Abstract
Background
Clinical outcome in patients with diffuse large B cell lymphomas (DLBCL) is highly variable and poorly predictable. Microarray studies showed that patients with DLBCL with a germinal centre B cell‐like (GCB) phenotype have a better prognosis than those with an activated B cell‐like (ABC) phenotype. The BMI1 proto‐oncogene was identified as one of the genes present in the signature of the ABC type of DLBCL, associated with a poor prognosis.
Objectives
(1) To investigate, in primary nodal DLBCL, the expression of BMI1 and its association with clinical outcome and DLBCL signature; (2) to look for an association between BMI1 expression and the expression of its putative downstream targets p14ARF and p16INK4a.
Results
BMI1 expression was found to be associated with poor clinical outcome, but not clearly with an ABC‐like phenotype of DLBCL. Expression of BMI1 was frequently, but not always, related to low levels of expression of p14ARF and p16INK4a.
Conclusion
Expression of BMI1 is associated with an unfavourable clinical outcome of primary nodal DLBCL.
Diffuse large B cell lymphomas (DLBCL) are heterogeneous in terms of clinical behaviour, histological features and differences in response to treatment. Currently, the clinical parameters comprised in the International Prognostic Index are used as a prognostic indicator in DLBCL.1 Also, various cellular and molecular factors that have prognostic significance in DLBCL have been identified.2,3,4,5,6,7,8,9
Neoplastic cells in DLBCL cases originate from germinal centre B (GCB) cells or their descendents.10 Recent studies based on microarray analysis showed that part of DLBCL phenotypically resemble non‐neoplastic GCB cells, but that part of DLBCL show an expression profile more consistent with an activated B cell (ABC)‐like phenotype.11,12 Furthermore, DLBCL with a GCB‐like phenotype have a considerably better prognosis than DLBCL with an ABC‐like phenotype.4BMI1 was identified as one of the genes that distinguish the GCB‐like from ABC‐like DLBCL (supplement of studies by Alizadeh et al11 and de Boer et al13), with high expression levels in ABC‐like DLBCL.
BMI1 belongs to the Polycomb group of genes that are important regulators of mammalian lymphopoiesis (reviewed in Raaphorst et al14) Furthermore, BMI1 has been shown to be essential for self‐renewal of haematopoietic and neural stem cells, in part through inhibition of genes regulating senescence.15 Initially, BMI1 was identified as a proto‐oncogene in the development of lymphomas.16 Further studies showed that overexpression of BMI1 in transgenic mice results in down regulation of the cell cycle inhibitors p19ARF and p16INK4a, and that BMI1 cooperates with c‐Myc in tumorigenesis by inhibiting c‐Myc‐induced apoptosis through p14ARF.15,17,18
p14ARF and p16INK4a are alternatively spliced products from the INK4a/ARF (CDKN2A) locus, with no structural homology. p14ARF is involved in the induction of apoptosis via p53, whereas p16INK4a is involved in the inhibition of cell cycle progression through cyclin D1 and CDK4/6.19 Both these processes can be controlled by BMI1 via the INK4a/ARF locus.20,21
In non‐neoplastic lymphoid tissues, BMI1 is primarily expressed in resting cells. In follicle center B cells, the non‐neoplastic counterpart of at least part of DLBCL, BMI1, was mainly expressed in centrocytes, but not in dividing centroblasts.22 This was in contrast with aggressive B cell lymphomas in which we have previously observed that BMI1 is frequently expressed in dividing neoplastic cells, suggesting that aberrant Polycomb group expression contributes to malignant transformation in these lymphomas.23,24 We also showed that BMI1 is preferentially expressed in aggressive B cell lymphomas (DLBCL, Burkitt's lymphomas and mantle cell lymphomas), and not in indolent lymphomas (follicular and small lymphocytic lymphomas).23 However, in this study we did not investigate whether BMI1 expression predicts clinical outcome.
Thus, BMI1 is a proto‐oncogene that, when aberrantly expressed in mice, is involved in the pathogenesis of lymphomas, possibly by disruption of the p14ARF and/or by p16INK4a regulated pathways.24,25 Furthermore, inhibition of apoptosis by disruption of the p14ARF pathway may result in reduced sensitivity to chemotherapy‐induced cell death and poor outcome in patients with BMI1‐positive lymphomas.
Therefore, we investigated in primary nodal DLBCL with extensive follow‐up whether BMI1 expression indeed correlates with an ABC‐like phenotype and whether expression of BMI1 predicts poor clinical outcome. Furthermore, to get insight into the possible pathogenic function of BMI1, we investigated whether expression of BMI1 is related to decreased levels of p14ARF and/or p16INK4a expression.
Materials and methods
Clinical material
Selection of formalin‐fixed, paraffin wax‐embedded tissue blocks of 60 biopsy specimens of primary nodal DLBCL and the clinical data of these patients were described previously.8 Table 1 summarises the patient characteristics. Most patients (n = 60) received polychemotherapy, consisting of CHOP (cyclophosphamide, doxorubicin, vincristine, prednisone) regimens or variants, either alone (n = 31) or in combination with involved field radiation (n = 24); in five cases, only involved field radiation was given. The institutional review board of the VU Medical Centre (Amsterdam, The Netherlands) approved the study. Informed consent was provided according to the Declaration of Helsinki.
Table 1 Patient characteristics obtained by univariate survival analysis.
| Patient characteristics | Number | Deaths with lymphoma | 5‐Year survival* | p Value† |
|---|---|---|---|---|
| IPI‡ | ||||
| 1 | 16 | 5 | 58 | NS |
| 2 | 21 | 12 | 44 | |
| 3 | 14 | 7 | 43 | |
| 4 | 8 | 6 | 0 | |
| B symptoms | ||||
| Yes | 19 | 13 | 32 | 0.02 |
| No | 41 | 18 | 52 | |
| Stage‡ | ||||
| 1+2 | 25 | 8 | 63 | 0.03 |
| 3+4 | 34 | 22 | 34 | |
| ABC/GCB profile§ | ||||
| GCB | 22 | 8 | 59 | 0.06 |
| ABC | 36 | 22 | 38 |
ABC, activated B cell; GCB, germinal centre B cells; IPI, International Prognostic Index.
*As estimated from the Kaplan–Meier curves.
†As determined by χ2 test, unless stated otherwise.
‡Data concerning stage and therefore IPI could not be retrieved in one patient.
§Data concerning the ABC/GCB profile could not be retrieved in two patients.
Antibodies used in this study
Monoclonal BMI1 antibody (clone 6C9) was generated previously,26 polyclonal p14ARF was obtained from Abcam, Cambridge, UK, and monoclonal p16Ink4a (clone E6H4) was a kind gift from Dr R Ridder. Specificity of these antibodies was determined by western blot analysis and bands were found at the predicted height (not shown).
Immunohistochemistry
Slides of paraffin wax‐embedded biopsy specimens were used for the detection of BMI1, p14ARF and p16INK4a. Sections were deparaffinised with xylene, endogenous peroxidase was blocked, and antigens were retrieved in either citrate buffer (10 mmol/l, pH 6.0) or TRIS/EDTA buffer (10 mmol/l/1 mmol/l, pH 9.0) in either microwave or autoclave.
Methods for detection of BMI1 were described previously.22 For p14ARF and p16INK4a detection, slides were rinsed in phosphate‐buffered saline and primary antibody was applied at optimal dilution. After 1 h at room temperature (20°C), slides were incubated with highly sensitive EnVison horseradish peroxidase system (Dako, Glostrup, Denmark). Bound antibodies were visualised by incubation with diaminobenzidine/H2O2. Slides were counterstained with haematoxylin.
Quantification of immunohistochemical staining
For all stainings, percentages of positive tumour cells were evaluated semiquantitatively in approximately the same area; scattered reactive lymphocytes served as an internal positive control. Two observers analysed all stainings independently. In case of disagreement, the staining results were reanalysed by the observers until consensus was reached.
To determine cut‐off levels with the most discriminative power, cut‐off was tested at 25%, 50% and 75% in relation to prognosis; the statistically most significant cut‐off (at 25% for all stainings) was used in this study.
Phenotypic analysis of GCB‐like DLBCL versus ABC‐like DLBCL
Previously, we divided cases in a GCB‐like or an ABC‐like subtype,27 by an algorithm modified from the one previously described by Hans et al4.
Statistical analysis
Survival time was measured from the time of initial diagnosis until death with disease or until the end of follow‐up. Patients who died of causes unrelated to the disease were censored at the time of death (n = 2).
Survival curves were constructed with the Kaplan–Meier method. Differences between the curves were analysed using the log‐rank test. Multivariate analysis was performed using the Cox proportional hazards model.28 Qualitative variables were analysed by Pearson χ2 test or by the Fisher exact test, where appropriate. The Kruskal–Wallis test to compare group means and the Spearman test was used to test correlations between different variables. All p values are based on two‐tailed statistical analysis, considering p values <0.05 as significant. Analyses were performed using SPSS V.10.1.
Results
Aberrant expression of BMI1 in DLBCL as compared with normal follicle centre cells
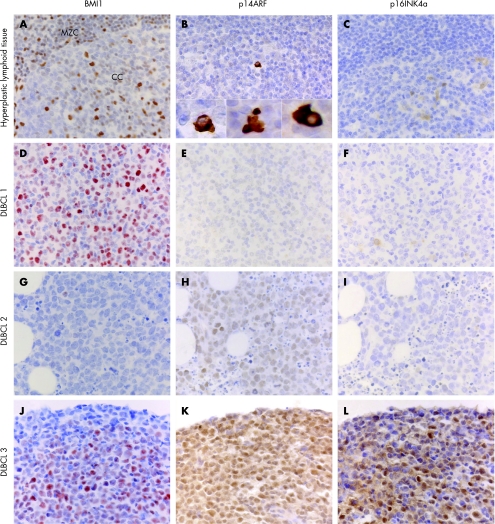
As described previously, BMI1 expression in non‐neoplastic germinal centres is almost completely restricted to non‐dividing cells.22,29 In DLBCL, BMI1 was detected in DLBCL in all patients, but in highly varying percentages of dividing lymphoma cells. The percentage of BMI1‐positive tumour cells ranged from <5% to almost 100% (fig 1); 25% of the cases had <25% whereas 75% had >25% BMI1‐positive tumour cells.
Figure 1 Expression of BMI1, p14ARF and p16INK4a in non‐neoplastic lymphoid tissue and in three primary nodular DLBCL. (A–C) hyperplastic lymphoid tissue. BMI1 staining is observed in resting mantle zone cells, centrocytes and T cells including germinal centre T cells, but not in centroblasts.29p14ARF and p16INK4a expression is detectable only in sporadic cells. Some of the p14ARF‐positive cells show morphological features of apoptosis (fig B insets). (D–L) BMI1, p14ARF and p16INK4a staining patterns in three different DLBCL cases. DLBCL #1: this case shows BMI1‐positive staining in >25% of tumour cells, but no expression of p14ARF and p16INK4a. DLBCL #2: expression of p14ARF is observed in the absence of BMI1 and p16INK4A expression. DLBCL #3: this case shows expression of BMI1, p14ARF and p16INK4a.
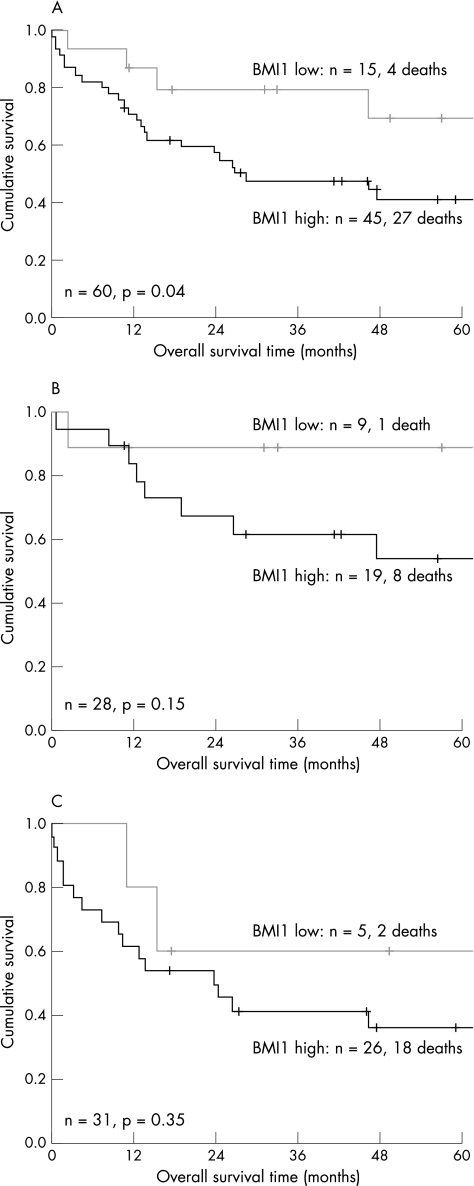
Low levels of BMI1 expression are related to a favourable clinical outcome
Using log‐rank and Cox regression analysis, different thresholds of percentages of BMI1‐positive tumour cells were tested; the threshold with optimal discriminative power with regard to overall survival was 25% (table 2, fig 2A). Using multivariate analysis including GCB/ABC phenotype and BMI1 expression as categorical variables did not yield additional significant prognostic value for expression of BMI1 above GCB versus ABC phenotype (fig 2B,C). However, expression of BMI1 tended to be related to poor outcome, especially in GCB‐like DLBCL (90% 5‐year overall survival time v 55% 5‐year overall survival time for BMI1 negative and positive cases, respectively).
Table 2 Patient characteristics in relation to BMI1 expression.
| Patient characteristics | % BMI1‐positive tumour cells | p Value* | |
|---|---|---|---|
| <25% (n = 15) | ⩾25% (n = 45) | ||
| Median age (range) | 62.3 (24–94) | 63 (34–94) | NS† |
| Sex | |||
| M | 12 | 27 | NS |
| F | 3 | 18 | |
| Stage‡ | |||
| 1+2 | 5 | 20 | NS |
| 3+4 | 10 | 24 | |
| B symptoms | |||
| Yes | 5 | 14 | NS |
| No | 10 | 31 | |
| IPI§ | |||
| 1+2 | 8 | 29 | NS |
| 3+4 | 7 | 15 | |
| Complete remission | |||
| Yes | 13 | 29 | NS |
| No | 2 | 16 | |
| Relapse¶ | |||
| Yes | 4 | 9 | NS |
| No | 9 | 20 | |
| Death | |||
| Yes | 4 | 27 | 0.04 |
| No | 11 | 18 | |
F, female; IPI, International Prognostic Index; M, male.
*As determined by χ2 test, unless stated otherwise.
†As determined by Mann–Whitney U test.
‡Data concerning the staging could not be retrieved in one case.
§Data concerning IPI could not be retrieved in one case.
¶These cases involve patients who did reach complete remission.
Figure 2 Comparison of overall survival in patients with primary nodal DLBCL according to BMI1 expression. High numbers of BMI1‐positive tumour cells are correlated with poor prognosis (A). BMI1 expression has no additional significant prognostic value above GCB (B) versus ABC (C) phenotype.
BMI1 expression is not restricted to activated B cell‐like DLBCL
Microarray studies have shown that BMI1 is one of the genes that distinguishes a GCB‐like DLBCL from an ABC‐like DLBCL. Using immunohistochemical analysis, cases with many BMI1 positive tumour cells more often showed an ABC‐like phenotype, although this relation was not significant (table 3).
Table 3 Tumour characteristics in relation to percentages of BMI1 positive tumour cells.
| Tumour characteristics | BMI1‐positive tumour cells (%) | p Value* | |
|---|---|---|---|
| <25% (n = 15) | ⩾25% (n = 45) | ||
| ABC/GCB profile | |||
| GCB | 9 | 19 | NS |
| ABC | 5 | 26 | |
| p14ARF | |||
| Negative | 2 | 8 | NS |
| Positive | 0 | 11 | |
| p16INK4a | |||
| Negative | 2 | 19 | NS |
| Positive | 0 | 6 | |
Results of interpretable cases are given. The number of cases for p14ARF and p16INK4a stainings is low as only cases were included, and for these it was confirmed that there are no genetical aberrations present in the CDKN2A locus. An ABC/GCB profile could not be defined in one case due to uninterpretable staining of one of the individual markers.
*As determined by χ2 test.
Aberrant expression of p16INK4a and p14ARF in DLBCL as compared with normal follicle centre cells
To determine whether BMI1 expression is related to (lowered) p14ARF or p16INK4a expression, we subsequently investigated p14ARF and p16INK4a expression in normal lymphoid tissue and DLBCL cases. In non‐neoplastic lymphoid tissue p14ARF and p16INK4a are expressed in a limited number of cells of the germinal centre (fig 1). Nuclear expression of p16INK4a was found mainly in centroblasts. Interestingly, cytoplasmic p14ARF expression was also detected in sporadic interfollicular cells with apoptotic characteristics, consistent with its apoptosis‐inducing function.20
In contrast, in p14ARF and p16INK4a positive tumours, expression was observed in the nucleus and cytoplasm of most neoplastic cells of DLBCL. In addition, in DLBCL, p14ARF‐positive cells did not show apoptotic characteristics (fig 1).
Expression of BMI1 in DLBCL is not consistently related to low levels of p16INK4a or p14ARF expression
Expression of p14ARF and p16INK4a was detected as nuclear staining in the tumour cells in 21 of 42 and in 22 of 59 cases, respectively. Expression of p14ARF and p16INK4a showed no direct correlation with expression of BMI1. However, p16INK4a tended to be more frequently absent in cases with many BMI1‐positive tumour cells: 29 of 43 patients with high numbers of BMI1‐positive tumour cells showed no expression of p16INK4a as compared with 8 of 16 patients with few BMI1‐positive tumour cells (p = 0.24; not shown). Also, when the analysis was restricted to cases without a proven genetic loss in the CDKN2A locus as determined by CGH analysis (n = 27; Oudejans JJ et al, manuscript in preparation), no correlation between expression of p14ARF and p16INK4a, and BMI1 was observed (table 3).
Discussion
In this study, we have shown that BMI1 and its presumed target genes p14ARF and p16INK4a are frequently aberrantly expressed in malignant cells of DLBCL when compared with normal follicle centre B‐lymphocytes, and that expression of BMI1 is related to an unfavourable clinical outcome in primary nodal DLBCL.
It was previously shown and confirmed by us that DLBCL with an ABC‐like phenotype have a relatively unfavourable clinical outcome and that one of the genes involved in the designation of this profile is BMI14 (supplement of studies by Alizadeh et al11, de Boer et al13 and Muris et al27). Although we found that BMI1 expression is indeed more frequently present in ABC‐like DLBCL, BMI1 expression was not restricted to this group of DLBCL. This might be explained by the fact that, also in the microarray study, BMI1 alone does not determine the GCB‐like versus ABC‐like phenotype.11,13 Expression of BMI1 was also not significantly correlated with expression of MUM‐1 (data not shown), which was previously shown to be related to a poor prognosis.27 This indicates that the prognostic effect of BMI1 expression is probably not directly related to MUM‐1 expression.
A possible explanation for the observed relationship between BMI1 expression and prognosis is that BMI1 inhibits apoptosis via down regulation of the p14ARF gene. Experimental model systems demonstrated that BMI1 cooperates with c‐Myc to induce lymphomas by preventing c‐Myc and Ink4a/ARF induced apoptosis,18 thus inhibiting the intrinsic apoptosis pathway and thereby explaining poor response to chemotherapy. However, inhibition of apoptosis by high expression of BMI1 would imply down regulation of p14ARF, which was observed only in some BMI1‐positive cases. In non‐neoplastic lymphoid tissues, expression of p14ARF in reactive lymph nodes was observed in a few cells frequently, with morphological features of apoptosis (see fig 1). This would be consistent with the normal tumour‐suppressive, apoptosis‐inducing function of p14ARF. However, in p14ARF‐positive DLBCL, diffuse positive staining was observed in cells without morphological features of apoptosis, suggesting that the normal apoptosis‐inducing effect of p14ARF is disrupted, consistent with a previous paper by Sanchez‐Aguilera et al.30 Thus, in some cases, p14ARF is expressed despite expression of BMI1, but its apoptosis‐inducing effect might be counteracted by downstream apoptosis‐inhibiting proteins like Bcl‐2 and XIAP.
Alternatively, BMI1 could be involved in down regulation of the tumour suppressor protein p16INK4a, resulting in suppression of senescence and cell cycle control,15 causing enhanced proliferation of the tumour cells. Indeed, in our study we also found BMI1‐positive cases that showed absence or low levels of p16INK4a expression. However, some BMI1‐positive patients also showed expression of p16INK4a in most rapidly proliferating cells. Similar to p14ARF, expression of p16INK4a in non‐neoplastic reactive lymphoid tissues is restricted to very few follicle centre cells. Thus, the normal cell cycle suppressive function of p16INK4a in these lymphomas is probably disrupted. Co‐expression of BMI1 and p14ARF/p16INK4a may be explained by phosphorylation of BMI1 and subsequent de‐repression of the INK4a/ARF locus, as recently shown by Voncken et al31
Recently, Glinsky et al32 also investigated the expression of BMI1 and its putative targets in relation to prognosis. Expression of these 11 BMI1 target genes was correlated to poor prognosis in patients with lymphoma. Thus, BMI1 seems to be an important gene in normal and neoplastic tissue, because of its role in regulating the expression of various genes involved in cell cycle control.
We conclude that BMI1 and p14ARF and p16INK4a are aberrantly expressed in some primary nodal DLBCL. Expression of BMI1 is related to unfavourable outcome. Down regulation of tumour suppressor genes p14ARF and p16INK4a by BMI1 might explain the relationship between BMI1 expression and poor outcome in some DLBCL. However, strong expression of p14ARF and p16INK4a in some DLBCL suggests that the tumour‐suppressive effect of these proteins is counteracted by downstream mechanisms.
Take‐home messages
Expression of BMI1 in diffuse large B cell lymphomas correlates with poor clinical outcome.
Strong expression of p14ARF and p16INK4a in some DLBCL suggests that the tumour‐suppressive effect of these proteins is counteracted by downstream mechanisms.
Expression of BMI1 is not restricted to activated B cell like DLBCL.
Acknowledgements
We thank the following persons for their help in collecting tumour material and clinical data: Professor R Willemze, Professor HC Kluin‐Nelemans, Dr LH Siegenbeek van Heukelom, Dr NM Jiwa, Dr MA MacKenzie and Dr JH Krieken. We also thank Dr R Ridder for kindly providing the p16INK4a antibody.
Abbreviations
ABC - activated B cell
DLBCL - diffuse large B cell lymphomas
GCB - germinal centre B cells
Footnotes
Competing interests: None.
References
- 1.The International Non‐Hodgkin's Lymphomas Prognostic Factors Project A predictive model for aggressive non‐Hodgkin's lymphoma. N Engl J Med 1993329987–994. [DOI] [PubMed] [Google Scholar]
- 2.Kramer M H, Hermans J, Parker J.et al Clinical significance of Bcl‐2 and p53 protein expression in diffuse large B‐cell lymphoma: a population‐based study. J Clin Oncol 1998142131–2138. [DOI] [PubMed] [Google Scholar]
- 3.Barrans S L, Carter I, Owen R G.et al Germinal center phenotype and bcl‐2 expression combined with the International Prognostic Index improves patient risk stratification in diffuse large B‐cell lymphoma. Blood 2002991136–1143. [DOI] [PubMed] [Google Scholar]
- 4.Hans C P, Weisenburger D D, Greiner T C.et al Confirmation of the molecular classification of diffuse large B‐cell lymphoma by immunohistochemistry using a tissue microarray. Blood 2004103275–282. [DOI] [PubMed] [Google Scholar]
- 5.Lossos I S, Jones C D, Warnke R.et al Expression of a single gene, BCL‐6, strongly predicts survival in patients with diffuse large B‐cell lymphoma. Blood 200198945–951. [DOI] [PubMed] [Google Scholar]
- 6.Chang C C, Liu Y C, Cleveland R P.et al Expression of c‐Myc and p53 correlates with clinical outcome in diffuse large B‐cell lymphomas. Am J Clin Pathol 2000113512–518. [DOI] [PubMed] [Google Scholar]
- 7.Yamaguchi M, Seto M, Okamoto M.et al De novo CD5+ diffuse large B‐cell lymphoma: a clinicopathologic study of 109 patients. Blood 200299815–821. [DOI] [PubMed] [Google Scholar]
- 8.Muris J J, Meijer C J, Cillessen S A.et al Prognostic significance of activated cytotoxic T‐lymphocytes in primary nodal diffuse large B‐cell lymphomas. Leukemia 200418589–596. [DOI] [PubMed] [Google Scholar]
- 9.Muris J J, Cillessen S A, Vos W.et al Immunohistochemical profiling of caspase signaling pathways predicts clinical response to chemotherapy in primary nodal diffuse large B‐cell lymphomas. Blood 20051052916–2923. [DOI] [PubMed] [Google Scholar]
- 10.Kuppers R, Klein U, Hansmann M L.et al Cellular origin of human B‐cell lymphomas. N Engl J Med 19993411520–1529. [DOI] [PubMed] [Google Scholar]
- 11.Alizadeh A A, Eisen M B, Davis R E.et al Distinct types of diffuse large B‐cell lymphoma identified by gene expression profiling. Nature 2000403503–511. [DOI] [PubMed] [Google Scholar]
- 12.Shipp M A, Ross K N, Tamayo P.et al Diffuse large B‐cell lymphoma outcome prediction by gene‐expression profiling and supervised machine learning. Nat Med 2002868–74. [DOI] [PubMed] [Google Scholar]
- 13.de Boer W P, Oudejans J J, Meijer C J.et al Analysing gene expressions with GRANK. Bioinformatics 2003192000–2001. [DOI] [PubMed] [Google Scholar]
- 14.Raaphorst F M, Otte A P, Meijer C J. Polycomb‐group genes as regulators of mammalian lymphopoiesis. Trends Immunol 200122682–690. [DOI] [PubMed] [Google Scholar]
- 15.Jacobs J J, Kieboom K, Marino S.et al The oncogene and Polycomb‐group gene BMI1 regulates cell proliferation and senescence through the ink4a locus. Nature 1999397164–168. [DOI] [PubMed] [Google Scholar]
- 16.van Lohuizen M, Verbeek S, Scheijen B.et al Identification of cooperating oncogenes in E mu‐myc transgenic mice by provirus tagging. Cell 199165737–752. [DOI] [PubMed] [Google Scholar]
- 17.Alkema M J, Bronk M, Verhoeven E.et al Identification of BMI1‐interacting proteins as constituents of a multimeric mammalian polycomb complex. Genes Dev 199711226–240. [DOI] [PubMed] [Google Scholar]
- 18.Jacobs J J, Scheijen B, Voncken J W.et alBMI1 collaborates with c‐Myc in tumorigenesis by inhibiting c‐Myc‐induced apoptosis via INK4a/ARF. Genes Dev 1999132678–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruas M, Peters G. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim Biophys Acta 19981378F115–F177. [DOI] [PubMed] [Google Scholar]
- 20.Park I K, Morrison S J, Clarke M F. Bmi1, stem cells, and senescence regulation. J Clin Invest 2004113175–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowe S W, Sherr C J. Tumor suppression by Ink4a‐Arf: progress and puzzles. Curr Opin Genet Dev 20031377–83. [DOI] [PubMed] [Google Scholar]
- 22.Van Galen J C, Dukers D F, Giroth C.et al Distinct expression patterns of polycomb oncoproteins and their binding partners during the germinal center reaction. Eur J Immunol 2004341870–1881. [DOI] [PubMed] [Google Scholar]
- 23.van Kemenade F J, Raaphorst F M, Blokzijl T.et al Coexpression of BMI1 and EZH2 polycomb‐group proteins is associated with cycling cells and degree of malignancy in B‐cell non‐Hodgkin lymphoma. Blood 2001973896–3901. [DOI] [PubMed] [Google Scholar]
- 24.Raaphorst F M. Of mice, flies, and man: the emerging role of polycomb‐group genes in human malignant lymphomas. Int J Hematol 200581281–287. [DOI] [PubMed] [Google Scholar]
- 25.Valk‐Lingbeek M E, Bruggeman S W, van Lohuizen M. Stem cells and cancer; the polycomb connection. Cell 2004118409–418. [DOI] [PubMed] [Google Scholar]
- 26.Hamer K M, Sewalt R G, den Blaauwen J L.et al A panel of monoclonal antibodies against human polycomb group proteins. Hybrid Hybridomics 200221245–252. [DOI] [PubMed] [Google Scholar]
- 27.Muris J J F, Meijer C J L M, Vos W.et al Immunohistochemical profiling based on Bcl‐2, CD10 and MUM1 expression improves risk stratification in patients with primary nodal diffuse large B‐cell lymphoma. J Pathol 2006208714–723. [DOI] [PubMed] [Google Scholar]
- 28.Cox D R. Regression models and life tables. J Roy Statist Soc Ser B Methodol 197234187–220. [Google Scholar]
- 29.Raaphorst F M, van Kemenade F J, Fieret E.et al Cutting edge: polycomb gene expression patterns reflect distinct B cell differentiation stages in human germinal centers. J Immunol 20001641–4. [DOI] [PubMed] [Google Scholar]
- 30.Sanchez‐Aguilera A, Sanchez‐Beato M, Garcia J F.et al p14(ARF) nuclear overexpression in aggressive B‐cell lymphomas is a sensor of malfunction of the common tumor suppressor pathways. Blood 2002991411–1418. [DOI] [PubMed] [Google Scholar]
- 31.Voncken J W, Niessen H, Neufeld B.et al MAPKAP kinase 3pK phosphorylates and regulates chromatin association of the polycomb group protein Bmi1. J Biol Chem 20052805178–5187. [DOI] [PubMed] [Google Scholar]
- 32.Glinsky G V, Berezovska O, Glinskii A B. Microarray analysis identifies a death‐from‐cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest 20051151503–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]