Abstract
Background
Differential counting of peripheral blood cells is an important diagnostic tool. Yet, this technique requires highly trained staff, is labour intensive and has limited statistical reliability. A recent development in this field was the introduction of automated peripheral blood differential counting systems. These computerised systems provide an automated morphological analysis of peripheral blood films, including a preclassification of both red and white cells (RBCs and WBCs, respectively).
Aims
To investigate the ability of two automated microscopy systems to examine peripheral blood smears.
Methods
Two automated microscopy systems, the Cellavision Diffmaster Octavia (Octavia) and Cellavision DM96 (DM96), were evaluated.
Results
The overall preclassification accuracy values for the Octavia and the DM96 systems were 87% and 92%, respectively. Evaluation of accuracy (WBC analysis) showed good correlation for both automated systems when compared with manual differentiation. Total analysis time (including post classification) was 5.4 min/slide for the Octavia and 3.2 min/slide for the DM96 (100 WBC/slide) system. The DM96 required even less time than manual differentiation by an experienced biomedical scientist.
Conclusions
The Octavia and the DM96 are automated cell analysis systems capable of morphological classification of RBCs and WBCs in peripheral blood smears. Classification accuracy depends on the type of pathological changes in the blood sample. Both systems operate most effectively in the analysis of non‐pathological blood samples.
Differential counting of blood cells is an important diagnostic tool for successful treatment and management of patients. Reliable and efficient analysis of patient samples is therefore crucial. Current automated cell counters are based on laser‐light scatter and flow‐cytochemical principles, and offer a leucocyte, red cell and platelet count, including a five‐part leucocyte differential and a panel for screening red cell abnormalities. A drawback of automated cell counters is that they offer very limited morphological information and are unable to reliably classify immature and abnormal cells. When abnormalities are suspected in a sample, results are automatically marked. In these cases or when quantification of the pathological cells is needed, a blood film is prepared for microscopical examination and cell classification. At Geïntegreerd Klinisch Chemisch Laboratorium (GKCL), Albert Schweitzer Ziekenhuis Dordrecht, The Netherlands, the microscopical review rate is about 21% of all processed blood samples, resulting in a vast amount of labour‐intensive work.
The examination of peripheral blood films is time consuming, requires highly trained staff and remains subject to significant statistical variance.1 In the past decades, efforts have been made to develop automated morphological analysis systems. The automated analysis of cell images was first described by Prewitt and Mendlesohn2 in 1966. Subsequently, other authors showed that digital image processing can be used for automated leucocyte recognition (reviewed by Bentley3).
Over the years, a limited number of digital analysis systems became commercially available—for example, the Hematrak 590 (Geometric data) and the Micro 21 (Cellavision) systems.4 Ideally, these automated systems should be able to analyse a peripheral blood smear morphologically. Results should be reproducible and with limited analytical error, and these systems should be faster or at least as fast as a biomedical scientist. Coupled with automated sample handling, a “walk‐away” system can be created that can partially replace the biomedical scientist. In addition, these systems should be capable of storing relevant morphological data and distribute images to other workstations for review purposes (telehaematology). However, the existing systems seemed to be slow compared with manual differential. Owing to poor automation and cell pattern recognition algorithms, these systems were not truly walk‐away systems and required frequent intervention by a biomedical scientist.5
More recently two new systems, the Cellavision Diffmaster Octavia (hereafter termed Octavia)and the Cellavision DM96 (hereafter termed DM96), were introduced in 2001 and 2004, respectively. Evaluations of these systems by Swolin et al6 and Kratz et al7 showed a good correlation with the manual differentiation of normal cells and blasts. The systems were able to preclassify 89% and 82%, respectively, of all leucocytes correctly with good reproducibility.
In this study, we evaluated two automated digital microscopy systems: the Octavia and its successor, the DM96. Precision and accuracy of the white cell (WBC) classification were tested for all major peripheral blood cell categories and red blood cell (RBC) morphology. Further, several performance characteristics were studied.
Materials and methods
Patient samples
Venous blood was collected using K3 EDTA as anticoagulant. After collection, samples were stored at room temperature until further analysis. Blood smears were prepared and stained (May–Grünwald Giemsa) within 4 h of sampling using an automated slidemaker (SP‐100, Sysmex, Kobe, Japan). To evaluate the accuracy of analysis, 200 blood samples were selected at random from the routine workload of the Department of Clinical Chemistry, Albert Schweitzer Hospital, Dordrecht, The Netherlands. All samples were screened by a Sysmex XE‐2100 cell counter and “flagged” for microscopical review. Within these 200 blood samples, various clinical conditions were present—for example, iron deficiency, vitamin B12 or folic acid deficiency, bacterial infections, viral infections, acute leukaemia, chronic leukaemia, non‐Hodgkin's lymphoma, myelodysplastic syndrome and polycythaemia vera. The WBC count in these patients ranged from 1.72 to 402×109/l.
Automated microscopy systems: Octavia and DM96
The Octavia and the DM96 are automated digital cell morphology systems for automated analysis of peripheral blood smears. Smears can be examined for, WBC distribution and morphology, RBC morphology and platelet concentration and morphology.
The Octavia consists of a motorised microscope (Olympus BX50WI, Olympus Europe, Hamburg, Germany) with a 100× objective, a Sony DXC‐9100P video camera (Sony, Tokyo, Japan), an automated motorised stage holder (eight‐slide capacity), hardware for motor and light control, and a computer, operating under Windows NT 4.0 with Cytologica software (V.3.0) (Cytologica, CellaVision AB, Lund, Sweden). Sample loading and immersion oil application are performed manually. When started by the operator, slides are processed automatically by the system.
The DM96 consists of a slide‐scanning unit and a computer operating under Windows XP with the Cellavision Blood Differential software (V.1.2). The slide‐scanning unit consists of a motorised microscope (10×, 50× and 100× objectives, Olympus U‐CMAD3), a digital charge‐coupled device camera (Basler A301FC, Basler Europe, Ahrensburg, Germany), an automatic immersion oil unit, a slide feeder unit with a barcode reader, a magazine feeder unit, a control unit that controls motors, sensors, oil applying and illumination, and casing. Slides are loaded onto the DM96 system in magazines (12 slides per magazine), and up to eight magazines can be loaded at the same time. Slides are processed immediately after loading on the magazine feeder unit.
The Octavia system locates the erythrocyte monolayer, and scans and photographs leucocytes or other objects at high magnification. On the other hand, the DM96 locates the erythrocyte monolayer and each potential leucocyte at low magnification, and subsequently takes images at a higher magnification. In both systems images are analysed using a neural network based on a large database of cells. Features such as colour, size and shape are used for each calculation. Finally, all images of WBCs are presented on the screen together with a preclassification. In addition, a composition image of the RBC morphology is made. The Octavia preclassifies the WBCs into the following classes: segmented neutrophils, band neutrophils, eosinophils, basophils, lymphocytes, monocytes, blast cells, promyelocytes, myelocytes, metamyelocytes, unidentified cells, erythroblasts, giant platelets, smudge cells and artefacts. The DM96 additionally recognises the classes lymphocyte (variant form), plasma cells and thrombocyte aggregation. Aberrant WBCs—for example, hairy cells or large granular lymphocytes—are not preclassified as a separate identity by the system. However, the operator can reclassify these cells and place them in a specific (user‐defined) cell class, or the operator can add a specific cell comment. Also, the RBC morphology is precharacterised by the systems which distinguish polychromasia, hypochromasia, anisocytosis, microcytosis, macrocytosis and poikilocytois as categories, using a scale from 0 to 3. The default settings can be adjusted by the operator. Comments about the presence of target cells, schistocytosis, sickle cells, spherocytosis, elliptocytosis, ovalocytosis, tear drop cells, stomatocytosis, acanthocytosis, echinocytosis, Howell–Jolly bodies, Pappenheimer bodies, basophilic stippling, parasites and 10 user‐defined characteristics can be added by the operator, but these RBC abnormalities are not precharacterised by the system.
The operator identifies and verifies the suggested classification of each cell. These systems are intended to be used by skilled operators specially trained in the use of the device and in classification of blood cells. When the slide is processed by the system, the data can be studied and verified directly by the operators. Subsequently, results are reported to the laboratory information system.
Blood smear analysis
Each blood smear was independently analysed by two experienced biomedical scientists, each classifying 200 WBCs, and using standard microscopy techniques according to the laboratory standard operating procedures, which are based on the VHL (Dutch association of laboratory haematology) guidelines (http://www.de‐vhl.nl/documents/Diffboekje.doc). Subsequently, all samples were analysed using the Octavia and the DM96 systems. Both machines were set to analyse 400 WBCs; however, in leucopenia samples this number was not always reached. Only samples in which 200 cells were collected were included in the final study. Owing to technical errors four samples were excluded, leaving 196 samples in the final evaluation. All 196 samples were used for evaluation of accuracy unless indicated otherwise. Technical errors were found in three samples where the slide process failed—for example, no monolayer could be found by the machine (Octavia n = 2 and DM96 n = 1) and one sample where the number of WBCs counted was not sufficient. No other errors were reported.
Experience of the biomedical scientists
Nine biomedical scientists of the laboratory of clinical chemistry of the Albert Schweitzer hospital, Dordrecht, the Netherlands, participated in study. All biomedical scientists work in the haematology section of the laboratory of clinical chemistry and are experienced in morphological haematology and participate in internal and external quality control procedures on a routine basis.
Study design
For evaluation of the test methods the following items were tested: the ability to preclassify WBCs, accuracy, within run imprecision and accuracy of RBC analysis and system throughput. Timing studies were also conducted: in an initial experiment three sets of eight slides were analysed by three different biomedical scientists. Total processing time was registered for each method. Secondly, a more elaborate time study was performed comparing total sample handling time by biomedical scientists performing manual differentiation with total handling time on the DM96. For this purpose, the DM96 was incorporated in routine procedure and operated by experienced biomedical scientists. Total handling times for both manual differentiation and DM96 were registered on eight consecutive days. Using manual differentiation slides were either only “screened” or, when needed, a 100 cell differential count was done. For the DM96 all slides were preclassified by the machine and subsequently viewed, corrected (when necessary) and authorised by the biomedical scientist.
Statistical analysis
Statistical analysis was carried out using “Analyse‐it”, a statistical software add‐in for Microsoft Excel (www.analyse‐it.com). The study design was based on the National Committee for Clinical Laboratory Stnadard document H20A.8 Results for evaluation of accuracy were analysed according to Bland and Altman.9
Results
Preclassification performed by the Octavia and the DM96
Preclassification—that is, the initial cell classification by the machine, was evaluated. Table 1 shows the percentage of the different cell types preclassified correctly according to the biomedical scientists. Of all normal leucocytes (segmented neutrophils, band neutrophils, eosinophils, basophils, lymphocytes and monocytes) analysed by the Octavia and DM96, in 89.7% and 95.1% of the cases the biomedical scientist agreed with the preclassification. The overall (all preclassified cell classes) preclassification accuracy for the Octavia and the DM96 were 87% and 92%, respectively.
Table 1 Pre‐classification agreement for the Octavia and the DM96*.
| Pre‐classification agreement (%) | % added from other categories | |||
|---|---|---|---|---|
| Cell class | Octavia | DM96 | Octavia | DM96 |
| Segmented neutrophils | 94.4 | 98.6 | 2.6 | 3.8 |
| Band neutrophils | 10.5 | 22.9 | 81.7 | 50.4 |
| Eosinophils | 95.4 | 93.5 | 2.5 | 4.5 |
| Basophils | 58.4 | 84.7 | 22.8 | 32.1 |
| Lymphocytes | 94.3 | 95.2 | 7 | 1.4 |
| Monocytes | 65 | 94 | 14.7 | 24 |
| Blast cells | 84.4 | 78.5 | 78.3 | 50.4 |
| Octavia | DM96 | |||
|---|---|---|---|---|
| Overall accuracy (%) | 87 | 92.0 | ||
| Abnormal called normal (%) | 4.4 | 1.6 | ||
| Normal misclassified as other normal (%) | 3.9 | 3.0 | ||
| Normal called abnormal (%) | 0.2 | 0.6 |
*For each cell class, the percentages of cells correctly pre‐classified by the systems, according to the biomedical scientists, are shown. This is the percentage of cells remaining in each cell class after classification by the biomedical scientist. In addition, the percentage of cells added from other categories of cells is shown. Overall pre‐classification accuracy is calculated as the sum of the number of cells per cell class remaining after classification by the biomedical scientist (final result) divided by the total number of cells after pre‐classification.
The Octavia preclassified 4.4% of the abnormal cells as normal cells, 3.9% of the normal cells were preclassified as other normal cells and 0.2% of the normal cells were pre‐classified as abnormal cells. This was 1.6%, 3.0% and 0.6% for the DM96. Preclassification was correct for 84.4% (Octavia) and 78.5% (DM96) of the blast cells (table 1).
Accuracy
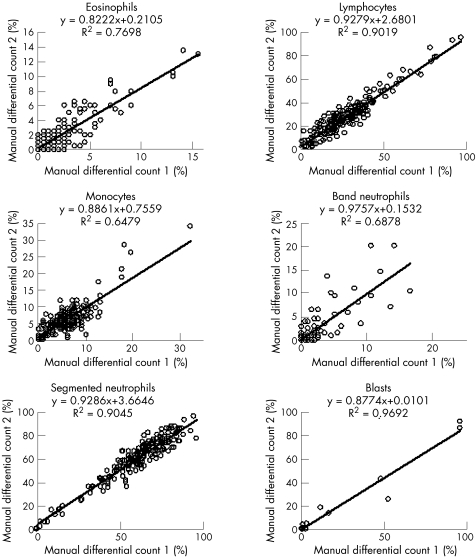
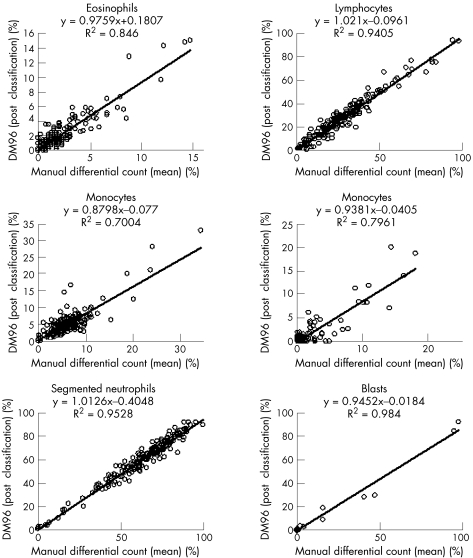
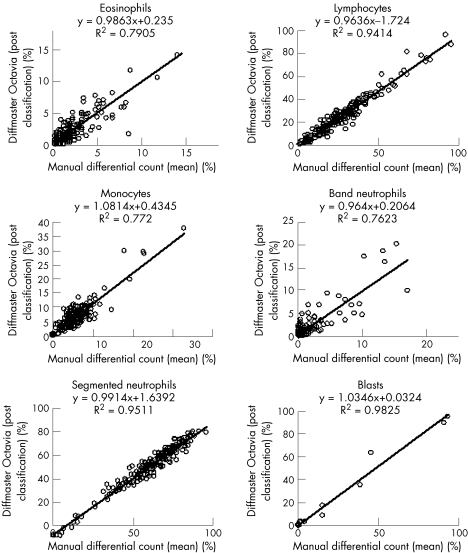
Evaluation of accuracy of the WBC classification was performed comparing the mean of the two manual differential counts by two biomedical scientists, with postclassification results of the Octavia and the DM96. Interindividual differences in manual differentiation were also analysed. This analysis was performed for all normal cell classes (segmented neutrophils, band neutrophils, eosinophils, basophils, lymphocytes and monocytes) and for blast cells. Results are shown in fig 1(A–C). For manual differentiation, regression coefficients ranged from 0.93 (R2 0.90) for lymphocyte counts to 0.89 (R2 0.65) for monocytes. Regression coefficients for the Octavia ranged from 0.96 (R2 0.94) for lymphocyte counts to 1.08 (R2 0.77) for monocytes and regression coefficients for the DM96 ranged from 1.02 (R2 0.94) for lymphocyte counts to 0.88 (R2 0.70) for monocytes. Regression coefficients for basophils showed considerable variation, due to the low number of cells in most samples and are therefore not shown.
Figure 1 Accuracy for eosinophils, lymphocytes, monocytes (n = 195), band neutrophils, segmented neutrohils and blast cells. Comparison of manual differential counts.
Figure 3 Accuracy for eosinophils, lymphocytes, monocytes, band neutrophils (n = 195), segmented neutrophils (n = 193) and blast cells. Comparison of the mean manual differential count with the DM96 counts.
Within‐run imprecision
Within‐run imprecision (reproducibility) for preclassification was evaluated for the Octavia and the DM96. Therefore four slides from four different samples were counted eight times, with 100 WBCs counted each time. A good reproducibility was observed in each cell category on both systems (table 2).
Table 2 Within‐run imprecision for the two systems, Octavia and DM96.
| Octavia | ||||||||
|---|---|---|---|---|---|---|---|---|
| Cell type | Slide 1 | Slide 2 | Slide 3 | Slide 4 | ||||
| Mean (%) | SD | Mean (%) | SD | Mean (%) | SD | Mean (%) | SD | |
| Segmented neutrophils | 91.7 | 1.4 | 65.7 | 0.7 | 90.7 | 1.4 | 54.6 | 4.6 |
| Band neutrophils | 2.0 | 1.3 | 2.0 | 0.4 | 4.7 | 1.5 | 4.2 | 1.4 |
| Eosinophils | 0 | 0 | 0 | 0 | 0.6 | 0.2 | 7.2 | 1.4 |
| Basophils | 0 | 0 | 0 | 0 | 0.3 | 0.4 | 2.3 | 1.2 |
| Lymphocytes | 2.1 | 0.8 | 21.4 | 0.2 | 1.6 | 1.3 | 15.8 | 2.2 |
| Monocytes | 3.9 | 0.7 | 9.8 | 0.7 | 1.9 | 0.5 | 5.1 | 1.2 |
| Blasts | 0 | 0 | 0 | 0 | 0 | 0 | 0.8 | 0.7 |
| DM96 | ||||||||
| Cell type | Slide 1 | Slide 2 | Slide 3 | Slide 4 | ||||
|---|---|---|---|---|---|---|---|---|
| Mean (%) | SD | Mean (%) | SD | Mean (%) | SD | Mean (%) | SD | |
| Segmented neutrophils | 90.8 | 0.8 | 66.2 | 1.2 | 90.8 | 1.0 | 52.6 | 2.2 |
| Band neutrophils | 3.7 | 0.8 | 3.5 | 0.5 | 6.4 | 0.5 | 2.0 | 0.8 |
| Eosinophils | 0 | 0 | 0 | 0 | 0.1 | 0.2 | 12.1 | 1.1 |
| Basophils | 0 | 0 | 0.2 | 0.3 | 0 | 0 | 3.9 | 0.6 |
| Lymphocytes | 3.2 | 0.4 | 23.4 | 1.8 | 2.2 | 0.4 | 16.8 | 2.4 |
| Monocytes | 2.3 | 0.5 | 5.6 | 1.3 | 0.4 | 0.3 | 0.6 | 0.2 |
| Blasts | 0 | 0 | 0 | 0 | 0 | 0 | 1.2 | 0.8 |
Throughput
Throughput for both systems was calculated from the runs in the study of evaluation of accuracy. Runtimes were obtained from the internal runtime records in the log files of the systems. Only those runs were included that were not interrupted in any way by technical or other interference. The system was set to analyse 400 WBCs. Results were as follows: average time of analysis per slide during the evaluation of accuracy was 13.6 min for the Octavia and 3.9 min for the DM96—that is, a throughput of 4–5 slides/h and 15–16 slides/h for the Octavia and the DM96, respectively.
For the DM96, this experiment was expanded with a comparison of throughput time when 100, 200, 300 or 400 WBCs were counted. In contrast with the evaluation of accuracy, here only normal samples (n = 12, WBC range 2.1–11.5×109/l, mean 7.2×109/l) were used. Average time of analysis was 1.51(40 slides/h), 1.95 (31), 2.50 (24) and 2.80 (21.4) min/slide, respectively.
RBC analysis
Evaluating the accuracy of red blood cell analysis, five categories of morphological changes were compared: polychromasia, hypochromasia, microcytosis, macrocytosis and poikilocytosis. Despite the fact that both automated systems also report anisocytosis, results on this category were not evaluated since in The Netherlands this category is replaced by microcytosis and macrocytosis.
Results were grouped as “normal+mild” or “moderate+severe” changes for all categories. Precharacterisation and postcharacterisation agreement in all categories ranged from 95–100% for normal+mild changes to 35–100% agreements for moderate to severe changes. Overall agreement for all categories ranged from 71–94% (table 3).
Table 3 Red blood cell morphology: agreement between pre‐characterisation and post‐characterisation results.
| Octavia | DM96 | |||||
|---|---|---|---|---|---|---|
| Normal to mild | Moderate to severe | Overall score | Normal to mild | Moderate to severe | Overall score | |
| Polychromasia | 99.5 (195) | 100 (2) | 86.8 | 99.5 (196) | — (1) | 93.9 |
| Hypochromasia | 100 (191) | 66.7 (6) | 91.9 | 100 (194) | 42.9 (3) | 93.9 |
| Microcytosis | 97.8 (189) | 66.7 (8) | 83.8 | 95.2 (181) | 77.8 (16) | 83.2 |
| Macrocytosis | 99.4 (185) | 35.3 (12) | 71.6 | 94.8 (185) | 34.8 (12) | 71.1 |
| Poikilocytosis | 100 (193) | 40.0 (4) | 88.3 | 98.3 (188) | 50.0 (9) | 88.8 |
Values are % agreement. Number of samples in parenheses.
Time‐efficiency studies
A time efficiency study was performed using three different sets, each containing 8 peripheral blood smears. To compare total processing time, slides in each set were manually differentiated (100 WBC count) by a biomedical scientist and total analysis time was registered. Each set of slides was also analysed on both systems (100 WBC count) and validated (post‐classification) by another biomedical scientist. In this way, each biomedical scientist analysed a different set of slides in the three settings (manual, Octavia and DM96). Total analysis time was defined as the time elapsed from sample loading to ready to report. When using the Octavia and the DM96 post‐classification was started as soon as preclassification of the first slide was finished.
Our results show that the DM96 took on average 25.7 min (95%CI 18.8 to 32.5) per set of eight slides for analysis, manual analysis took 33.3 min (95%CI 29.5 to 36.1) and the Octavia 43.0 min (95%CI 37.0 to 48.9; table 4). So, using the DM96 can save approximately 1 min/slide compared with the manual differential count, whereas it takes about 1.2 min more using the Octavia.
Table 4 Total processing time for the reference method, the Octavia and the DM96.
| Method | Mean (SD) | |||
|---|---|---|---|---|
| Reference method | Octavia | DM96 | ||
| Biomedical scientist 1 | 34 | 37 | 25 | 32.0 (6.2) |
| Biomedical scientist 2 | 36 | 47 | 20 | 34.3 (13.6) |
| Biomedical scientist 3 | 30 | 45 | 32 | 35.7 (8.1) |
| Mean (SD) | 33.3 (3.1) | 43.0 (5.3) | 25.7 (6.0) | |
| min/slide | 4.2 | 5.4 | 3.2 | |
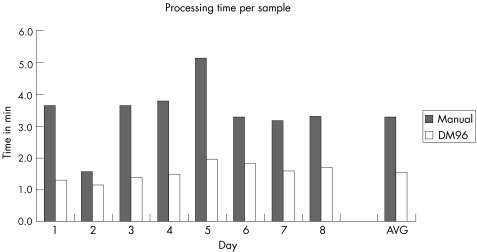
Finally, the operational efficiency of the DM96 was evaluated by comparing the total time of analysis on the DM96 to that of manual differentiation. Over 8 days total handing time per sample (100 WBC count) was measured in a routine workflow setting for manual differentiation and for another 8 days for the DM96. Results (fig 2) show that the actual sample handling time in routine morphological analysis using a DM96 is <50% than that of manual differentiation. Average time of analysis was 3.3 min/slide for the reference method and 1.6 min/slide for the DM96.
Figure 2 Accuracy for eosinophils, lymphocytes, monocytes (n = 195), band neutrophils, segmented neutrohils and blast cells. Comparison of the mean manual differential count with the Octavia results.
Figure 4 Operational efficiency: results for the manual differential count (n = 220) and the DM96 (n = 315). For each method, the mean time of analysis per sample is indicated; also indicated is the average (AVG) of all days.
Discussion
Nowadays, counting and classification of human leucocytes in laboratories is completely automated. Routinely a five‐part leucocyte differential count is performed, together with an analysis of the RBCs. Yet, microscopic analysis of blood smears is still an indispensable part of the laboratory workflow, as automated cell counters are not capable of advanced morphological analysis of RBCs and WBCs. Manual analysis of peripheral blood smears is time consuming and requires highly trained biomedical scientists. However, laboratory budgets are under continuous pressure and recruiting new biomedical scientists is increasingly difficult in many countries. Investing in the automation of blood smear analysis could therefore be beneficial to laboratory efficacy.
In this study, an evaluation of two different systems, capable of automated analysis of blood smears. The Octavia and the DM96 were tested for reliability, accuracy, throughput and efficiency. Although the overall preclassification accuracy (ie, the percentage of cells where the biomedical scientist agrees with the suggestion made by the machine) for both the Octavia and the DM96 is adequate, the DM96 scores better. This is owing to the use of upgraded software on the DM96. In case of blast cells, the Octavia preclassified 84.4% of the cells correctly, thereby scoring somewhat better than the DM96 (78.5%). So, for the Octavia of the cells initially placed in the category blasts about 6% more are classified correctly, according to the operator. However, less blast cells are preclassified as other cells (ie, initially missed and placed in other cell categories by the machine) by the DM96, as 46% of the cells finally reported as blast is added from other categories by the operator, whereas this figure is 78% for the Octavia.
Final WBC results of the Octavia and the DM96 (after verification by the operator) were compared with the gold standard—that is, manual differentiation. An appraisal of this gold standard was also performed, showing that the overall accuracy of manual differentiation does not exceed the accuracy of both automated systems. In other words: interbiomedical scientist variation of manual differentiation is in the same order as the variation between manual and automated differentiation.
For both systems the accuracy of RBC analysis was also evaluated. Characterisation by the machine (precharacterisation) was compared with the final judgement by the biomedical scientist (postcharacterisation). The percentages of agreement for normal to mild morphological changes are within satisfactory range (94.8–100%); the percentages of agreement are lower for mild to severe changes (for categories hypochromasia, microcytosis and macrocytosis). However, the number of samples precharacterised as mild to severe morphological changes are rather small. Moreover, for this evaluation the default RBC precharacterisation settings were used; this contributed to the percentage of disagreement. Settings can be adjusted (in contrast with the WBC settings) if considered necessary, in accordance with local standards.
Time‐efficiency studies showed that the DM96 is superior to the Octavia. This is mainly due to the fact that the DM96 uses three lenses instead of one lens on the Octavia. Whereas the Octavia system uses a 100× lens for both localisation and analysis, the DM96 uses the 10× and 50× objectives to locate and the 100× objective to analyse the leucocytes. In this way, a considerable increase in throughput can be realised. Introducing the DM96 into our routine laboratory practice reduced total analysis time per blood smear with approximately 50%.
During the evaluation period, about 10% of the samples processed on DM96 were reviewed manually on a standard microscope. In nearly all cases, these samples contained pathological cells and manual review was considered necessary. We expect this percentage to decline in the future, when the biomedical scientists become more familiar with “digital microscopy”.
Introduction of automated and computerised examination of blood smears offers additional advantages. Each WBC image analysed by the system is stored and therefore available for re‐evaluation. Especially in more complex cases this feature enables the operator to discuss the classification with colleagues or experts in the field at any time. Using so called “remote review software” that can be installed on any PC or laptop, it also allows verification and authorisation of peripheral blood smear analysis results from another location. This option can be interesting for satellite or small laboratories, which depend on expertise from main locations for morphological assessment. In this way, telehaematology comes within reach of routine haematological laboratories. In addition, review of these stored images can be used for quality control and training purposes.
Will these systems replace manual differentiation completely in the near future? Five‐part differential analysis alone will not enable complete classification peripheral RBCs and WBCs. However, combined with automated morphological assessment of each WBC using automated image analysis software, the future potential of these systems is very promising. Given the short time these systems are on the market, the results are already convincing. Future improvement of the image analysis algorithms will certainly increase the number of morphological categories that can be recognised automatically. Combining all information using a decision algorithm, this could well lead to an automated diagnosis of certain haematological malignancies—for example, lymphomas—in the future. Other future applications of the automated microscope may perhaps include analysis of bone marrow spreads, lymph node preparations, urine sediments and cerebrospinal fluid cell morphology.
Conclusion
Automated image analysis and classification has now reached a stage where it can meet the requirements of a modern routine haematological laboratory in terms of reliability and efficiency. Both the Octavia and DM96 are reliable and accurate systems that enable automated analysis of blood smears in the haematology laboratory. Both systems operate most effectively in screening routine blood samples. The automated systems reduced work pressure by efficiently analysing and reporting non‐pathological smears, rendering more time for analysis of pathological slides.
Take-home messages
Automated image analysis meets the requirements of a modern routine haematolgical laboratory
The Octavia and the DM96 are reliable and accurate systems that enable automated blood smear analysis
The introduction of the DM96 can reduce work pressure by efficiently processing non‐pathological smears, giving more time for analysis of pathological slides
Acknowledgements
We thank Annemiek de Pijper‐van Poppel, Barbara van de Breevaart‐Ardonne, Bea Koetsier, Carla Jongmans, Hanny van der Giessen‐van den Hooft, Peggy Brosens, Pleunie de Lint‐van Driel, Ria de Jong‐Kwikkers, Ria Teuns for their technical assistance and for performing the morphological analysis. We thank Dr P.B. Berendes and Dr J. Prins for critical reading of the document.
Abbreviations
DM96 - Cellavision DM96
Octavia - Cellavision Diffmaster Octavia
RBC - red blood cell
SSU - slide scanning unit
WBC - white blood cell
Footnotes
Competing interests: None declared.
References
- 1.Rumke C L. Imprecision of ratio‐derived differential leukocyte counts. Blood Cells 198511311–315. [PubMed] [Google Scholar]
- 2.Prewitt J M, Mendelsohn M L. The analysis of cell images. Ann N Y Acad Sci 19661281035–1053. [DOI] [PubMed] [Google Scholar]
- 3.Bentley S A. Automated differential white cell counts: a critical appraisal. Baillieres Clin Haematol 19903851–869. [DOI] [PubMed] [Google Scholar]
- 4.Tatsumi N, Pierre R V. Automated image processing. Past, present, and future of blood cell morphology identification. Clin Lab Med 200222299–315, viii. [DOI] [PubMed] [Google Scholar]
- 5.Houwen B. The differential cell count. Lab Hematol 2001789–100. [Google Scholar]
- 6.Swolin B, Simonsson P, Backman S.et al Differential counting of blood leukocytes using automated microscopy and a decision support system based on artificial neural networks‐‐evaluation of DiffMaster Octavia. Clin Lab Haematol 200325139–147. [DOI] [PubMed] [Google Scholar]
- 7.Kratz A, Bengtsson H, Casey J E.et al Performance evaluation of the Cellavision DM96 system. Am J Clin Pathol 2005124770–781. [DOI] [PubMed] [Google Scholar]
- 8.National Committee for Clinical Laboratory Standards H20‐A reference leukocyte differential count (proportional) and evaluation of instrumental methods; approved standard. Natl Committee Clin Lab Stand 1992171–33. [Google Scholar]
- 9.Bland J M, Altman D G. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 19861307–310. [PubMed] [Google Scholar]