Abstract
Based on theoretical evidence, it has been proposed that HIV-1 may encode several selenoprotein modules, one of which (overlapping the env gp41-coding region) has highly significant sequence similarity to the mammalian selenoprotein glutathione peroxidase (GPx; EC 1.11.1.9). The similarity score of the putative HIV-1 viral GPx homolog relative to an aligned set of known GPx is 6.3 SD higher than expected for random sequences of similar composition. Based on that alignment, a molecular model of the HIV-1 GPx was constructed by homology modeling from the bovine GPx crystal structure. Despite extensive truncation relative to the cellular GPx gene, the structural core and the geometry of the catalytic triad of selenocysteine, glutamine, and tryptophan are well conserved in the viral GPx. All of the insertions and deletions predicted by the alignment proved to be structurally feasible. The model is energetically favorable, with a computed molecular mechanics strain energy close to that of the bovine GPx structure, when normalized on a per-residue basis. However, considering the remote homology, this model is intended only to provide a working hypothesis allowing for a similar active site and structural core. To validate the theoretical predictions, we cloned the hypothetical HIV-1 gene and found it to encode functional GPx activity when expressed as a selenoprotein in mammalian cells. In transfected canine kidney cells, the increase in GPx activity ranged from 21% to 43% relative to controls (average 30%, n = 9, P < 0.0001), whereas, in transfected MCF7 cells, which have low endogenous GPx activity, a near 100% increase was observed (average 99%, n = 3, P < 0.05).
As various genome projects have continued to expand the number of entries in nucleic acid sequence databases, there has been an increasing demand for computational biology and computational chemistry methods capable of solving several fundamental problems (1). The latter include the prediction of (i) the existence, location and architecture of genes, (ii) the functions of the encoded proteins, and, ultimately, (iii) the structures of the encoded proteins. Advances in comparative sequence analysis, including methods for the identification of remote homologs (1, 2), coupled with advances in protein structure prediction and molecular mechanics (3–5), have now brought all of these objectives within reach, at least when there is some degree of homology between a novel gene and known examples in databases. The ability to identify remote homologs and predict their protein structures is still a major challenge for computational chemists and biologists.
The need for such advanced computational methods should not be underestimated, because their use can lead to the identification of genes whose existence or function is not obvious and which can be missed even after extensive analysis by conventional methods. To illustrate exactly such a case, we present here an example involving a complex retroviral genome, that of the HIV type 1 (HIV-1). In 1994, based on theoretical evidence, Taylor et al. (6) proposed that HIV-1 might encode several selenoprotein modules. Subsequently, one of the proposed HIV-1 selenoprotein genes, potentially expressed by a ribosomal frameshift from the env coding region (thus named env-fs), was demonstrated to have highly significant sequence similarity to the mammalian selenoprotein glutathione peroxidase (GPx; EC 1.11.1.9), which contains a catalytic selenocysteine (Sec) residue, encoded by the UGA codon in RNA (7). There is no mystery as to why this potential gene was not identified earlier, because the UGA codon generally serves as a stop signal for protein synthesis, and only rarely as a Sec codon (8); furthermore, like the retroviral pol gene, the putative HIV-1 GPx gene lacks a start codon, and thus does not appear to be an “open” reading frame. However, it does have a well conserved potential −1 frameshift sequence (A AAA) consistent with the established “P-site slippage” mechanism (7, 9).
In the current study, based on a revised multiple sequence alignment (Fig. 1), we have generated a full three-dimensional (3-D) protein model of the hypothetical HIV-1 selenium (Se)-dependent GPx, refined the structure by using molecular mechanics, and applied various structural and energetic criteria to assess the model. This application of homology modeling is a further example of an original method for “structural evaluation of distant homology,” previously described by Bhat and Taylor (10), that involves evaluation of a proposed homology by examining the consistency between the 3-D model based on the sequence alignment and existing biochemical or other data relevant to mechanism and structure. However, because of the remote homology involved, such models are not expected to be accurate at the atomic level, but instead are expected simply to reflect the potential for a similar protein fold and, as in this case, a common structural core and active-site geometry.
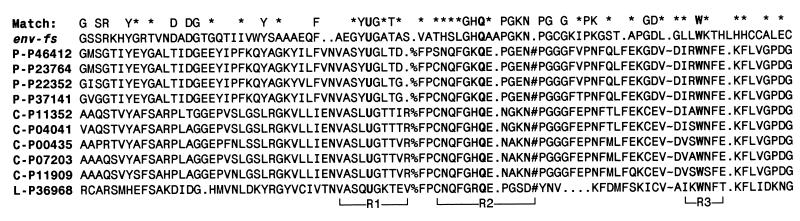
Figure 1.
Sequence alignment of HIV-1 env-fs sequence vs. Se-dependent GPx. Sequences are single-letter amino acid code, with U as the symbol for Sec, encoded by the UGA codon in RNA. GPx sequences are listed by Swiss-Prot database accession number, after a prefix of either P- (plasma), C- (cellular), or L- (phospholipid hydroperoxide), identifying three major families of Se-dependent GPx sequences. The N terminal of mature GPx protein begins at the fourth residue in the alignment. Env-fs amino acids identical to one or more of the aligned GPx sequences are shown as letters in the “match” line above the alignment; similar residues are indicated by an asterisk. Three active-site regions that are adjacent in 3-D space are shown below the alignment, labeled R1–R3. All three regions and their essential catalytic amino acids, Sec, Gln, and Trp (U, Q, and W, respectively, shown highlighted in bold), are represented in the truncated env-fs sequence. There are three large internal deletions in env-fs relative to the GPx sequences (numbered 1–3 in Fig. 2), at the locations indicated by the symbols %, #, and ∼ in the alignment, involving 19, 11, and 41 residues, respectively. As computed by using the alignment shown, i.e., omitting the three internal GPx regions where there are major deletions in the putative HIV-1 GPx homolog, the total similarity score of the env-fs sequence to the aligned GPx sequences is 6.3 SD above the average similarity score computed for 100 optimally aligned randomly shuffled sequences of identical composition. This significance score rises to 6.7 SD if the comparison is made only to the plasma GPx sequences, to which the HIV sequence is most similar overall; however, it also has features of the cellular and phospholipid hydroperoxide types of GPx.
To validate the predictions of the theoretical studies, we have cloned this hypothetical HIV-1 gene, expressed it as a selenoprotein in several mammalian cell lines, and assayed for functional GPx enzyme activity in the transfected cells. The theoretical analysis was also useful in the design of this expression vector, because the structural analysis enabled us to predict where to engineer a start codon in the HIV-1 sequence that would correspond to the N terminal of the mature bovine GPx protein.
Materials and Methods
Sequence Analysis.
The statistical significance of the similarity between the hypothetical HIV-1 env-fs protein (putative GPx) as a query sequence and a prealigned set of mammalian GPx sequences was assessed by using the blockalign program of Zhang et al. (11). This program produces an optimal, gapped alignment of a probe sequence against an existing sequence alignment. By random shuffling of the query sequence, the average similarity score for optimally aligned random sequences of identical composition can be calculated. Briefly, the method involves (i) the optimal alignment of the query sequence against the prealigned family (GPx), which also yields a similarity score, followed by (ii) repeated random shuffling and realignment of the query sequence and (iii) calculation of the average similarity score and SD for optimally aligned random sequences of identical composition to the query sequence. The significance of the sequence similarity between the query and the given sequence alignment can then be expressed as a shuffling statistic (Z score), calculated as the distance of the actual score from the average random score, in SD. For the alignment of Fig. 1, the blosum62 amino acid similarity matrix was used, with gap creation and gap extension penalties of 6 and 2, respectively; 100 random shufflings were used for the significance calculation. Sec residues were treated as Cys for the purpose of these calculations, because no amino acid similarity matrix has yet been developed that includes Sec; as discussed previously (11), this approximation will lead to a slight underestimation of the significance of alignments involving Sec residues in conserved positions.
Molecular Modeling.
The sybyl program (Tripos Associates, St. Louis) was used to build the putative HIV-1 viral GPx homology model, starting from the bovine cellular GPx x-ray crystal structure (12), file 1GP1 from the Protein Data Bank (http://www.rcsb.org/pdb). Based on the alignment of Fig. 1 (in which the bovine GPx sequence is shown as no. P00435), the model was constructed by using the biopolymer protein_loop option of sybyl, which uses a knowledge-based approach to rebuild regions where insertions and deletions are predicted by the alignment. Both insertions and deletions were handled by deleting one or more residues on either side of the affected region, and then building a new loop that included those residues, plus or minus the required number of residues (e.g., for a single residue insertion, delete 2, rebuild 3). The resulting model was refined by molecular mechanics energy minimization using the Kollman all-atom force field as implemented in sybyl. A notable difference between Cys and Sec residues is the increased acidity of the selenol group, which has a pKa around 6, as opposed to approximately 7 for the thiol group of Cys. Thus, the Sec residue was modeled as the deprotonated Se− species. Force field parameters and partial atomic charges for the Sec residue were estimated by analogy to the Kollman parameters for the Cys residue, combined with the results of ab initio molecular orbital calculations on Cys, Sec, methyl thiol, and methyl selenol, obtained by using the gaussian 94 program (Gaussian, Pittsburgh, PA). Kollman partial charges were assigned to the Sec residue in such a way that the net change from the Cys charges was zero, to preserve electrical neutrality (charge conservation). The molecular orbital calculations on Cys and Sec indicated that changing S to Se had little effect on the charge on the amino acid α-carbon; thus, the partial charges on the Sec backbone atoms, including the α-carbon, were set to the same values as those for Cys (which are the same for all amino acids in this force field). The total charge on the neutral side chain is then 0.016, as calculated by Kollman for Cys. Adding a −1 formal charge to the side chain for the ionized state gives a total charge for the side chain of −0.984. The partial charges derived from the MP2/6-311 + +G** ab initio calculation for the C, two H, and Se atoms of deprotonated methyl selenol (which summed to −1.089) were then scaled to sum to the required total (−0.984), resulting in the following partial charges: Se, −0.851; βC, −0.293; and βH, 0.08. For comparison, the computed Se− charge based on a similar STO-3G calculation would have been −0.706, a significant underestimation of the negative charge as calculated by using the higher basis set. van der Waals parameters for Se were set to r = 2.29 and ɛ = 0.276 (13). Other parameters and force constants were taken to be identical to those for S, except for the following equilibrium values: angle HC CT Se, 110.5°; angle CT CT Se, 108.2°; bond CT Se, 1.95 Å (HC and CT are Kollman force field atom types, an H and C, respectively). The full set of force field parameters and charges are posted on our web site, http://bioinfo.chem.uga.edu/homepage/wtaylor. For the electrostatic potential energy term, the default distance-dependent dielectric model was used (i.e., dielectric constant = r, where r is the distance in angstroms between nuclei). A ΔE of 0.001 kcal/mol was used as the termination criterion in the final energy minimization.
The model was initially minimized by using a set of distance constraints to maintain the geometry of the three residues involved in the catalytic triad, Sec (U), Gln (Q), and Trp (W). In previous computer simulations of various GPx active-site intermediates modeled from the bovine GPx crystal structure, the Q and W residues were shown to act as H-bond donors to the ionized Se atom (14). Thus, the interatomic distances from the Se atom to the Trp N1 (3.5 Å), from Se to the Gln amide N (3.4 Å), and between the Gln and Trp nitrogens (3.9 Å) were constrained for the first round of energy minimization, after which the constraints were removed for the final minimization of the model. For comparison, a monomer from the bovine GPx structure was minimized similarly but without constraints.
Construction of HIV-1 GPx Eukaryotic Expression Vector.
Because the hypothetical HIV-1 GPx protein encoded by env-fs is predicted to be expressed by a −1 frameshift (6, 7), the primary gene product would be an env gp120-GPx fusion protein, with an expected molecular mass of about 130 kDa. The GPx homology region is the C-terminal 89 amino acids of that predicted fusion product, in which form it might function as a minor env glycoprotein isoform on the outer membrane surface of virions or infected cells (11). However, a potential HIV-1 protease site was predicted immediately upstream of the GPx homology region (6, 7), so the GPx module may also be cleaved and released as an independent small (9 kDa) protein, functioning inside the cell and possibly in the HIV-1 virion. It is this low molecular mass isoform of the putative HIV-1 GPx protein that we have cloned, incorporating a Met start codon immediately before the Gly residue that is at the N-terminal end of env-fs in the GPx alignment shown in Fig. 1. The DNA fragment corresponding to the putative HIV-1 GPx homolog was PCR amplified from the env gp41-coding region of the pBH10 clone of HIV-1, obtained from the National Institutes of Health AIDS Research and Reference Reagent Program, Rockville, MD. The PCR primers used for the HIV-1 env-GPx fragment were: 5′ sense, 5′-TTTGCTAGCATGGGAAGCAGCAGGAAGCACTATG-3′, containing an NheI site, and 3′ reverse complement 5′-GCAAGCTTCTAGCATTCCAAAGCACAGC-3′, containing a HindIII site. Note that in the first primer listed above the required ATG start codon has been incorporated just before the start of the coding sequence (GGAAGC … ).
The established mechanism of Sec insertion in eukaryotic selenoproteins involves an RNA stem-loop structure called a selenocysteine insertion sequence (SECIS) element in the 3′-untranslated region (3′-UTR) of the mRNA. Via a looping back interaction with the ribosome, the SECIS element facilitates the decoding of in-frame UGA codons in the upstream coding region as Sec (8). Incorporation of a segment of the 3′-UTR spanning the SECIS element of a known eukaryotic selenoprotein has been shown to be sufficient for Sec incorporation in heterologous selenoprotein genes, and even in peptides encoded by synthetic oligonucleotides with in-frame UGA codons (15, 16). Because the mechanisms and location of downstream RNA structure elements that HIV may use for recoding the UGA stop codon as a sense codon for Sec are still unknown (11), the putative HIV-1 GPx expression construct was designed to include the SECIS element of the rat 5′-deiodinase (5′-DI) gene (15), to ensure the translation of the upstream in-frame UGA codon of the HIV-1 env-fs peptide as Sec during expression in mammalian cells. A region of the 3′-UTR of the rat type I 5′-DI gene was PCR amplified from pCDM8-G21, provided by Marla Berry, Harvard Medical School, Boston, MA. The PCR primers used for the 5′-DI SECIS were: 5′ sense, 5′-GCAAGCTTCGAGTAACTCTGTTCCACTG-3′, containing a HindIII site, and 3′ reverse complement, 5′-CCGGATCCCGGATTATAATCGTTAGC-3′, containing a BamHI site.
To generate the eukaryotic expression vector pGD, the HIV-1 env-GPx and 5′-DI SECIS fragments were subcloned into the pEGFP-C1 vector (CLONTECH), by using the following cloning sites: NheI, HindIII (GPx) and HindIII, BamHI (SECIS). The sequence of the entire transcribed region of the resultant plasmid pGD was verified by DNA sequencing. Sequencing and oligonucleotide syntheses were performed at the University of Georgia Molecular Genetics Instrumentation Facility. Restriction enzymes were obtained from Promega; other chemicals were obtained from Sigma unless otherwise specified.
Cell Culture and Transfection.
The MCF-7 human breast cell line was obtained from the American Type Culture Collection (Manassas, VA). The MDCK (canine kidney) cell line was a gift from Fengxiang Gao, Centers for Disease Control and Prevention, Atlanta, GA. Both cell types were grown under 5% CO2 at 37°C, in DMEM/F12 medium supplemented with 2 mM l-glutamine, 5 μg/ml each of insulin, transferrin, and gentamycin, and 0.5% newborn calf serum (Atlanta Biologicals, Norcross, GA). MDCK cells were transiently transfected by using Lipofectamine (GIBCO), on six-well plates, with each well seeded with 2.5 × 105 cells and 10 μg of plasmid DNA. After transfection, cells were grown in medium containing 20 nM sodium selenite for 72 h and then harvested for the GPx activity assay. MCF-7 cells were stably transfected by the same method, with 10 μg of plasmid used for 3 × 105 cells. Transfected cells were selected for antibiotic resistance to G418 at 400 μg/ml, by changing G418-containing medium supplemented with 100 nM sodium selenite every 3 days for 21–30 days. Positive clones were pooled and propagated in the same medium until confluence and harvested for the GPx activity assay.
GPx Enzyme Activity Assay.
Both transiently transfected MDCK cells and stably transfected MCF-7 cells were harvested and resuspended in PBS. Cells were lysed by sonication at 4°C at low power. Cell lysates were centrifuged at 25,000 × g for 25 min at 4°C. GPx activity in the supernatant was measured by spectrophotometric determination of NADPH utilization (as described in ref. 17), in the presence of standardized concentrations of glutathione and t-butyl hydroperoxide as substrates. The specific enzymatic activities are presented as means ± SD, in units per mg of protein. Concentration was measured by the Lowry method with BSA as the standard.
Results
Alignment and Similarity Assessment of HIV-1 env-fs vs. GPx Sequences.
As detailed previously, the putative viral GPx module is encoded overlapping the HIV-1 env gp41-coding region in the −1 reading frame (6, 7); thus the gene was tentatively named env-fs, for env frameshift. It contains a single UGA codon (potentially encoding Sec) near the middle of a small but highly conserved ORF, lacking a start codon but having a conserved −1 frameshift signal near its 5′ end. The translated HIV-1 env-fs sequence contains a common variant of the GPx active-site consensus sequence spanning the catalytic Sec, shown as a U in Fig. 1. A strong similarity between the entire HIV-1-encoded env-fs sequence and an aligned set of GPx sequences has been demonstrated: as previously aligned by Taylor et al. (7), the similarity score of this HIV sequence vs. an aligned group of GPx sequences is 5 SD above the average similarity score of randomly shuffled sequences of identical composition. We examined alternative alignments of the C-terminal region of env-fs to GPx in the light of the bovine GPx crystal structure and its active site (12, 14), and identified the viral sequence potentially matching a third GPx active-site region with a conserved Trp (W), shown as region 3 (R3) in the revised alignment of Fig. 1. Thus, this alignment is different in the C-terminal region from that published previously (7). The proposed homology and the alignment shown are strongly supported by the fact that the HIV-1 Trp codon in question is highly conserved, being found in over 98% of all reported group M (“main”) HIV-1 sequences. Because of the degeneracy of the genetic code, this conservation cannot be explained by any known coding or structural features of HIV-1, which strongly suggests that this is a functional gene (see ref. 11 for a detailed explanation).
The significance score for the env-fs vs. GPx similarity increases to 6.3 SD based on the revised alignment of Fig. 1, which reveals a total of three major internal deletions (of 19, 11, and 41 residues) in the HIV-1 sequence relative to the mammalian GPx sequences, as well as a truncation of 30 residues at the C terminus (not shown). These deletions in env-fs, which comprise a total of over 50% of the sequence of the mammalian GPx homologs, lie between or downstream of the three conserved active-site regions (YUG, GHQ, and W) in the primary sequence.
The putative HIV-1 GPx is most similar to the plasma GPx sequences, having 23 of 89 residues (26%) identical to aligned plasma GPx residues in Fig. 1, whereas only 19 of 89 (18%) residues are identical to those of the aligned bovine cellular GPx sequence (no. P00435 in Fig. 1).
Molecular Modeling.
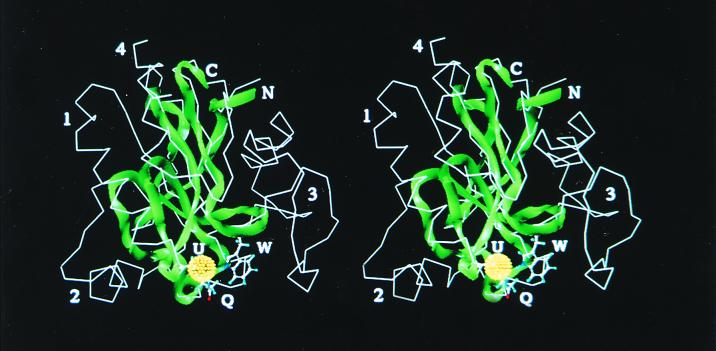
To assess the proposed homology in structural terms (10), a 3-D molecular model of the HIV-1 viral GPx (Fig. 2) was made by homology modeling from the bovine GPx structure (12), based on the alignment of Fig. 1. Despite the presence of four major deletions in the HIV primary sequence relative to known GPx genes (labeled 1–4 in Fig. 2), modeling of the putative HIV-1 GPx homolog is structurally feasible, because these deletions lie outside of the active-site domain, and several of the internal deletions, despite their large size (up to 41 residues), are located in places where their excision permits the backbone to be easily reconnected despite the loss of numerous residues. For example, from an examination of deleted region 3 as shown in Fig. 2, it can be seen that this domain begins and ends at approximately the same location (a little above the active-site W label in the figure), and thus coils back on itself, so that its deletion permits the resulting structure to be annealed without disruption of the structural core. A similar situation exists for deletion 1, which consists of a helix and part of a β sheet that folds back on the helix. The deletion labeled 4 in Fig. 2 is the C-terminal region not included in Fig. 1; this deletion involves the loss of an outer strand of the core β sheet, which is thereby reduced from 5 to 4 strands, along with the C-terminal helix seen at the upper center of Fig. 2, which lies on the β sheet in the bovine GPx structure. Despite these major deletions, the truncated GPx homolog encoded by HIV-1 includes the structural β sheet core of the enzyme, and the entire active-site region including the catalytic triad, the residues of which are located in three separate regions (R1-R3 in Fig. 1) of the protein chain that come together in 3-D space to form the active site (labeled U, Q, and W in Fig. 2). Significantly, the second and third deletions involve noncatalytic regions whose function in the bovine GPx is in multimerization to form GPx tetramers. The absence of these domains suggests that the HIV-1 GPx homolog probably does not function as a multimer, for which there is a precedent, because the phospholipid hydroperoxide GPx is known to act as a monomeric enzyme (18).
Figure 2.
Stereo view of the aligned protein backbones of the putative HIV-1 GPx homology model (ribbon rendition) and the x-ray crystal structure of the bovine GPx monomer (line rendition). The active site is in the lower foreground, with the side chains of the catalytic triad (Sec, Gln, and Trp) shown in a ball and stick rendition and labeled U, Q, and W, respectively; the Se atom is shown as a dot sphere. The C and N terminals of the HIV-1 peptide are labeled. The regions labeled by numbers 1–3 correspond to the major internal deletions shown as %, #, and ∼ in the sequence alignment of Fig. 1. The region labeled 4 is a C-terminal deletion, not shown in the alignment. These four deletions are domains of the bovine GPx that have no equivalent in the highly truncated HIV-1 GPx homolog. The deleted regions include a helix involved in dimer formation (no. 2) and another region (no. 3) that is involved in dimer and tetramer formation. No. 1 is an internal deletion between the U and the Q. Despite these deletions, the structural and catalytic core of the enzyme is conserved in the truncated HIV-1 homolog.
Minimization of the protein model as described in Materials and Methods was readily accomplished, by using a simple distant-dependent dielectric model for solvation, rather than explicit waters; this approximation was also used in the only major molecular dynamics study of GPx published to date (14), and is adequate for our purposes. In the final unconstrained model, the critical distances between the Se atom and the 2 H-bonding NH donors were still very close to those in the bovine GPx structure (Se to Trp N1, 3.2 vs. 3.5 Å; Se to Gln N, 3.3 vs. 3.4 Å). A comparison of the active-site geometry of the fully minimized model vs. the bovine GPx crystal structure was made by means of an rms fit of the three heavy (non-H) atoms of the residues of the catalytic triad involved in H-bonding (Sec Se, Trp N1, Gln amide N). This fit gave an rms value of 0.42 Å, suggesting that the required active-site geometry could be conserved in the HIV-1 GPx homolog despite the extensive deletions and low sequence identity (18%) relative to the bovine sequence.
Energy minimization of the protein model with the Kollman all-atom force field led to a substantially negative molecular mechanics “strain” energy of −1410 kcal/mol for 89 residues (−15.8 kcal/mol⋅residue), which on a per-residue basis is reasonably close to, but not quite as low as, the value obtained by minimizing the bovine GPx crystal structure with the same parameters, −3620 kcal/mol for 185 residues (−19.6 kcal/mol⋅residue). A closer examination of the nonbonded energy terms (which are the major contributors to the energy) suggests that the volume packing of side chains in the model is not quite as good as that for the crystal structure (total van der Waals energy of −2.9 kcal/mol⋅residue for the model vs. −4.0 kcal/mol⋅residue for the bovine GPx), but that the electronic stabilization of the structure is almost as good (total electrostatic energy of −16.9 kcal/mol⋅residue for the model vs. −18.2 kcal/mol⋅residue for the bovine GPx; the total H bond energy is essentially identical, −0.39 vs. −0.40 kcal/mol⋅residue). This electrostatic stabilization arises in part because of a favorable distribution of ionizable acidic and basic residues that, in the model, participate in a number of salt bridges between distant residues, which can potentially stabilize the structure, particularly by anchoring the N and C termini of the protein chain. The less favorable van der Waals energy term in the model is actually to be expected because of the somewhat arbitrary computer-generated side-chain conformations in the models, which can be only partially remedied by manual reorientation.
Cloning, Expression, and Functional Assay of the Putative HIV-1 GPx Homolog.
The expression construct pGD was designed to include the SECIS element of the rat 5′-DI gene, to ensure the expression of the HIV-1 env-fs peptide as a selenoprotein in mammalian cells, such as the MCF7 and MDCK canine kidney cell lines, which contain all of the necessary cellular machinery for selenoprotein expression. The question is, does that expressed peptide encode functional GPx enzyme activity, or is it a “nonsense” peptide, the expression of which would lead to a decline in cellular GPx activity by competing for limited cellular pools of tRNASec? In cells transfected with the pGD construct, we observed a consistent and significant increase in GPx activity relative to cells transfected with an “empty” construct, the parent pC1 plasmid lacking the env-fs and 5′-DI SECIS inserts. In transiently transfected canine kidney (MDCK) cells, an increase in GPx activity ranging from 21% to 43% relative to controls was observed (average 30%, n = 9, P < 0.0001), whereas in stably transfected MCF7 cells, which have low endogenous GPx activity, a near 100% increase could be demonstrated (average 99%, n = 3, P < 0.05). However, the absolute increase in GPx activity was greater in MDCK cells, probably because MCF7 cells have low levels of tRNASec and/or other cellular factors required for selenoprotein synthesis, consistent with their low endogenous GPx activity. The concentration of sodium selenite in the medium required for optimal GPx expression was also different in the two cell types, being 100 nM in MCF7 cells, as opposed to only 20 nM in MDCK cells. These results are detailed in Table 1.
Table 1.
GPx activity in cells transfected with HIV-1 construct (pGD) vs. controls transfected with “empty” construct (pC1)
| Cells/construct | Transfection | SeO32−, nM | Activity, units/mg protein |
|---|---|---|---|
| MDCK (n = 9) | |||
| pC1 | Transient | 20 | 54.3 ± 4.20 |
| pGD | Transient | 20 | 70.8 ± 6.13* |
| MCF7 (n = 3) | |||
| pC1 | Stable | 100 | 4.96 ± 2.26 |
| pGD | Stable | 100 | 9.86 ± 1.30** |
, P < 0.0001, compared with pC1;
, P < 0.05, compared with pC1.
Discussion
By means of comparative sequence analysis, molecular modeling, and functional assay of the expressed protein, we have assessed the twofold hypothesis that a novel HIV-1 gene, env-fs, is encoded in an overlapping reading frame of the env gene, and that the protein it codes for is a highly truncated Se-dependent GPx module (6, 7). Although the pairwise identity of the putative HIV-1 GPx homolog to individual GPx sequences is very low, by using a sensitive multiple sequence comparison method, we have demonstrated a highly significant sequence similarity, with a Z score of >6 SD (Fig. 1). Furthermore, the essential sequence elements (residues of the GPx catalytic triad) are highly conserved within HIV-1 subtypes, as discussed previously (7, 11).
The proposed homology model is supported by the following factors:
(i) Despite several extensive deletions and truncations, the sequence encoded by HIV-1 includes the structural core and the complete catalytic center of the GPx enzyme.
(ii) The deletions are structurally feasible, because the deleted regions project away from the active site and in several cases consist of noncatalytic domains involved in dimer and tetramer formation.
(iii) The geometry of the GPx active site, e.g., distances between critical active-site atoms and residues, is not substantially changed in the model relative to the bovine GPx crystal structure.
(iv) There is no exceptional molecular mechanics strain energy associated with the HIV-1 GPx model, which appears to have an energetically favorable distribution of charged residues leading to the electronic stabilization of the structure.
Thus, overall, the molecular modeling results suggest that the proposed homology is structurally feasible and consistent with the known structural requirements for Se-dependent GPx catalysis (14), supporting the hypothesis that the hypothetical HIV-1 gene env-fs encodes a truncated but potentially functional Se-dependent GPx homolog. Using a standard enzyme assay, we have validated the theoretical results by cloning the putative viral peptide shown as env-fs in Fig. 1, and demonstrating that it encodes functional GPx activity when expressed as a selenoprotein in mammalian cells.
The significance of these results can best be understood in the context of current knowledge regarding the biological roles of Se in the immune system and in the regulation of HIV-1 transcription. Se is an essential trace mineral that serves as a potent dietary antioxidant, in addition to other biological functions. Many of its cellular actions, mediated by selenoproteins such as GPx and thioredoxin reductase (TDR), are intimately linked to the redox status of the cell, and to the redox regulation of genes that are important for various immune cell functions (19). Significantly, oxidative stress has been widely documented in AIDS patients (20, 21), and is known to be an activator of HIV-1 replication in vitro (22, 23). In light of those observations, and the established redox-related roles of Se and selenoproteins, it is not surprising that Se has been found to be of critical importance in HIV-1 infection. Se status has consistently been correlated with various indicators of HIV-1 disease progression, and Se deficiency is highly correlated with HIV-related mortality (reviewed in refs. 7 and 24; 25–27).
GPx and TDR are two of the most widely distributed selenoproteins, and both have been demonstrated to be potent regulators of NF-κB, which is the primary cellular factor involved in the regulation of HIV-1 transcription. Remarkably, GPx and TDR appear to have opposed functions, i.e., GPx inhibits NF-κB by reducing oxidant tone (23, 28), whereas TDR is involved in the reductive activation of an essential Cys residue required for the DNA-binding activity of NF-κB (29). Thus, via these known cellular selenoproteins, Se is a potent regulator of both NF-κB activity and HIV-1 transcription. It is therefore reasonable that HIV-1 could have evolved to directly participate in those regulatory processes, by encoding its own selenoprotein.
Finally, it must be noted that our results are inconsistent with the conclusions of a recent study by Gladyshev et al. (30), who examined selenoprotein expression in HIV-1-infected and uninfected T cells, and concluded that there was no evidence of HIV-encoded selenoproteins. However, they did note that in HIV-infected cells there was a decline in levels of cellular selenoproteins, and an increase in “low molecular mass” Se compounds. The latter observation is of interest, because several potential HIV selenoproteins are predicted to be of low molecular mass (7–9 kDa), consistent with what Gladyshev et al. observe in HIV-infected T cells. One such product is the 9-kDa isoform of the HIV-1 GPx homolog that we have cloned and found to be active in our functional assays reported above. Furthermore, the primary observation of Gladyshev et al., that levels of cellular selenoproteins decline in HIV-infected cells, is also highly consistent with the predictions of the HIV selenoprotein theory, being expected as a consequence of HIV selenoprotein expression (6, 24).
In conclusion, we have presented compelling theoretical and functional evidence that strongly suggests that HIV-1 encodes a Se-dependent GPx gene. The fact that this gene remained undetected in the HIV-1 sequence for 10 years shows that the identification of genes, even in known mRNA sequences, is not always a trivial matter. Our ability to predict the function of the encoded protein despite its low similarity and extensive truncation relative to individual GPx sequences shows that, even in cases of very distant homology, it is sometimes possible to predict protein function from sequence. Because of that remote relationship, particularly when combined with the extensive truncation of the HIV-1 GPx homolog relative to the cellular enzyme, we emphasize that our model for the 3-D structure of the viral homolog is not expected to be accurate at the atomic level. However, the model does help to explain how it is possible for such a highly truncated homolog to possess GPx enzyme activity. Its degree of correspondence to the actual structure must await verification, pending an experimental structure determination. In that regard, we have expressed the Cys mutant of the protein in Escherichia coli, and purified the protein to homogeneity for structural and functional studies; significantly, the purified 9-kDa protein also has low but measurable GPx enzyme activity, despite the mutation of the Sec residue to Cys, necessary for bacterial expression of the protein (unpublished data).
Most significantly, the existence of an HIV-encoded selenoprotein gene represents the basis for a novel pathogenic mechanism (involving in part competition with cellular selenoprotein synthesis as discussed above) that can help to explain why the effects of HIV-1 infection are exacerbated in individuals who are Se deficient (24–27). Our hypotheses regarding the potential roles of viral selenoproteins in HIV-1 pathogenesis have been discussed in more detail elsewhere and will not be repeated here because of space limitations (6, 7, 11, 24, 31).
Acknowledgments
We thank Dr. Karl Kirshner, University of Georgia, for assistance with the gaussian calculations, Dr. Marla Berry, Harvard Medical School, for providing a vector containing the rat 5′-DI gene SECIS element, and finally, Dr. Marianna K. Baum, University of Miami School of Medicine, for her continued support. This work was supported in part by U.S. Public Health Service Grant RO1 DA11378 (National Institute on Drug Abuse).
Abbreviations
- 3-D
three-dimensional
- GPx
glutathione peroxidase
- Sec
selenocysteine
- SECIS
selenocysteine insertion sequence
- 5′-DI
5′-deiodinase
- TDR
thioredoxin reductase
- HIV-1
HIV type 1
References
- 1.Rawlings C J, Searls D B. Curr Opin Genet Dev. 1997;7:416–423. doi: 10.1016/s0959-437x(97)80158-x. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rost B, Sander C. Annu Rev Biophys Biomol Struct. 1996;25:113–136. doi: 10.1146/annurev.bb.25.060196.000553. [DOI] [PubMed] [Google Scholar]
- 4.Westhead D R, Thornton J M. Curr Opin Biotechnol. 1998;9:383–389. doi: 10.1016/s0958-1669(98)80012-8. [DOI] [PubMed] [Google Scholar]
- 5.Liwo A, Lee J, Ripoll D R, Pillardy J, Scheraga H A. Proc Natl Acad Sci USA. 1999;96:5482–5485. doi: 10.1073/pnas.96.10.5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor E W, Ramanathan C S, Jalluri R K, Nadimpalli R G. J Med Chem. 1994;37:2637–2654. doi: 10.1021/jm00043a004. [DOI] [PubMed] [Google Scholar]
- 7.Taylor E W, Bhat A, Nadimpalli R G, Zhang W, Kececioglu J. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;15:393–394. doi: 10.1097/00042560-199708150-00013. [DOI] [PubMed] [Google Scholar]
- 8.Low S C, Berry M J. Trends Biochem Sci. 1996;21:203–208. [PubMed] [Google Scholar]
- 9.Gramstat A, Prufer D, Rohde W. Nucleic Acids Res. 1994;22:3911–3917. doi: 10.1093/nar/22.19.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhat A A, Taylor E W. J Mol Mod. 1996;2:46–50. [Google Scholar]
- 11.Zhang W, Ramanathan C S, Nadimpalli R G, Bhat A A, Cox A G, Taylor E W. Biol Trace Elem Res. 1999;70:97–116. doi: 10.1007/BF02783852. [DOI] [PubMed] [Google Scholar]
- 12.Epp O, Ladenstein R, Wendel A. Eur J Biochem. 1983;133:51–69. doi: 10.1111/j.1432-1033.1983.tb07429.x. [DOI] [PubMed] [Google Scholar]
- 13.Allinger N L, Yuh Y H, Lii J-H. J Am Chem Soc. 1989;111:8551–8582. [Google Scholar]
- 14.Aumann K D, Bedorf N, Brigelius-Flohe R, Schomburg D, Flohe L. Biomed Environ Sci. 1997;10:136–155. [PubMed] [Google Scholar]
- 15.Berry M J, Banu L, Harney J W, Larsen P R. EMBO J. 1993;12:3315–3322. doi: 10.1002/j.1460-2075.1993.tb06001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kollmus H, Flohe L, McCarthy J E. Nucleic Acids Res. 1996;24:1195–1201. doi: 10.1093/nar/24.7.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beutler E. Red Cell Metabolism. New York: Grune and Stratton; 1984. pp. 74–76. [Google Scholar]
- 18.Sunde R A. In: Handbook of Nutritionally Essential Minerals. O'Dell B L, Sunde R A, editors. New York: Dekker; 1997. pp. 493–556. [Google Scholar]
- 19.McKenzie R C, Rafferty T S, Beckett G J. Immunol Today. 1998;19:342–345. doi: 10.1016/s0167-5699(98)01294-8. [DOI] [PubMed] [Google Scholar]
- 20.Israel N, Gougerot-Pocidalo M A. Cell Mol Life Sci. 1997;53:864–870. doi: 10.1007/s000180050106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allard J P, Aghdassi E, Chau J, Salit I, Walmsley S. Am J Clin Nutr. 1998;67:143–147. doi: 10.1093/ajcn/67.1.143. [DOI] [PubMed] [Google Scholar]
- 22.Israel N, Gougerot-Pocidalo M A, Aillet F, Virelizier J L. J Immunol. 1992;149:3386–3393. [PubMed] [Google Scholar]
- 23.Sappey C, Legrand-Poels S, Best-Belpomme M, Favier A, Rentier B, Piette J. AIDS Res Hum Retroviruses. 1994;10:1451–1461. doi: 10.1089/aid.1994.10.1451. [DOI] [PubMed] [Google Scholar]
- 24.Taylor E W, Nadimpalli R G, Ramanathan C S. Biol Trace Element Res. 1997;56:63–91. doi: 10.1007/BF02778984. [DOI] [PubMed] [Google Scholar]
- 25.Look M P, Rockstroh J K, Rao G S, Kreuzer K A, Barton S, Lemoch H, Sudhop T, Hoch J, Stockinger K, Spengler U, Sauerbruch T. Eur J Clin Nutr. 1997;51:266–272. doi: 10.1038/sj.ejcn.1600401. [DOI] [PubMed] [Google Scholar]
- 26.Constans J, Pellegrin J L, Sergeant C, Simonoff M, Pellegrin I, Fleury H, Leng B, Conri C. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;10:392. doi: 10.1097/00042560-199511000-00015. [DOI] [PubMed] [Google Scholar]
- 27.Baum M K, Shor-Posner G, Lai S, Zhang G, Lai H, Fletcher M A, Sauberlich H, Page J B. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;15:370–376. doi: 10.1097/00042560-199708150-00007. [DOI] [PubMed] [Google Scholar]
- 28.Hori K, Hatfield D, Maldarelli F, Lee B J, Clouse K A. AIDS Res Hum Retroviruses. 1997;13:1325–1332. doi: 10.1089/aid.1997.13.1325. [DOI] [PubMed] [Google Scholar]
- 29.Gorlatov S N, Stadtman T C. Proc Natl Acad Sci USA. 1998;95:8520–8525. doi: 10.1073/pnas.95.15.8520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gladyshev V N, Stadtman T C, Hatfield D L, Jeang K-T. Proc Natl Acad Sci USA. 1999;96:835–839. doi: 10.1073/pnas.96.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor, E. W., Cox, A. G., Zhao, L., Ruzicka, J. A., Bhat, A. A., Zhang, W., Nadimpalli, R. G. & Dean, R. D. (2000) J. Acquir. Immune Defic. Syndr. Hum. Retrovirol., in press. [DOI] [PubMed]