Although it has been more than five years since the draft of the human genome was first announced,w1 only in recent months has there been substantial progress in the identification of genes that are implicated in susceptibility or cause of myocardial infarction (MI). There are many reasons for this lag, particularly related to the point that myocardial infarction is a “complex” trait. As opposed to “simple” Mendelian traits which are infrequent, deterministic and inherited in an autosomal dominant, recessive, or X‐linked mode, complex traits are relatively common, probabilistic, and involve gene–gene and gene–environmental interactions. The technology and methodology to identify complex traits have rapidly evolved, and still can be considered to be in flux, with a progression from linkage studies using genome wide scanning to whole genome single nucleotide polymorphism (SNP) association studies. With this refinement in approaches, several genes are now implicated for susceptibility for myocardial infarction and one is already the subject of a dedicated randomised pharmacologic intervention trial for “personalised” medicine. This paper will review the progress in this burgeoning and important field, to provide the reader with a perspective on how genetic information will someday radically alter our approach and facilitate the prevention of heart attacks.
THE TOOLS
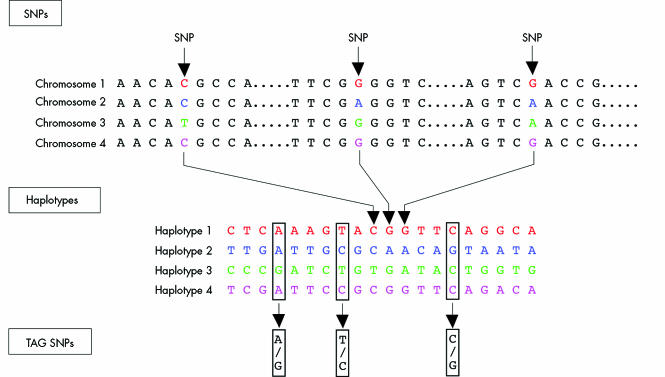
With a human genome that consists of 3.1 billion base pairs, finding the particular alleles that are promoting or protecting an individual to have an MI appears, on the surface, to be especially daunting. But the number of genes that comprise the genome is just over 25 000 and although the function of many has yet to be determined, the chromosomal map of the location of these genes is virtually complete. With 3.1 billion nucleotide pairs (cytosine, adenine, guanine, thymine), fortunately the inter‐subject variability is rather limited to only 10 million SNPs—or 0.3% of the base pairs that are in “play”. So Mother Nature has helped to simplify the hunt by having considerably less genes than what was anticipated (most experts had projected 80–100 000), and far less variability than might have otherwise occurred. Recent insights have made this even simpler, with the realisation that there are haplotype blocks, that thousands of nucleotides are inherited as a unit, and that these blocks can be identified using a “tag” SNPw2 (fig 1). The ongoing International Haplotype Map (“HapMap”) project is characterising the haplotypes and tag‐SNPs for the human genome and it is expected that knowledge of only 250–350 000 tag‐SNPs (depending on racial ancestry) will be all that is necessary to identify each individual genomically. Thus, in essence, only 0.008% of the nucleotides comprising the human genome can be used to codify each individual. Comparing the critical SNPs of this rather small subset among affected patients and suitable controls will undoubtedly accelerate the definition of the genomic basis of MI.
Figure 1 Tag single nucleotide polymorphisms (SNPs) identify haplotype blocks. A, adenine; C, cytosine; G, guanine; T, thymine. Reproduced with permission from The International Hap Map Consortium.W2
Besides genomics, there are several other vital tools in the kit. The human transcriptome, comprised of the gene expression profile of the 25 000 genes, represents critical substrate that can be gleaned through microarrays. By systematically assessing the genes that are hyper‐ or hypo‐expressed in diseased compared with unaffected specimens, gene chip processing of arterial tissue has already been implicated in identifying genes that are associated with atherosclerosis.w3 Similarly, study of the human proteome, which consists of about 100 000 proteins (a 4:1 ratio to genes due to alternative splicing), is a means of identifying downstream evidence for a particular gene's involvement.w4 Finally, ascertainment of metabolic profiles, the study of “metabolomics”, involving approximately 10 000 metabolites, has also been effective in finding “fingerprints” associated with coronary artery disease.w5 The transcriptome, proteome, and metabolome information can all be used to corroborate the functional significance and mechanism of a putative disease‐causing gene.
“OLD” PRIMARY APPROACHES
For the first decade of the hunt for MI genes, case–control association studies were performed. Such studies typically looked at a particular SNP in a relatively small number of cases and controls and concluded that an association was or was not present. Such an approach was intrinsically biased, picking an SNP of interest to the investigators, and severely limited—an “ice pick” view of one base pair among three billion with dreadful lack of statistical power. Overall, such studies were not replicated and gave the field a bad name for reporting false positive and potentially false negative data.1
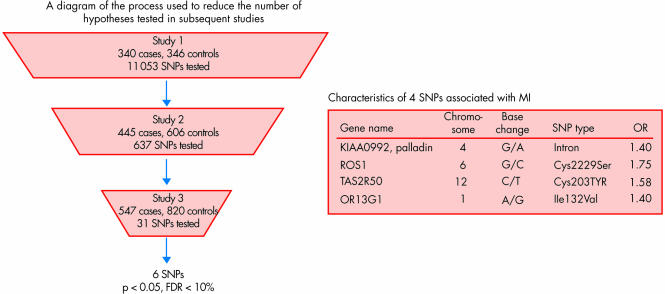
The next stage of investigation involved “high throughput” genotyping studies whereby instead of one or a few SNPs, hundreds could be assessed at once. This provided a slightly broader window into the genome, but still relatively limited to hundreds of SNPs at best. Furthermore, the problem of statistical power was compounded by correction for multiple comparisons, and the list of SNPs being assessed still represented a biased, non‐systematic approach. Despite these limitations, the chance of identifying important SNPs was increased similar to buying hundreds of tickets for the lottery contest, and indeed many such SNPs were replicated. Our work with the thrombospondin family of genes, for example, has been an example of a success for association with premature MI. Multiple groups have independently confirmed the association of thrombospondin 4 and 2 with risk of MI2 and functional genomic work has strongly supported the underlying mechanism.w6 w7 Other genes encoding for matrix proteins, such as connexin‐37 and stromelysin‐1, have also been validated in this way.3 Notwithstanding some success with high‐throughput genotyping, this approach is rather limited and cannot be considered adequate with respect to more recently accepted standards. Shiffman and colleagues have recently raised the bar for high‐throughput genotyping technology in analysing 11 053 SNPs in 6891 genes for three different cohorts of MI patients and controls (fig 2).4 Four SNPs were significantly associated with MI, replicated in all of the populations assessed, with odds ratios ranging from 1.4 to 1.8.4
Figure 2 High‐throughput gene association study in three cohorts yielding four single nucleotide polymorphisms (SNPs). FDR, false discovery rate.
LINKAGE STUDIES IN MULTIPLEX FAMILIES AND RICH PEDIGREES
A far more attractive technique is to systematically assess the genome which is possible through genome‐wide scanning. This refers to the use of microsatellite markers, about 400 such markers evenly spaced across the genome. Each microsatellite consists of tandem repeats of nucleotides (for example, TGTGTGTGTGTG….) selected approximately every megabase (Mb = 1 million base pairs). Instead of picking a candidate gene of interest, this approach lets the genome do the talking. Affected sibling pairs with MI can be scanned in such a way to determine whether a “linkage peak” exists, such that there is evidence of sharing of a particular allele or locus. The signal is much like a frequency distribution, and the significance of the linkage peak is expressed as a logarithm of the odds (LOD) score. An LOD score of > 3.5 corresponds to a p value of < 10−6. There have been several genome‐wide scans performed for coronary artery disease or MI (table 1), each identifying particular loci or linkage peaks of interest.5,6,7w8–12 Thus far only two scans have yielded specific genes that are implicated for susceptibility for MI. These are 5‐lipo‐oxygenase activation protein (FLAP), leukotriene‐4 hydrolase (LTA4), and myocyte enhancing factor (MEF2A). While such linkage studies are characteristically performed in hundreds of families and include affected sibling pairs (“multiplex” families), and unaffecting siblings in some analyses, a genome wide scan can also be done in a rich pedigree. Such a technique was recently used to identify the Notch‐1 gene as a cause of familial aortic valve calcification,w13 and was used to identify MEF2A in a single large family. In contrast to genome‐wide scanning of multiplex families, however, the rich pedigree approach is only helpful in defining a Mendelian gene; it relies on assumption of an inheritance pattern of autosomal, X‐linked or mitochondrial. The information generated can then be used to test whether other variants of the mutation or polymorphism account for a complex trait, which does not follow the characteristic or “simple” inheritance pattern.
Table 1 Genome wide scans for coronary artery disease or myocardial infarction.
| Study | Population | No. of families | Mean age (years) | Locus/loci | Gene |
|---|---|---|---|---|---|
| Sibling pairs | |||||
| Pajukanta et al 2000w8 | Finnish | 156 | <55 | 2q21 and Xq23 | – |
| Francke et al 2001w10 | Mauritian | 99 | 47 | 16p13 | – |
| Broeckel et al 2002w11 | European | 513 | 52 | 14q32 | – |
| Harrap et al 2002w12 | Australian | 61 | 62 | 2q36 | – |
| Wang et al 20045 | American | 428 | 44 | 1p34–36 | – |
| Hauser et al 2004w9 | Euro‐Amer | 438 | <56 | 3q13 | – |
| Helgadottir et al 20046 | Icelandics | 296 | – | 13q12–13 | FLAP |
| Helgadottir et al 200514 | Icelandics | 296 | – | 12q22 | LTA4H |
| Rich pedigree | |||||
| Wang et al 20037 | American | 1 | 58 | 15q26 | MEF2A |
WHOLE GENOME ASSOCIATION STUDIES
The next step in progression of being more systematic and requiring considerable more genotyping involves characterisation of tens of thousands to millions of SNPs. The first attempt to do this was performed by Ozaki and colleagues in Japan who analysed more than 65 000 SNPs in nearly 14 000 genes.8 This led to the discovery of lymphotoxin‐α (LTA), such that 2 SNPs in the third coding region (exon) had a highly significant association with MI (p < 10−6 and odds ratio 1.8). Interestingly, the LTA discovery set the stage for finding a key SNP in galectin‐2, the principal ligand for LTA, for a very strong association with MI (p < 10−6).9
Hinds and associates at Perlegen took this field forward with their recent report of 1 586 383 SNPs in 71 individuals of diverse racial ancestry.10 It is now possible to do a genome wide association study with 100 000 to conceivably 2 000 000 or more SNPs. Indeed, this was used in the recent discovery of complement factor H as the basis for age‐related macular degeneration, the leading cause of adult blindnessw14–18 which has been replicated by four other groups using other techniques including genome wide linkage scanning. By focusing on tag‐SNPs (fig 1), it is thought that only 250 000 for European‐Americans or 350 000 for Africans will be necessary to provide full disclosure genomic information for alleles that have a > 5% frequency.11 Use of this approach will further accelerate the identification of genes that induce MI susceptibility.
ORTHOLOGOUS GENES
Beyond direct human genomic or proteomic studies, orthologous genes for MI have been discovered from mouse and rat studies. A recent example is the multi‐histocompatibility (MHC) complex in inbred rats with a class II transactivator (MHC2TA) haplotype associated with increased expression that led to identification of a promoter SNP in the same gene in humans with increased susceptibility to rheumatoid arthritis, multiple sclerosis, and MI.w19 A recent study of Ox40L in mice demonstrating increased susceptibility to atherosclerosis led to the finding of higher susceptibility to MI in two separate patient cohorts.w20 Besides these examples, the original discovery of the importance of the 5‐lipooxygenase pathway was in certain strains of mice which were particularly susceptible to atherosclerosisw21; this was later confirmed to be an important pathway in the FLAP and LTA4 genome wide scanning human genetic studies. A murine model of coronary artery plaque rupture, histologically very similar to the picture in man, consisting of a double knockout of both the apoE and SRB‐1 genes,12 is an important foundation for testing MI gene discovery for confirmation in experimental models.
LESSONS FROM GENES RECENTLY IMPLICATED FOR MYOCARDIAL INFARCTION SUSCEPTIBILITY
5‐lipooxygenase activation protein (FLAP)
From a genome‐wide scan of 296 Icelandic multiplex families, Helgadottir and associates from DeCode Genetics identified a linkage peak at 13q12–13 that, through fine mapping of the locus, was attributed to a 4 SNP marker FLAP haplotype present in over 10% of MI patients and accounting for a twofold risk of MI and stroke.6 FLAP is a critical contributor to the conversion of 5‐lipooxygenase in the leukotriene pathway that ultimately yields leukotriene B4, one of the most potent white cell pro‐inflammatory mediators. This “gain‐of‐function” FLAP haplotype has now been confirmed in a British population for MI,6 a Scottish cohort for stroke,w22 and a Cleveland population for MI (unpublished data).
The knowledge that FLAP was implicated as the putative genomic basis for MI in patients led to the hypothesis that a FLAP blocker would have a particular, individualised, tailored benefit. Such gene‐targeted pharmacotherapy, the first in the field of cardiovascular medicine, was tested in a phase II placebo‐controlled, crossover trial of 191 patients with the at‐risk FLAP haplotype.13 This demonstrated that patients with the at‐risk haplotype had a significant 26% decrease in white cell leukotriene B4 production.13 A large‐scale phase III trial testing a FLAP blocker in patients with the at‐risk haplotype of high inflammatory biomarkers for reduction clinical events (prevention of a second MI or death) was initiated in late 2005.
Leukotriene A4 hydrolase (LTA4)
In the same genome‐wide scan of Iceland MI families, another locus on chromosome 12 corresponded to the gene LTA4, an intermediate in the same pathway that stems from 5‐LO to leukotriene B4. In a subsequent case–control study of 1553 patients with MI and 863 controls, a haplotype I LTA4 was associated with an odds ratio for MI of 1.45 (p = 0.0091).14 However, this at‐risk haplotype was present in 3.2% of African‐Americans with an odds ratio of 3.47 (p = 0.000022) but was much more common in European Americans, at 14.8% frequency, with an odds ratio of only 1.16. This led the investigators to conclude that the at‐risk haplotype of LTA4 has ethnic specificity. Indeed, in the exhaustive study by Hinds et al of almost 1.6 million SNPs in individuals of African, European‐American, or Asian ancestry, it was found that about 30% of SNPs and haplotype blocks across the genome are race‐specific, while approximately 70% are “cosmopolitan”, meaning there does not appear to be any partitioning according to ancestry.10 Patients with the LTA4 at‐risk haplotype were included with the FLAP blocker study (13% of the enrolled patients carried this abnormal haplotype) and appear to have a favourable biomarker response to an inhibitor of the pathway. Whether this will translate to reduction of adverse clinical events, and whether this will be a true gene‐specific advantage of therapy (as opposed to broad benefit without respect to genotype) remains to be determined in clinical trials.
Myocyte enhancing factor 2A (MEF 2A)
In late 2003 the Cleveland Clinic group reported the first gene that directly related to MI and coronary artery disease—MEF 2A.7 This was the outgrowth of a rich pedigree, a nuclear family of 21 individuals with a pattern of autosomal dominant inheritance. A genome wide scan for this family yielded a significant locus (LOD score of 4.2) at the telomere region of chromosome 15. At this locus consisting of 93 genes, the only gene known to have any prior connection with cardiovascular disease was MEF2A, a signal transduction factor that had shown to be critical for normal myocardial development. In the proband of this family, a 21 base pair deletion of the stop codon (exon 11) was present and this same mutation co‐segregated with all affected members of the family. Of note, the MEF2A deletion was not present in over 200 controls who had undergone coronary angiography and were free of any coronary disease. Transfection of the mutation into endothelial cells, with immunohistochemical staining, demonstrated that the transcription factor could not localise in the nucleus, giving a striking cytoplasmic pattern compared with wild‐type typical nuclear localisation. These findings from a large family with 248 living members and further genotyping in the extended family providing additional confirmation, together with the functional defect identified, strongly implicated MEF2A as an MI causing gene. Further support was lent by identifying point mutations in exon 7 among nearly 2% of 200 patients with coronary artery disease but not in 200 control individuals who had undergone coronary angiography.15 The point mutations also were associated with diminished transcription activity of MEF2A, suggesting they, too, are functionally important.
Subsequently, Weng et al published a report that challenged the MEF2A finding by demonstrating that the 21 base pair deletion was also found in individuals without known coronary artery disease, and approximately 0.15% of the population.16 That report, however, was not definitive, because only one family was adequately characterised and the individuals did not undergo coronary angiography or were not old enough to be ruled out for subsequent development of coronary disease. These investigators also identified a point mutation in MEF 2A present only in a patient with coronary disease and not controls, but excluded it as functionally significant on the basis of computational analysis.16 However, the mutation identified (S360L) is located in the critical transcriptional activation domain and may well affect the transactivation function of MEF2A.17
A recent report added to the strength of evidence for MEF2A as an MI causing gene. Gonzalez and associates from Spain analysed a Pro279Leu exon 7 variant in MEF2A and found it to have an odds ratio of 3.1 (p = 0.009) in 483 cases compared with 1189 controls; the frequency of the exon 7 SNP was 2.3% and 0.8%, respectively. Of note, all of the controls were less than 50 years old, such that some of these individuals may still have developed an MI at a more advanced age, and the odds ratio may represent an under‐estimate.18
In summary, there is still controversy as to whether MEF2A can be unequivocally considered as a disease‐causing gene for MI. Using transgenic mice, the precise mechanism by which the 21 base pair deletion mutation affects this transcription factor and potentially induces coronary artery disease or plaque rupture is being assessed. Further work by independent groups will determine if the initial replication will be confirmed by others. Most genes do not have absolute, deterministic phenotypic effects such that incomplete or variable penetrance is one possible explanation for the disparity noted to date. Another explanation relates to the heterogeneity of individuals, or population stratification, such that the phenotypic manifestations are also indexed to particular ethnic, ancestral and demographic characteristics.19
KEY POINTS FROM RECENT MYOCARDIAL INFARCTION GENES
There are a number of valuable lessons from the genes that have recently been identified to influence MI susceptibility. First, replication of the findings in multiple populations is ideal. This was the case with age‐related macular degeneration (AMD) in five different cohortsw14–18 and may be viewed as the platinum reference standard in the field of complex traits. It is not infrequent that a group cannot replicate a gene which is later confirmed by other groups, such as occurred with lymphotoxin‐α.w23 Second, functional data, ideally with mouse models that recapitulate the human phenotype (coronary plaque rupture) are needed to support the putative gene identification. Transfection of a mutation into cells can be viewed as supportive, but not nearly as important as a biologic system. Third, underlying linkage peaks one can find genes that constitute the genomic basis for a complex trait. Such was the case for FLAP and LTA4 and has been now consistently shown in a variety of complex traits including asthma, obesity, insulin‐dependent diabetes, inflammatory bowel disease, and many others. Usually a haplotype of a gene is identified as the one that confers an “at‐risk” effect, while another haplotype may be identified that renders protection from the condition, further anchoring the importance of the gene's effect. Fourth, identification of a gene for MI has led to a clinical programme to inhibit the “gain‐of‐function” gene (FLAP) and this may be prototypic of gene‐specific therapeutic programmes of the future. Fifth, a Mendelian gene discovered through an autosomal dominant inheritance pattern in a single family appears to have relevance to sporadic MI. In the case of MEF2A, in which the original mutation was an in‐frame deletion in the stop codon that fully blocked transcription, point mutations in other exons have been found in sporadic cases of MI and coronary artery disease. Accordingly, it is possible to identify genes for MI, a complex trait, via classic Mendelian genetics.
Sixth, we have already seen the importance of ancestry with the example of LTA4. Genes for MI will likely vary, to at least some extent, with certain genes being more important in particular racial or ethnic backgrounds.
MYOCARDIAL INFARCTION IS A RESTRICTIVE PHENOTYPE
One of the most important lessons to date has been acknowledgement that MI and coronary artery disease are phenotypically distinct. In many of the studies to date, no significance for gene association has been found for coronary artery disease, but a highly significant relationship has been found for MI. Examples of our work include the findings on thrombospondins 1, 2 and 4 of an association with premature MI that did not yield any significance for coronary atherosclerotic disease, and for chromosome 1p 34–36 linkage (table 1) which was only significant for MI but not coronary artery disease. Interestingly, for all the genome wide scans performed to date, none has found the gene underlying the linkage peak for coronary artery disease, but FLAP and LTA4 were identified using MI as the phenotype.
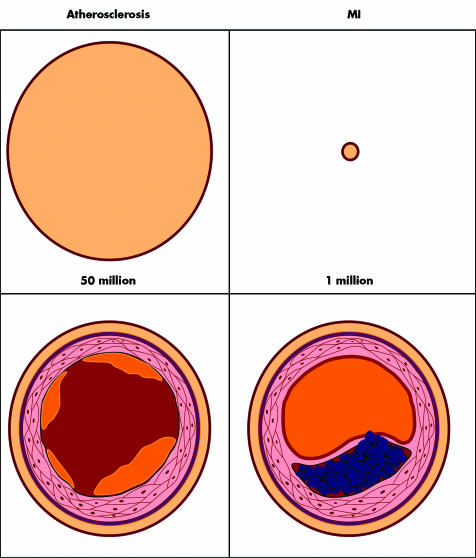
There is good rationale for partitioning the phenotype of MI and coronary artery disease (fig 3). MI is a much more restrictive phenotype which is relatively rare and has discrete clinical manifestations that “cross the line” to myocardial necrosis. Pathophysiologically, it requires a plaque rupture, fissure or erosion. On the other hand, coronary artery disease is exceptionally common, and in some form or other (including fatty streaks) is nearly pervasive in the adult population. The clinical manifestations are especially broad, ranging from asymptomatic to unstable angina. Arbitrary definitions can be used by angiography such as a 50% or 70% stenosis, or, alternatively, abnormal results of a functional test such as exercise with nuclear scintigraphy. Using coronary angiography in families with MI, Fischer et al demonstrated remarkable correlation of the location of epicardial artery involvement among sibling pairs.20 For the purpose of defining the genetic basis, MI is a far better phenotype and the yield has been notably better to date as compared with the phenotype of coronary artery disease.
Figure 3 Atherosclerotic coronary artery disease and myocardial infarction (MI). The top panels represent the number of patients affected in the United States; the bottom panels show cross‐section schematics of the underlying coronary artery pathobiology.
THE FUTURE OF MYOCARDIAL INFARCTION GENETICS
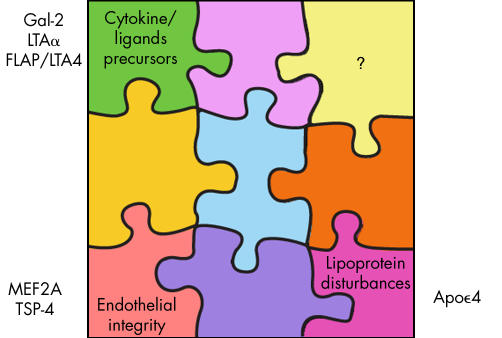
At this point we have only solved a few pieces of the puzzle (fig 4), which may be considered “corner” pieces because they encompass key categories of cytokine precursors, factors influencing vessel wall integrity, or those associated with lipoprotein disturbances. One of the most exciting aspects of genomic medicine is that by approaching the human genome systematically and without bias, it will ultimately tell us the “secret” pathways or genes that we would otherwise not have attributed to MI. Several examples have already been identified such as lymphotoxin‐α, or galectin‐2, or FLAP and LTA4. Before genome wide association or linkage studies, we would never have expected such genes to be implicated. On the basis of identifying the genes, tailored treatments can be designed such as we have already seen with the FLAP blocker programme. More importantly, by diagnosing the genetic susceptibility at an early age, efforts can be initiated far sooner in the individual's lifetime with respect to healthy lifestyle, including more rigorous attention to diet and exercise, along with more aggressive risk factor control. This could even extend to much earlier pharmacologic intervention with medications such as statins or aspirin in appropriate individuals deemed at high risk at the genomic level. Ultimately, in the years ahead, patients who are interested in the information will be able to undergo a panel of MI genes for determination of their risk and a genetic consult for tailored prevention.
Figure 4 The myocardial infarction genetics puzzle.
The genetics of heart attack: key points
Myocardial infarction (MI) is a complex trait: common, probabilistic with gene–gene and gene–environment interactions
Genome‐wide scans of affected families with MI demonstrate linkage for multiple chromosomal loci
5‐Lipooxygenase activating protein (FLAP), leukotriene‐4 hydrolase (LTA4), and myocyte enhancing factor (MEF2A) have been identified to be associated with MI by such linkage studies
The first gene‐specific trial for secondary prevention of MI was performed with a FLAP blocker in patients with gain‐of‐function FLAP gene variant
MI is a much more restrictive phenotype than coronary artery disease and it has therefore been more useful to define its genomic basis
The process of discovery of MI genes will be considerably faster now. First, the human genome map has been extensively annotated, such that a finding of a locus of interest will more quickly lead to identifying the gene responsible. Second, the ability to perform whole genome association studies transforms the field, providing a comprehensive and near “full disclosure” assessment. The International HapMap and the efforts to identify the tag‐SNPs will greatly facilitate the hunt, and allow for rapid and precise detection of the critical haplotypes of this complex trait. Third, the cost of the technology has fortunately plummeted. For 1 cent per genotype, in 1000 cases and 1000 controls, a total of 10 million SNPs assessed would cost $200 million. Now the projected cost of a genotype in the next year or two is reduced to 0.01 cent and the number of tag‐SNPs assessed 250 000 instead of 10 million. For the same 1000 cases and 1000 controls the cost of a whole genome association study would be reduced to $500 000, or only 2–3% of what it would take to do a less efficient study a short time ago. Ultimately, inexpensive complete resequencing of the human genome may be available at a low cost for individuals, but this may not be necessary if the promise of tag‐SNPs is fulfilled.
Some years ago we realised that treating a patient in the midst of a heart attack was like a “fire drill”. To actually salvage myocardium is exceptionally challenging once the event has been initiated. This is especially the case for the average patient who lingers more than two hours before seeking medical attention. In the future, advances in genomic medicine will pave the way for much improved prevention of MI through genetic screening of individuals deemed at increased risk. Such insight should greatly catapult us forward in our efforts to triumph over this exceptionally important complex trait.
Additional references appear on the Heart website—http://www.heartjnl.com/supplemental
Additional references appear on the Heart website—http://www.heartjnl.com/supplemental
Supplementary Material
Footnotes
Supported by National Heart Lung and Blood Institute P50 HL 077107
In compliance with EBAC/EACCME guidelines, all authors participating in Education in Heart have disclosed potential conflicts of interest that might cause a bias in the article
Additional references appear on the Heart website—http://www.heartjnl.com/supplemental
References
- 1.Hirschhorn J N, Lohmueller K, Byrne E.et al A comprehensive review of genetic association studies. Genetics in Medicine 2002445–61.A detailed review emphasising false positive studies of single nucleotide polymorphisms. [DOI] [PubMed] [Google Scholar]
- 2.McCarthy J J, Parker A, Moliterno D J.et al for the GENEQUEST Investigators. Large‐scale association analysis for identification of genes underlying premature coronary heart disease: cumulative perspective from analysis of 111 candidate genes. J Med Genet 200441334–341.First high‐throughput study of variants for myocardial infarction which would now be considered of limited scope. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamada Y, Izawa H, Ichihara S.et al Prediction of the risk of myocardial infarction from polymorphisms in candidate genes. N Engl J Med 20023471916–1923.Extensive genotyping of patients with sporadic myocardial infarction demonstrating four gender‐specific variants that were statistically significant. [DOI] [PubMed] [Google Scholar]
- 4.Shiffman D, Ellis S G, Rowland C M.et al Identification of four gene variants associated with myocardial infarction. Am J Human Genet 200577596–605.Recent whole genome SNP study of 6891 genes identifying four variants associated with myocardial infarction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Q, Rao S, Shen G ‐ Q.et al Premature myocardial infarction novel susceptibility locus on chromosome 1p34–36 identified by genome‐wide linkage analysis. Am J Human Genet 200474262–271.The GeneQuest genome wide linkage study that demonstrated a very substantial chromosome linkage peak for myocardial infarction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helgadottir A, Manolescu A, Thorleifsson G.et al The gene encoding 5‐lipoxygenase activating protein (FLAP) confers risk of myocardial infarction and stroke. Nat Genet 200436233–239. [DOI] [PubMed] [Google Scholar]
- 7.Wang L, Fan C, Topol S E.et al Mutation of MEF2A in an inherited disorder with features of coronary artery disease. Science 20033021578–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ozaki K, Ohnishi Y, Iida A.et al Functional SNPs in the lymphotoxin‐alpha gene that are associated with susceptibility to myocardial infarction. Nat Genet 200232650–654. [DOI] [PubMed] [Google Scholar]
- 9.Ozaki K, Inoue K, Sato H.et al Functional variation in LGALS2 confers risk of myocardial infarction and regulates lymphotoxin‐alpha secretion in vitro. Nature 200442972–75. [DOI] [PubMed] [Google Scholar]
- 10.Hinds D A, Stuve L L, Nilsen G B.et al Whole‐genome patterns of common DNA variation in three human populations. Science 20053071072–1079.First edition of the haplotype map with analysis of more than 1.5 million SNPs in 71 individuals of diverse geographic ancestry. [DOI] [PubMed] [Google Scholar]
- 11.Hirschhorn J N, Daly M J. Genome‐wide association studies for common diseases and complex traits. Nat Rev Genet 2005695–108. [DOI] [PubMed] [Google Scholar]
- 12.Braun A, Trigatti B L, Post M J.et al Loss of SR‐BI expression leads to the early onset of occlusive atherosclerotic coronary artery disease, spontaneous myocardial infarctions, severe cardiac dysfunction, and premature death in apolipoprotein E‐deficient mice. Circ Res 200290270–276. [DOI] [PubMed] [Google Scholar]
- 13.Hakonarson H, Thorvaldsson S, Helgadottir A.et al A randomized, placebo‐controlled, crossover trial of an inhibitor of the 5‐lipoxygenase activating protein (FLAP), DG‐031, shows reduction in concentrations of biomarkers of risk of myocardial infarction in patients with coronary artery disease and at‐risk variants of the FLAP and leukotriene A4 (LTA4) hydrolase genes. JAMA 20052932245–2256.First gene‐specific trial in cardiovascular medicine testing a FLAP blocker in patients with gain‐of‐function FLAP genetic variant. [DOI] [PubMed] [Google Scholar]
- 14.Helgadottir A, Manolescu A, Helgason A.et al A variant of the gene encoding leukotriene A4 hydrolase confers ethnic specific risk of myocardial infarction. Nat Genet 20063868–74. [DOI] [PubMed] [Google Scholar]
- 15.Bhagavatula M R K, Fan C, Shen G ‐ Q.et al Transcription factor MEF2A mutations in patients with coronary artery disease. Hum Mol Genet 2004133181–3188.Demonstration of point MEF2A mutations in 1.6% of sporadic MI individuals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weng L, Kavaslar N, Ustaszewskaet al Lack of MEF 2A mutations in coronary artery disease. J Clin Invest 20051151016–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q, Rao S, Topol E J. Miscues on the “lack of MEF2A mutations” in coronary artery disease. J Clin Invest 20051151399–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez P, Garcia‐Castro M, Reguero J R.et al The Pro279Leu variant in the transcription factor MEF2A is associated with myocardial infarction. J Med Genet 200643167–169.Replication and extension of MEF2A findings via point mutation and its association with myocardial infarction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freedman M L, Reich D, Penney K L.et al Assessing the impact of population stratification on genetic association studies. Nat Genet 200436388–393. [DOI] [PubMed] [Google Scholar]
- 20.Fischer M, Broeckel U, Holmer S.et al Distinct heritable patterns of angiographic coronary artery disease in families with myocardial infarction. Circulation 2005111855–862.Angiographic study of siblings affected with MI that demonstrates remarkable concordance of location of atherosclerosis in the coronary anatomy. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.