Abstract
Background
Chronic fatigue syndrome (CFS) is an illness with unknown aetiology and pathophysiology. The difference in incidence by sex observed for CFS indicates a role for oestrogen and oestrogen receptors in disease development. Furthermore, an immunomediated pathogenesis has been suggested for CFS, providing an additional connection to oestrogen, which displays immunomodular functions.
Aims
To investigate a possible association of oestrogen receptor (ER) mRNAs and two ERβ single‐nucleotide polymorphisms (SNPs) with CFS.
Methods
Messenger RNA levels of ERα, ERβ wt and ERβ cx were investigated in peripheral blood mononuclear cells from 30 patients with CFS and 36 healthy controls by quantitative real‐time polymerase chain reaction. Two ERβ SNPs were scored in the same material.
Results
The CFS group showed significantly lower mRNA expression levels of ERβ wt compared with the healthy control group. No differences were observed for ERα or ERβ cx between patients and controls. There were no significant differences in frequency for the investigated ERβ SNPs between cases and controls.
Conclusions
The reduced ERβ wt expression level observed in this study is consistent with an immune‐mediated pathogenesis of CFS. Additionally, the observation that ERβ wt expression is decreased in CFS could provide an entry point to identify interesting, potentially disease‐causing, candidate molecules for further study. A possible connection between oestrogen, oestrogen receptors and CFS should be evaluated further.
Fatigue is a central component of many diseases and illnesses. Fatigue of unknown aetiology and pathophysiology lasting for >6 months, with at least four out of eight specified symptoms, is termed chronic fatigue syndrome (CFS).1 The symptoms are impairment of cognition and memory, recurring sore throat, tender lymph nodes, mild muscle pain, joint ache, headaches of a new type, unrefreshing sleep and post‐exertional malaise.1 Prevalence rates of 0.2–0.5% have been reported in the Western world, with a predominance of women (2–4 times higher rates compared with men).2,3 The actual target tissues for CFS remain elusive. However, peripheral blood mononuclear cells (PBMCs) have been shown to act as indicators for abnormal biological processes occurring throughout the body.4,5
Oestrogen is a steroid hormone that has an important role in various physiological processes including sexual development and in the reproductive cycle.6 Oestrogens have been shown to be potential immunomodulators. Several autoimmune diseases such as rheumatoid arthritis and multiple sclerosis affect an excess of females.7 Furthermore, both multiple sclerosis and rheumatoid arthritis generally improve during pregnancy, suggesting that oestrogen could have an immunosuppressive role in these contexts.7 Oestradiol and cyclic progestin treatment has improved the health status of premenopausal patients with CFS, and one study showed improved health during pregnancy,8,9 when oestrogen levels are naturally high. Both CFS and oestrogen have been linked to a Th2 type response of the immune system.7,10,11
Oestrogen exerts its effects by binding to the oestrogen receptors (ERs). 6,12 Oestrogen receptors have been implicated in several diseases presenting unequal proportions of men and women, such as breast cancer and osteoporosis.13 The oestrogen receptors belong to the nuclear receptor superfamily.12 There are two oestrogen receptors, ERα and ERβ, which have unique and overlapping roles. For ERβ, there exists a human splice variant, ERβ cx, which differs at the C‐terminal end of the protein.14 It is unclear what regulates ERβ wt/ERβ cx ratios. Possibilities include differential promoter usage and differential mRNA stability. Human ERβ is expressed from two alternative, tissue‐specific, first exons, 0N and 0K.15,16 In this context, Hirata et al have shown that 0N is coupled to ERβ wt and 0K to ERβ cx in the testis.15
Frequency differences between patient and control groups in naturally occurring base‐pair changes, referred to as single‐nucleotide polymorphisms (SNPs), indicate a linkage of the particular genomic region with the disease under study. Association of SNPs in ERβ with disease has been reported in, for example, patients with anorexia nervosa and bulimia.17,18 These diseases also have a female predominance. ERβ SNPs have also been studied in relation to prostate cancer,19 Alzheimer's disease,20 pre‐eclampsia,21 efficacy of hormone replacement therapy,22 hypertension,23 Parkinson's disease,24,25 breast cancer,26 as well as other conditions.13
Based on the unequal sex distribution for CFS and the reported improvement in health status on oestrogen treatment, we hypothesised that differential expression of oestrogen receptors could occur in CFS. In this study, we investigate this hypothesis by exploring the possible associations between oestrogen receptor mRNA expression levels and/or genetic variants and CFS.
Methods
Study population
The study cohort consisted of 30 patients with CFS and 36 voluntary healthy controls (table 1).
Table 1 Study cohort information including clinical patient data such as illness duration, illness onset type and classification according to the International Classification of diseases, 10th revision, system.
| Patients with CFS * (n = 30) | Controls (n = 36) | |
|---|---|---|
| Age (years), mean (range) | 40 (26–54) | 44 (26–65) |
| Men/women | 9/21 | 10/26 |
| Premenopausal (<52 years)/postmenopausal (>52 years)† | 18/3 | 19/7 |
| Illness duration (years)‡, median (range) | 4.5 (1.5–25) | – |
| Illness onset type‡, number (male/female) | ||
| Gradual | 15 (4/11) | – |
| Sudden | 9 (2/7) | – |
| CFS classification (ICD‐10)‡ | ||
| Non‐infectious | 11 (2/9) | – |
| Infectious | 13 (4/9) | – |
CFS, chronic fatigue syndrome; ICD‐10, International Classification of diseases, 10th revision.
*Patients with CFS were diagnosed according to the 1994 case definition.1
†The mean age for menopause in Sweden is 52 years.
‡Clinical data were available for 24 patients.
Sample preparation
PBMCs were isolated from all patients and controls immediately after blood withdrawal (Venglect evacuated blood collection tubes, heparin, Terumo, Leuven, Belgium) and after obtaining written informed consent (ethical approval 130/02, Karolinska University Hospital, Huddinge, Stockholm, Sweden). Ten million PBMCs were lysed (TRIzol Reagent, Invitrogen, Carlsbad, California, USA) and stored at −80°C. Total RNA was extracted by phase separation and quantified by spectrophotometry (NanoDrop Technologies, Wilmington, Delaware, USA). Twelve samples, one from each extraction batch, were also analysed using the 2100 Bioanalyzer instrument (Agilent Technologies, Palo Alto, California, USA). Complementary DNA was synthesised from total RNA using random primers and the SuperScript™ III system, including a DNase treatment step (Invitrogen). Two separate cDNA synthesis reactions were performed on each RNA sample.
Quantification of mRNA expression
Messenger RNA expression levels were quantified using the ABI real‐time polymerase chain reaction system (7700, Applied Biosystems, Foster City, California, USA). TaqMan assays were used for all transcripts. Primers and probes were either designed using the Primer Express software V.2 (Applied Biosystems, table 2) or purchased from Applied Biosystems (Assays‐on‐Demand Hs01100359 (ERβ wt), Hs01105520 (ERβ cx), Hs99999901_s1 (18S) and 4310884E (glyceraldehyde‐3‐phosphate dehydrogenase)). Samples were run in triplicate. Comparative analysis, by the ABI Prism 7700 Sequence Detection System (Applied Biosystems), was used to calculate mRNA levels, and the two‐sided t test with unequal variance was used for statistical analysis. Data calculations and statistics were performed in Excel. Serial dilutions of pSG5 hER‐plasmid (ERα, ERβ wt and ERβ cx) of known concentrations were run for absolute quantification of ER mRNA expression levels.
Table 2 Primers and probes used for real‐time quantitative‐polymerase chain reaction assays.
| Transcript | Forward 5′‐3′ | Reverse 5′‐3′ | Probe 5′‐3′ |
|---|---|---|---|
| ERα | GCTAGGAAGTGGGAATGATGAAAG | TCTGGCGCTTGTGTTTCAAC | TGGGATACGAAAAGACCGAAGAGGAGGG |
| ERβ 0K | GCTCAGGTTACAGTCATCCCAAT | CAAGAAGAGGCACAAAGGTCATT | TGGTTCTGAAGCCATTATACTTGCCCACG |
| ERβ 0N | AAGCACGTGTCCGCATTTTAG | TCTCAAAGATTCGTGGGCAAGT | AGGCCGGTGTGTTTATCTGCAAGCCATTAT |
ER, oestrogen receptor.
The probes are dual‐labelled oligonucleotides with 5′‐FAM and 3′‐TAMRA.
ERβ SNP analysis
Synthesised PBMC cDNA samples were used for analysis of ERβ SNPs rs4986938 (ERβ wt) and rs928554 (ERβ cx). The SNP analysis was performed as described elsewhere.18
Results
ERβ wt mRNA levels are lower in patients with CFS compared with controls
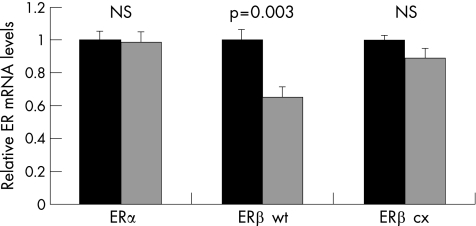
The CFS group showed significantly lower mRNA expression levels for ERβ wt compared with the control group (fig 1), using either of the control genes for normalisation. This was also true when subdividing the patient and control groups according to sex (pfemale<0.007 and pmale<0.02). The results were repeated with a subsequent cDNA synthesis starting with the same RNA samples. ERα and ERβ cx mRNA levels did not differ between patients with CFS and controls (fig 1). There were no differences in oestrogen receptor mRNA levels between the sexes, and no correlation with age (data not shown).
Figure 1 Average mRNA expression levels, with bars for the standard error of mean, comparing the entire patient group with the control group for ERα, ERβ wt and ERβ cx using 18S rRNA for normalisation. Significantly reduced levels were observed for ERβ wt, whereas no differences were observed for ERα or ERβ cx. Similar results were observed using glyceraldehyde‐3‐phosphate dehydrogenase for normalisation (results not shown).
ER mRNA levels were also compared between CFS subgroups (table 1). Reduced ERβ cx mRNA expression levels were observed in the subgroup of patients with shorter illness duration (⩽2.5 years) compared with the group with longer duration (⩾9 years). Overall, the ERα and ERβ cx mRNA expression levels in PBMCs were about 100‐fold higher (femtograms) than the ERβ wt mRNA level.
Of the two known promoters for ERβ, only expression from the 0N promoter was detected in PBMCs (data not shown).
ERβ SNPs are not associated with CFS in this cohort
As our cohort is relatively small, we chose to score only the rs4986938 (ERβ wt) and rs928554 (ERβ cx) SNPs, where the allele frequencies for the rare alleles are >35%. No significant difference in allele or genotype frequencies was observed between CFS and control groups (data not shown). However, it is worth noting that the ERβ rs4986938 AA genotype was more often found in the CFS group (data not shown).
Discussion
Significantly reduced levels of ERβ wt mRNA in PBMCs were found in patients with CFS compared with controls (fig 1). Based on this observation, we extended our studies to include determination of the possible association of ERβ SNPs and promoter levels with CFS. The levels of ERβ wt mRNA are low in PBMCs. However, the actual target tissues for CFS are unknown, and it is possible that these tissues express higher amounts of ERβ wt mRNA, still maintaining the differential expression observed in this study. Lower levels of ERβ cx mRNA were identified in patients with CFS with shorter illness duration compared with patients with longer duration. However, the groups and the difference between them are small, so the significance of this finding is at present unclear.
A recent study by Phiel et al shows the presence of ERα and ERβ mRNA levels in fractionated T and B lymphocytes.27 However, the assay used in the study does not discriminate between ERβ wt and ERβ cx. Our PBMC samples contain both T and B lymphocytes, and it would be of interest to investigate whether there are differences in the ERβ splice variant ratios between the lymphocyte fractions.
Interestingly, in this study, we find differences in the levels of the ERβ wt mRNA, but not in the splice variant ERβ cx, when comparing patients with CFS and controls. It is not clear how the relative levels of these transcripts are regulated. Our data do not support promoter usage as a means by which ERβ wt/ERβ cx ratios are regulated, as we detect expression from only one promoter (0N, data not shown). Consistent with our results, promoter 0N has been shown to be used in peripheral leucocytes.15 Tissue‐specific promoter usage has, however, been reported, and the presence of additional ERβ promoters must be considered in this context.15,16 It is possible that SNPs in the differing 3′ untranslated regions of ERβ wt (rs4986938) and ERβ cx (rs928554) transcripts regulate mRNA stability and ultimately determine ERβ wt:ERβ cx ratios. In this context, the ERβ rs4986938 AA genotype was over‐represented in the CFS group with lower ERβ wt expression. The possibility that this SNP regulates mRNA stability can be furthered explored by directly assaying the mRNA stability of mRNAs incorporating the G and A alleles.
Several gene expression studies have been performed in the search for differently transcribed genes between patients with CFS and controls.4,28,29,30,31,32 These studies confirmed the role of the immune system in CFS, as several genes involved in immunity and defence were shown to differ between patients with CFS and controls.
Oestrogens have been suggested as immunomodulatory factors. This is based on observations including the female predominance of certain autoimmune disorders,7 which might be explained by lower oestrogen secretion in men. Thus, CFS and oestrogen signalling are connected via the unequal sex distribution and their association with aspects of immunomodulation. However, none of the gene expression studies referred to above identified ERβ as a differentially expressed gene, which might reflect its low expression level.
At present, specific oestrogen receptor modulators are being developed as novel therapeutics for immune‐mediated diseases. In a study by Follettie et al, the specific ERβ agonist ERB‐041 was used to treat antigen‐induced arthritis in rats.33 ERB‐041 treatment led to sustained improvement and down regulation of genes known to be upregulated in rheumatoid arthritis.33 ERB‐041 also showed positive effects in animal models of inflammatory bowel disease.34
In conclusion, the difference in expression of ERβ wt mRNA between patients with CFS and controls observed in this study could contribute to some of the symptoms observed in CFS. Reduced levels of ERβ wt mRNA may be simply a marker for changed function of other cellular components, which are involved in CFS. The present finding of lower ERβ levels in patients with CFS compared with controls supports an immunopathogenesis for CFS. However, further work is needed to clarify whether reduced ERβ expression is a primary event in CFS or is caused by a down regulation secondary to altered oestrogen levels. The reduced ERβ wt mRNA levels in patients with CFS could provide an entry point to identify interesting and potentially disease‐causing candidate molecules for further study. Studies investigating ERβ wt protein levels and cellular effects will be required to confirm an involvement of ERβ wt in CFS. Future studies should involve evaluation of oestrogen levels and effects after oestradiol treatment in relation to CFS pathology.
Acknowledgements
We thank Dr Chunyan Zhao for the ERβ 0N and ERβ 0K primers and probes.
Abbreviations
CFS - chronic fatigue syndrome
PBMC - peripheral blood mononuclear cell
SNP - single‐nucleotide polymorphism
Footnotes
Funding: This work was supported by the Swedish Research Council, the Swedish Cancer Fund, and the Swedish Council for Working Life and Social Research. MN was supported by the Golje Memory Foundation.
Competing interests: None.
References
- 1.Fukuda K, Straus S E, Hickie I.et al The chronic fatigue syndrome: a comprehensive approach to its definition and study. International Chronic Fatigue Syndrome Study Group. Ann Intern Med 1994121953–959. [DOI] [PubMed] [Google Scholar]
- 2.Jason L A, Richman J A, Rademaker A W.et al A community‐based study of chronic fatigue syndrome. Arch Intern Med 19991592129–2137. [DOI] [PubMed] [Google Scholar]
- 3.Reyes M, Nisenbaum R, Hoaglin D C.et al Prevalence and incidence of chronic fatigue syndrome in Wichita, Kansas. Arch Intern Med 20031631530–1536. [DOI] [PubMed] [Google Scholar]
- 4.Vernon S D, Unger E R, Dimulescu I M.et al Utility of the blood for gene expression profiling and biomarker discovery in chronic fatigue syndrome. Dis Markers 200218193–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitney A R, Diehn M, Popper S J.et al Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci USA 20031001896–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nilsson S, Makela S, Treuter E.et al Mechanisms of estrogen action. Physiol Rev 2001811535–1565. [DOI] [PubMed] [Google Scholar]
- 7.Lang T J. Estrogen as an immunomodulator. Clin Immunol 2004113224–230. [DOI] [PubMed] [Google Scholar]
- 8.Studd J, Panay N. Chronic fatigue syndrome. Lancet 19963481384. [DOI] [PubMed] [Google Scholar]
- 9.Panay N, Studd J W. The psychotherapeutic effects of estrogens. Gynecol Endocrinol 199812353–365. [DOI] [PubMed] [Google Scholar]
- 10.Patarca R. Cytokines and chronic fatigue syndrome. Ann N Y Acad Sci 2001933185–200. [DOI] [PubMed] [Google Scholar]
- 11.Skowera A, Cleare A, Blair D.et al High levels of type 2 cytokine‐producing cells in chronic fatigue syndrome. Clin Exp Immunol 2004135294–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matthews J, Gustafsson J A. Estrogen signaling: a subtle balance between ER alpha and ER beta. Mol Interv 20033281–292. [DOI] [PubMed] [Google Scholar]
- 13.Nilsson M, Dahlman‐Wright K, Gustafsson J A. Nuclear receptors in disease: the oestrogen receptors. Essays Biochem 200440157–167. [DOI] [PubMed] [Google Scholar]
- 14.Ogawa S, Inoue S, Watanabe T.et al Molecular cloning and characterization of human estrogen receptor betacx: a potential inhibitor ofestrogen action in human. Nucleic Acids Res 1998263505–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirata S, Shoda T, Kato J.et al The multiple untranslated first exons system of the human estrogen receptor beta (ER beta) gene. J Steroid Biochem Mol Biol 20017833–40. [DOI] [PubMed] [Google Scholar]
- 16.Zhao C, Lam E W, Sunters A.et al Expression of estrogen receptor beta isoforms in normal breast epithelial cells and breast cancer: regulation by methylation. Oncogene 2003227600–7606. [DOI] [PubMed] [Google Scholar]
- 17.Eastwood H, Brown K M, Markovic D.et al Variation in the ESR1 and ESR2 genes and genetic susceptibility to anorexia nervosa. Mol Psychiatry 2002786–89. [DOI] [PubMed] [Google Scholar]
- 18.Nilsson M, Naessen S, Dahlman I.et al Association of estrogen receptor beta gene polymorphisms with bulimic disease in women. Mol Psychiatry 2004928–34. [DOI] [PubMed] [Google Scholar]
- 19.Sun Y H, Yang B, Wang X H.et al Association between single‐nucleotide polymorphisms in estrogen receptor beta gene and risk of prostate cancer. Zhonghua Wai Ke Za Zhi 200543948–951. [PubMed] [Google Scholar]
- 20.Pirskanen M, Hiltunen M, Mannermaa A.et al Estrogen receptor beta gene variants are associated with increased risk of Alzheimer's disease in women. Eur J Hum Genet 2005131000–1006. [DOI] [PubMed] [Google Scholar]
- 21.Maruyama A, Nakayama T, Sato N.et al Association study using single nucleotide polymorphisms in the estrogen receptor beta (ESR2) gene for preeclampsia. Hypertens Res 200427903–909. [DOI] [PubMed] [Google Scholar]
- 22.Almeida S, Franken N, Zandona M R.et al Estrogen receptor 2 and progesterone receptor gene polymorphisms and lipid levels in women with different hormonal status. Pharmacogenomics J 2005530–34. [DOI] [PubMed] [Google Scholar]
- 23.Ogawa S, Emi M, Shiraki M.et al Association of estrogen receptor beta (ESR2) gene polymorphism with blood pressure. J Hum Genet 200045327–330. [DOI] [PubMed] [Google Scholar]
- 24.Hakansson A, Westberg L, Nilsson S.et al Interaction of polymorphisms in the genes encoding interleukin‐6 and estrogen receptor beta on the susceptibility to Parkinson's disease. Am J Med Genet B Neuropsychiatr Genet 200513388–92. [DOI] [PubMed] [Google Scholar]
- 25.Westberg L, Hakansson A, Melke J.et al Association between the estrogen receptor beta gene and age of onset of Parkinson's disease. Psychoneuroendocrinology 200429993–998. [DOI] [PubMed] [Google Scholar]
- 26.Gold B, Kalush F, Bergeron J.et al Estrogen receptor genotypes and haplotypes associated with breast cancer risk. Cancer Res 2004648891–8900. [DOI] [PubMed] [Google Scholar]
- 27.Phiel K L, Henderson R A, Adelman S J.et al Differential estrogen receptor gene expression in human peripheral blood mononuclear cell populations. Immunol Lett 200597107–113. [DOI] [PubMed] [Google Scholar]
- 28.Kaushik N, Fear D, Richards S C.et al Gene expression in peripheral blood mononuclear cells from patients with chronic fatigue syndrome. J Clin Pathol 200558826–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Powell R, Ren J, Lewith G.et al Identification of novel expressed sequences, up‐regulated in the leucocytes of chronic fatigue syndrome patients. Clin Exp Allergy 2003331450–1456. [DOI] [PubMed] [Google Scholar]
- 30.Steinau M, Unger E R, Vernon S D.et al Differential‐display PCR of peripheral blood for biomarker discovery in chronic fatigue syndrome. J Mol Med 200482750–755. [DOI] [PubMed] [Google Scholar]
- 31.Whistler T, Unger E R, Nisenbaum R.et al Integration of gene expression, clinical, and epidemiologic data to characterize chronic fatigue syndrome. J Transl Med 2003110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gräns H, Nilsson P, Evengård B. Gene expression profiling in the chronic fatigue syndrome. J Intern Med 2005258388–390. [DOI] [PubMed] [Google Scholar]
- 33.Follettie M T, Pinard M, Keith J C., Jret al Organ messenger ribonucleic acid and plasma proteome changes in the adjuvant‐induced arthritis model: responses to disease induction and therapy with the estrogen receptor‐beta selective agonist ERB‐041. Endocrinology 2006147714–723. [DOI] [PubMed] [Google Scholar]
- 34.Harris H A, Albert L M, Leathurby Y.et al Evaluation of an estrogen receptor‐beta agonist in animal models of human disease. Endocrinology 20031444241–4249. [DOI] [PubMed] [Google Scholar]