterns of chromosomal imbalances in muscle invasive bladder cancer. Int J Oncol 2000171025–1029. [DOI] [PubMed] [Google Scholar]
- 14.Tomovska S, Richter J, Suess K.et al Molecular cytogenetic alterations associated with rapid tumor cell proliferation in advanced urinary bladder cancer. Int J Oncol 2001181239–1244. [DOI] [PubMed] [Google Scholar]
- 15.Evans A J, Gallie B L, Jewett M A.et al Defining a 0.5‐mb region of genomic gain on chromosome 6p22 in bladder cancer by quantitative‐multiplex polymerase chain reaction. Am J Pathol 2004164285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prat E, Bernues M, Caballin M R.et al Detection of chromosomal imbalances in papillary bladder tumors by comparative genomic hybridization. Urology 200157986–992. [DOI] [PubMed] [Google Scholar]
- 17.Bruch J, Schulz W A, Haussler J.et al Delineation of the 6p22 amplification unit in urinary bladder carcinoma cell lines. Cancer Res 2000604526–4530. [PubMed] [Google Scholar]
- 18.Richter J, Wagner U, Schraml P.et al Chromosomal imbalances are associated with a high risk of progression in early invasive (pT1) urinary bladder cancer. Cancer Res 1999595687–5691. [PubMed] [Google Scholar]
- 19.Simon R, Eltze E, Schafer K L.et al Cytogenetic analysis of multifocal bladder cancer supports a monoclonal origin and intraepithelial spread of tumor cells. Cancer Res 200161355–362. [PubMed] [Google Scholar]
- 20.Terracciano L, Richter J, Tornillo L.et al Chromosomal imbalances in small cell carcinomas of the urinary bladder. J Pathol 1999189230–235. [DOI] [PubMed] [Google Scholar]
- 21.Veltman J A, Fridlyand J, Pejavar S.et al Array‐based comparative genomic hybridization for genome‐wide screening of DNA copy number in bladder tumors. Cancer Res 2003632872–2880. [PubMed] [Google Scholar]
- 22.Feber A, Clark J, Goodwin G.et al Amplification and overexpression of E2F3 in human bladder cancer. Oncogene 2004231627–1630. [DOI] [PubMed] [Google Scholar]
- 23.Oeggerli M, Tomovska S, Schraml P.et al E2F3 amplification and overexpression is associated with invasive tumor growth and rapid tumor cell proliferation in urinary bladder cancer. Oncogene 2004235616–5623. [DOI] [PubMed] [Google Scholar]
- 24.Hurst C D, Fiegler H, Carr P.et al High‐resolution analysis of genomic copy number alterations in bladder cancer by microarray‐based comparative genomic hybridization. Oncogene 2004232250–2263. [DOI] [PubMed] [Google Scholar]
- 25.Wu Q, Hoffmann M J, Hartmann F H.et al Amplification and overexpression of the ID4 gene at 6p22.3 in bladder cancer. Mol Cancer 2005416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diep C B, Parada L A, Teixeira M R.et al Genetic profiling of colorectal cancer liver metastases by combined comparative genomic hybridization and G‐banding analysis. Genes Chromosomes Cancer 200336189–197. [DOI] [PubMed] [Google Scholar]
- 27.Koon N, Zaika A, Moskaluk C A.et al Clustering of molecular alterations in gastroesophageal carcinomas. Neoplasia 20046143–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marchio A, Meddeb M, Pineau P.et al Recurrent chromosomal abnormalities in hepatocellular carcinoma detected by comparative genomic hybridization. Genes Chromosomes Cancer 19971859–65. [PubMed] [Google Scholar]
- 29.Lin Y W, Sheu J C, Huang G T.et al Chromosomal abnormality in hepatocellular carcinoma by comparative genomic hybridisation in Taiwan. Eur J Cancer 199935652–658. [DOI] [PubMed] [Google Scholar]
- 30.Chen Y J, Yeh S H, Chen J T.et al Chromosomal changes and clonality relationship between primary and recurrent hepatocellular carcinoma. Gastroenterology 2000119431–440. [DOI] [PubMed] [Google Scholar]
- 31.Hwang H J, Kim G J, Lee G B.et al A comprehensive karyotypic analysis on Korean hepatocellular carcinoma cell lines by cross‐species color banding and comparative genomic hybridization. Cancer Genet Cytogenet 2003141128–137. [DOI] [PubMed] [Google Scholar]
- 32.Pang A, Ng I O, Fan S T.et al Clinicopathologic significance of genetic alterations in hepatocellular carcinoma. Cancer Genet Cytogenet 20031468–15. [DOI] [PubMed] [Google Scholar]
- 33.Patil M A, Chua M S, Pan K H.et al An integrated data analysis approach to characterize genes highly expressed in hepatocellular carcinoma. Oncogene 2005243737–3747. [DOI] [PubMed] [Google Scholar]
- 34.Crawley J J, Furge K A. Identification of frequent cytogenetic aberrations in hepatocellular carcinoma using gene‐expression microarray data. Genome Biol 20023Research0075.1–Research0075.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moinzadeh P, Breuhahn K, Stutzer H.et al Chromosome alterations in human hepatocellular carcinomas correlate with aetiology and histological grade—results of an explorative CGH meta‐analysis. Br J Cancer 200592935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richard F, Pacyna‐Gengelbach M, Schluns K.et al Patterns of chromosomal imbalances in invasive breast cancer. Int J Cancer 200089305–310. [PubMed] [Google Scholar]
- 37.Seute A, Sinn H P, Schlenk R F.et al Clinical relevance of genomic aberrations in homogeneously treated high‐risk stage II/III breast cancer patients. Int J Cancer 20019380–84. [DOI] [PubMed] [Google Scholar]
- 38.Harle M, Arens N, Moll I.et al Comparative genomic hybridization (CGH) discloses chromosomal and subchromosomal copy number changes in Merkel cell carcinomas. J Cutan Pathol 199623391–397. [DOI] [PubMed] [Google Scholar]
- 39.Casalone R, Mazzola D, Righi R.et al Cytogenetic and interphase FISH analyses of 73 basal cell and three squamous cell carcinomas: different findings in direct preparations and short‐term cell cultures. Cancer Genet Cytogenet 2000118136–143. [DOI] [PubMed] [Google Scholar]
- 40.Partheen K, Levan K, Osterberg L.et al Analysis of cytogenetic alterations in stage III serous ovarian adenocarcinoma reveals a heterogeneous group regarding survival, surgical outcome, and substage. Genes Chromosomes Cancer 200440342–348. [DOI] [PubMed] [Google Scholar]
- 41.Hauptmann S, Denkert C, Koch I.et al Genetic alterations in epithelial ovarian tumors analyzed by comparative genomic hybridization. Hum Pathol 200233632–641. [DOI] [PubMed] [Google Scholar]
- 42.Bernardini M, Lee C H, Beheshti B.et al High‐resolution mapping of genomic imbalance and identification of gene expression profiles associated with differential chemotherapy response in serous epithelial ovarian cancer. Neoplasia 20057603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aalto Y, Nordling S, Kivioja A H.et al Among numerous DNA copy number changes, losses of chromosome 13 are highly recurrent in plasmacytoma. Genes Chromosomes Cancer 199925104–107. [PubMed] [Google Scholar]
- 44.Stokke T, DeAngelis P, Smedshammer L.et al Loss of chromosome 11q21–23.1 and 17p and gain of chromosome 6p are independent prognostic indicators in B‐cell non‐Hodgkin's lymphoma. Br J Cancer 2001851900–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monni O, Joensuu H, Franssila K.et al DNA copy number changes in diffuse large B‐cell lymphoma—comparative genomic hybridization study. Blood 1996875269–5278. [PubMed] [Google Scholar]
- 46.Kuchiki H, Saino M, Nobukuni T.et al Detection of amplification of a chromosomal fragment at 6p21 including the cyclin D3 gene in a glioblastoma cell line by arbitrarily primed polymerase chain reaction. Int J Cancer 200085113–116. [DOI] [PubMed] [Google Scholar]
- 47.Hough R E, Goepel J R, Alcock H E.et al Copy number gain at 12q12–14 may be important in the transformation from follicular lymphoma to diffuse large B cell lymphoma. Br J Cancer 200184499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez‐Climent J A, Alizadeh A A, Segraves R.et al Transformation of follicular lymphoma to diffuse large cell lymphoma is associated with a heterogeneous set of DNA copy number and gene expression alterations. Blood 20031013109–3117. [DOI] [PubMed] [Google Scholar]
- 49.Lossos I S, Alizadeh A A, Diehn M.et al Transformation of follicular lymphoma to diffuse large‐cell lymphoma: alternative patterns with increased or decreased expression of c‐myc and its regulated genes. Proc Natl Acad Sci USA 2002998886–8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gordon K B, Thompson C T, Char D H.et al Comparative genomic hybridization in the detection of DNA copy number abnormalities in uveal melanoma. Cancer Res 1994544764–4768. [PubMed] [Google Scholar]
- 51.Speicher M R, Prescher G, du Manoir S.et al Chromosomal gains and losses in uveal melanomas detected by comparative genomic hybridization. Cancer Res 1994543817–3823. [PubMed] [Google Scholar]
- 52.Aalto Y, Eriksson L, Seregard S.et al Concomitant loss of chromosome 3 and whole arm losses and gains of chromosome 1, 6, or 8 in metastasizing primary uveal melanoma. Invest Ophthalmol Vis Sci 200142313–317. [PubMed] [Google Scholar]
- 53.Tschentscher F, Prescher G, Zeschnigk M.et al Identification of chromosomes 3, 6, and 8 aberrations in uveal melanoma by microsatellite analysis in comparison to comparative genomic hybridization. Cancer Genet Cytogenet 200012213–17. [DOI] [PubMed] [Google Scholar]
- 54.Vajdic C M, Hutchins A M, Kricker A.et al Chromosomal gains and losses in ocular melanoma detected by comparative genomic hybridization in an Australian population‐based study. Cancer Genet Cytogenet 200314412–17. [DOI] [PubMed] [Google Scholar]
- 55.Erol N, Oner U, Artan S.et al Chromosomal abnormalities, p53 and Bcl‐2 expression and clinical outcome in choroidal melanoma. Melanoma Res 200414473–478. [DOI] [PubMed] [Google Scholar]
- 56.Hughes S, Damato B E, Giddings I.et al Microarray comparative genomic hybridisation analysis of intraocular uveal melanomas identifies distinctive imbalances associated with loss of chromosome 3. Br J Cancer 20056310–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bastian B C, Kashani‐Sabet M, Hamm H.et al Gene amplifications characterize acral melanoma and permit the detection of occult tumor cells in the surrounding skin. Cancer Res 2000601968–1973. [PubMed] [Google Scholar]
- 58.Bastian B C, Olshen A B, LeBoit P E.et al Classifying melanocytic tumors based on DNA copy number changes. Am J Pathol 20031631765–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Dijk M, Sprenger S, Rombout P.et al Distinct chromosomal aberrations in sinonasal mucosal melanoma as detected by comparative genomic hybridization. Genes Chromosomes Cancer 200336151–158. [DOI] [PubMed] [Google Scholar]
- 60.Prescher G, Bornfeld N, Friedrichs W.et al Cytogenetics of twelve cases of uveal melanoma and patterns of nonrandom anomalies and isochromosome formation. Cancer Genet Cytogenet 19958040–46. [DOI] [PubMed] [Google Scholar]
- 61.Hoglund M, Gisselsson D, Hansen G B.et al Dissecting karyotypic patterns in malignant melanomas: temporal clustering of losses and gains in melanoma karyotypic evolution. Int J Cancer 200410857–65. [DOI] [PubMed] [Google Scholar]
- 62.Balazs M, Adam Z, Treszl A.et al Chromosomal imbalances in primary and metastatic melanomas revealed by comparative genomic hybridization. Cytometry 200146222–232. [DOI] [PubMed] [Google Scholar]
- 63.Namiki T, Yanagawa S, Izumo T.et al Genomic alterations in primary cutaneous melanomas detected by metaphase comparative genomic hybridization with laser capture or manual microdissection: 6p gains may predict poor outcome. Cancer Genet Cytogenet 20051571–11. [DOI] [PubMed] [Google Scholar]
- 64.Ozaki T, Schaefer K L, Wai D.et al Genetic imbalances revealed by comparative genomic hybridization in osteosarcomas. Int J Cancer 2002102355–365. [DOI] [PubMed] [Google Scholar]
- 65.Forus A, Weghuis D O, Smeets D.et al Comparative genomic hybridization analysis of human sarcomas: II. Identification of novel amplicons at 6p and 17p in osteosarcomas. Genes Chromosomes Cancer 19951415–21. [DOI] [PubMed] [Google Scholar]
- 66.Squire J A, Pei J, Marrano P.et al High‐resolution mapping of amplifications and deletions in pediatric osteosarcoma by use of CGH analysis of cDNA microarrays. Genes Chromosomes Cancer 200338215–225. [DOI] [PubMed] [Google Scholar]
- 67.Lau C C, Harris C P, Lu X Y.et al Frequent amplification and rearrangement of chromosomal bands 6p12–p21 and 17p11.2 in osteosarcoma. Genes Chromosomes Cancer 20043911–21. [DOI] [PubMed] [Google Scholar]
- 68.Man T K, Lu X Y, Jaeweon K.et al Genome‐wide array comparative genomic hybridization analysis reveals distinct amplifications in osteosarcoma. BMC Cancer 2004445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tarkkanen M, Elomaa I, Blomqvist C.et al DNA sequence copy number increase at 8q: a potential new prognostic marker in high‐grade osteosarcoma. Int J Cancer 199984114–121. [DOI] [PubMed] [Google Scholar]
- 70.Tarkkanen M, Kiuru‐Kuhlefelt S, Blomqvist C.et al Clinical correlations of genetic changes by comparative genomic hybridization in Ewing sarcoma and related tumors. Cancer Genet Cytogenet 199911435–41. [DOI] [PubMed] [Google Scholar]
- 71.Lim G, Karaskova J, Beheshti B.et al An integrated mBAND and submegabase resolution tiling set (SMRT) CGH array analysis of focal amplification, microdeletions, and ladder structures consistent with breakage‐fusion‐bridge cycle events in osteosarcoma. Genes Chromosomes Cancer 200542392–403. [DOI] [PubMed] [Google Scholar]
- 72.Schmidt H, Wurl P, Taubert H.et al Genomic imbalances of 7p and 17q in malignant peripheral nerve sheath tumors are clinically relevant. Genes Chromosomes Cancer 199925205–211. [PubMed] [Google Scholar]
- 73.Hu J, Rao U N, Jasani S.et al Loss of DNA copy number of 10q is associated with aggressive behavior of leiomyosarcomas: a comparative genomic hybridization study. Cancer Genet Cytogenet 200516120–27. [DOI] [PubMed] [Google Scholar]
- 74.Mairal A, Pinglier E, Gilbert E.et al Detection of chromosome imbalances in retinoblastoma by parallel karyotype and CGH analyses. Genes Chromosomes Cancer 200028370–379. [PubMed] [Google Scholar]
- 75.Herzog S, Lohmann D R, Buiting K.et al Marked differences in unilateral isolated retinoblastomas from young and older children studied by comparative genomic hybridization. Hum Genet 200110898–104. [DOI] [PubMed] [Google Scholar]
- 76.Lillington D M, Kingston J E, Coen P G.et al Comparative genomic hybridization of 49 primary retinoblastoma tumors identifies chromosomal regions associated with histopathology, progression, and patient outcome. Genes Chromosomes Cancer 200336121–128. [DOI] [PubMed] [Google Scholar]
- 77.Zielinski B, Gratias S, Toedt G.et al Detection of chromosomal imbalances in retinoblastoma by matrix‐based comparative genomic hybridization. Genes Chromosomes Cancer 200543294–301. [DOI] [PubMed] [Google Scholar]
- 78.Potluri V R, Helson L, Ellsworth R M.et al Chromosomal abnormalities in human retinoblastoma. A review. Cancer 198658663–671. [DOI] [PubMed] [Google Scholar]
- 79.Horsthemke B, Greger V, Becher R.et al Mechanism of i(6p) formation in retinoblastoma tumor cells. Cancer Genet Cytogenet 19893795–102. [DOI] [PubMed] [Google Scholar]
- 80.Cano J, Oliveros O, Yunis E. Phenotype variants, malignancy, and additional copies of 6p in retinoblastoma. Cancer Genet Cytogenet 199476112–115. [DOI] [PubMed] [Google Scholar]
- 81.Gallie B L, Campbell C, Devlin H.et al Developmental basis of retinal‐specific induction of cancer by RB mutation. Cancer Res 199959(Suppl)S1731–S1735. [PubMed] [Google Scholar]
- 82.Chen D, Pajovic S, Duckett A.et al Genomic amplification in retinoblastoma narrowed to 0.6 megabase on chromosome 6p containing a kinesin‐like gene, RBKIN. Cancer Res 200262967–971. [PubMed] [Google Scholar]
- 83.Orlic M, Spencer C E, Wang L.et al Expression analysis of 6p22 genomic gain in retinoblastoma. Genes Chromosomes Cancer 20064572–82. [DOI] [PubMed] [Google Scholar]
- 84.Grasemann C, Gratias S, Stephan H.et al Gains and overexpression identify DEK and E2F3 as targets of chromosome 6p gains in retinoblastoma. Oncogene 2005246441–6449. [DOI] [PubMed] [Google Scholar]
- 85.Schulten H J, Gunawan B, Enders C.et al Overrepresentation of 8q in carcinosarcomas and endometrial adenocarcinomas. Am J Clin Pathol 2004122546–551. [DOI] [PubMed] [Google Scholar]
- 86.Buschges R, Weber R G, Actor B.et al Amplification and expression of cyclin D genes (CCND1, CCND2 and CCND3) in human malignant gliomas. Brain Pathol 19999435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Plantaz D, Mohapatra G, Matthay K K.et al Gain of chromosome 17 is the most frequent abnormality detected in neuroblastoma by comparative genomic hybridization. Am J Pathol 199715081–89. [PMC free article] [PubMed] [Google Scholar]
- 88.Brinkschmidt C, Poremba C, Christiansen H.et al Comparative genomic hybridization and telomerase activity analysis identify two biologically different groups of 4s neuroblastomas. Br J Cancer 1998772223–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Woodfine K, Fiegler H, Beare D M.et al Replication timing of the human genome. Hum Mol Genet 200413191–202. [DOI] [PubMed] [Google Scholar]