Abstract
The chromosome end-replicating enzyme telomerase is composed of a template-containing RNA subunit, a reverse transcriptase (TERT), and additional proteins. The importance of conserved amino acid residues in Trt1p, the TERT of Schizosaccharomyces pombe, was tested. Mutation to alanine of the proposed catalytic aspartates in reverse transcriptase motifs A and C and of conserved amino acids in motifs 1 and B′ resulted in defective growth, progressive loss of telomeric DNA, and loss of detectable telomerase enzymatic activity in vitro. Mutation of the phenylalanine (F) in the conserved FYxTE of telomerase-specific motif T had no phenotype in vivo or in vitro whereas mutation of a conserved amino acid in RT motif 2 had an intermediate effect. In addition to identifying single amino acids of TERT required for telomere maintenance in the fission yeast, this work provides useful tools for S. pombe telomerase research: a functional epitope-tagged version of Trt1p that allows detection of the protein even in crude cellular extracts, and a convenient and robust in vitro enzymatic activity assay based on immunopurification of telomerase.
Telomeres, the ends of eukaryotic chromosomes, are specialized DNA-protein complexes that serve to protect chromosome ends from degradation and end-to-end fusions (1). In most species, the DNA component consists of short repetitive sequences and varies in total length from less than 50 bp in hypotrichous ciliated protozoa and ≈300 bp in yeasts to thousands of base pairs in mammalian cells. The strand running 5′ to 3′ from the centromere to the telomere is commonly rich in G- and T-nucleotides and forms a 3′ single-stranded overhang at the very end of the telomere. Conventional DNA-dependent DNA polymerases are incapable of replicating these 3′ overhangs because the opposite strand is missing and cannot serve as a template for polymerization (2). Cells that lose their telomeres cease dividing and enter a stage known as senescence. In most organisms, this “end replication problem” is solved by the action of the enzyme telomerase (1).
Telomerase is a ribonucleoprotein complex in which a portion of the RNA subunit serves as template for the DNA polymerization reaction catalyzed by a protein subunit (3). Therefore, telomerase is a reverse transcriptase (RT). All telomerase catalytic protein subunits identified so far include a domain with amino acid sequence homology to RTs from other sources, such as retroviruses and retrotransposons, and are referred to as members of the TERT (telomerase reverse transcriptase) polymerase subclass (4, 5). Seven motifs that possess amino acid residues highly conserved throughout all RTs are found within this RT-domain (4–6). These motifs presumably contribute to a common tertiary folding as part of the “right hand” model seen in crystal structures of other reverse transcriptases like the HIV-1 RT (7), and some of the conserved residues are proposed to have specific roles in catalysis of the DNA polymerization reaction (8). An additional motif specific to TERT proteins, the T-motif, precedes the RT-motifs (9). The functions of these motifs are just beginning to be explored, although a contribution of the T-motif to binding of the RNA subunit has been discussed (10, 11) and supported experimentally in Tetrahymena (T. Bryan, K. Goodrich, and T.R.C., unpublished work).
In the fission yeast Schizosaccharomyces pombe, TERT is encoded by the trt1+ gene. Deletion of trt1+ causes progressive telomere shortening and the appearance of many elongated, nondividing cells (9). Interestingly, a subpopulation of trt1− cells can survive by two distinct pathways (12). Some survivors maintain their telomeres, presumably by a recombinational mode similar to a RAD52p-dependent mechanism based on homologous recombination previously observed in telomerase-negative Saccharomyces cerevisiae strains (13). Other survivors escape the need for telomerase by circularization of all three chromosomes, a phenotype also observed on mutation of the S. pombe homologs of the Ataxia telangiectasia mutated gene (14).
Mutation of TERT and comparison of phenotypes in vivo and in vitro has previously been reported only in S. cerevisiae. TERTs are only modestly conserved between species, and the S. pombe TERT (Trt1p) is no more closely related to S. cerevisiae Est2p (27% identity in RT motifs) than it is to human TERT (30% identity) (9). Thus, structure-function relationships established for the budding yeast may not be directly applicable to the fission yeast. Here we study the effects of point mutations in the RT-domain and T motif of the S. pombe TERT in vivo and compare the results to the catalytic activity of the mutant enzymes in vitro.
Materials and Methods
Growth of S. pombe Strains.
Strains CF199 (h−, leu1-32, ade6-M210, ura4-D18, his3-D1), CF382 (h+/h−, leu1-32/leu1-32, ade6-M210/ade6-M216, ura4-D18/ura4-D18, his3-D1/his3-D1, trt1+/trt1−∷his3+, taz1+/taz1−∷ura4+), CF797 (h−, leu1-32, ade6-M210, ura4-D18, his3-D1, trt1−∷his3+ [pNR210-trt1+ (HSV1-tk, ade6+, trt1+)]), and CF830 (h−, leu1-32, ade6-M210, ura4-D18, his3-D1, trt1∷his3 [pKAN1-Cmyc9trt1+ (kanMX, Cmyc9trt1+)], where Cmyc9 represents a C-terminal nine copies of the c-myc epitope tag) were grown in YES (yeast extract supplements) rich medium or PMG (pombe minimal glutamate) medium with required supplements. Geneticin disulfate (Sigma) was added to YES at a final concentration of 100 μg/ml, 5-fluoro-2′ deoxyuridine at 50 μM when required.
Plasmid Construction.
Plasmid pBS-trt1+ has the S. pombe genomic KpnI fragment bearing the trt1+ gene cloned into the KpnI site of pBluescript II SK(+). Plasmid pKAN-trt1+ contains the same trt1+ fragment and the kanMX4 marker. Plasmid pNR210-trt1+ was made by insertion of the same trt1+ fragment into the KpnI site of pNR210, which contains the ade6+ marker and HSV1-tk driven by the alcohol dehydrogenase gene promoter (a gift of N. Rhind and P. Russell, Scripps Research Institute, La Jolla, CA). To construct genes encoding fusion proteins of Trt1p with epitope tags, a NotI restriction site was engineered immediately after the start-codon or immediately before the stop-codon of trt1+ in pBS-trt1+ by site-directed mutagenesis. DNA fragments encoding several different tags were cloned into the NotI sites, but most proved to be unsuitable, either leading to a loss of telomerase activity in vivo or giving insufficient signal to acceptably detect the low-copy expression of Trt1p fusion proteins on immunoblots. Insertion of a myc9 epitope tag (a gift of K. Nasmyth, Institute of Molecular Pathology, Vienna) into the NotI-sites immediately before the trt1+ stop codon gave pBS-Cmyc9trt1+. Excising the Cmyc9trt1+ gene with KpnI and placing it into the KpnI site of pKAN1 gave pKAN1-Cmyc9trt1+.
Site-Directed Mutagenesis.
Point mutations in the trt1+ gene were introduced by using the QuickChange Site-directed Mutagenesis Kit (Stratagene) or by using PCR mutagenesis. An appropriate restriction fragment containing the mutation was substituted for the corresponding fragment of pBS-trt1+, giving pBS-trt1mut. The swapped fragments were sequenced completely to assure the presence of the mutation(s) as well as the absence of any second-site mutations. Mutated genes were recloned into the pKAN1 vector, giving pKAN1-trt1mut.
Extract Preparation.
Cells in log phase growth were harvested, were resuspended in TMG(300) [10 mM Tris⋅HCl, pH 8.0/1 mM MgCl2/10% (vol/vol) glycerol/300 mM NaCl] containing protease inhibitors (0.5 mM phenylmethanesulfonyl fluoride/1 mM benzamidine/1 μg/ml pepstatin A/5 μg/ml chymostatin/5 μg/ml leupeptin), 0.1 mM DTT, and 1 mM EDTA, and were lysed by vortexing with glass beads. Extracts were cleared by centrifugation for 10 min at 5,600 × g and twice for 10 min at 16,000 × g. Total protein concentrations were determined by using the Bio-Rad Protein Assay.
Immunopurification.
Agarose beads conjugated with monoclonal anti-c-myc (9E10) antibody (Santa Cruz Biotechnology) were equilibrated in TMG(200) [10 mM Tris⋅HCl, pH 8.0/1 mM MgCl2/10% (vol/vol) glycerol/200 mM NaCl] containing 0.1 mM DTT and protease inhibitors as listed above. Cell extracts were adjusted to a total protein concentration of 5 mg/ml, 0.1% (vol/vol) Tween 20, and 0.2% (vol/vol) RNase inhibitor RNasin (Promega). Adjusted extract (500 μl) was added to 60 μl of equilibrated beads (50% slurry) and was incubated at 4°C for 6 h with gentle shaking. Beads were washed three times with TMG(200) plus 0.1% Tween-20 and 0.1 mM DTT, were washed once with TMG(50) [10 mM Tris⋅HCl, pH 8.0/1 mM MgCl2/10% (vol/vol) glycerol/50 mM NaCl] plus 0.1 mM DTT, and were resuspended in 60 μl TMG(50) plus 0.5 mM DTT and 0.2% (vol/vol) RNasin.
In Vitro Telomerase Activity Assay.
Agarose beads from 30-μl suspension after immunoprecipitation were incubated in 6 μl of reaction buffer (75 mM Tris⋅HCl, pH 8.0/90 mM NaCl/7.5% glycerol/5 mM MgCl2/0.1 mM spermidine/0.1 mM DTT) containing dATP, dTTP, and dCTP at 200 μM each, 12.5 μM [α-32P]dGTP (800 Ci/mmol), and 5 μM oligonucleotide primer for 25 min at 30°C. To disrupt elongated primer-enzyme complexes, 2.2 μl of stop buffer [100 mM Tris⋅HCl, pH 7.5/200 mM EDTA/2.5% (wt/vol) SDS/1% (wt/vol) proteinase K] were added, and samples were incubated for 15 min at 37°C; 85 μl of ddH2O were then added. 32P-5′-end labeled 100-mer oligonucleotide was added as a precipitation control before phenol/chloroform extraction. Reaction products were separated on a 10% PAGE/8 M urea sequencing gel and were visualized and quantitated by using a phosphorimager. For RNase controls, agarose beads were preincubated for 10 min at 30°C in 2 μl of TMG(50) plus 1 mM DTT with or without RNase A (Sigma) at a final concentration of 5 mg/ml, before the addition of 6 μl reaction buffer. Primer oligonucleotide PBoli 14 (5′-TGT GGT GTG TGG GTG TG-3′) and primers based on S. pombe telomeric sequence repeats were gel-purified.
Results
Introduction of Mutated trt1+ Genes by Plasmid Shuffle.
Seven amino acid residues in motifs T to C of the RT-domain of S. pombe Trt1p, which are strictly conserved between TERT sequences from eight different species, were mutated to alanine by site-directed mutagenesis (Fig. 1, top). Mutated genes on the pKAN1-trt1mut vectors were tested in vivo in a genomic trt1−∷his3+ background. The deletion of trt1+ in this strain was originally covered with a wild-type trt1+ gene on the pKAN1 vector. First, this strain was transformed with pNR210-trt1+,which carries the ade6+ and HSV1-tk marker genes. Selection for only the ade6+ marker leads to the loss of pKAN1-trt1+, and the resulting strain was labeled CF797. Now it was possible to replace the wild-type trt1+ gene with the mutated trt1mut genes at a defined moment in time by plasmid shuffle, using negative selection against the thymidine kinase (HSV1-tk) marker on pNR210-trt1+ with 5-fluoro-2′ deoxyuridine (15). After transformation of pKAN1-trt1mut plasmids, selection on plates containing kanamycin and 5-fluoro-2′ deoxyuridine led to the replacement of pNR210-trt1+ with pKAN1-trt1mut. Resulting colonies were grown at 32°C for 3 days and then were used to inoculate liquid precultures in YES plus geneticin for growth curves.
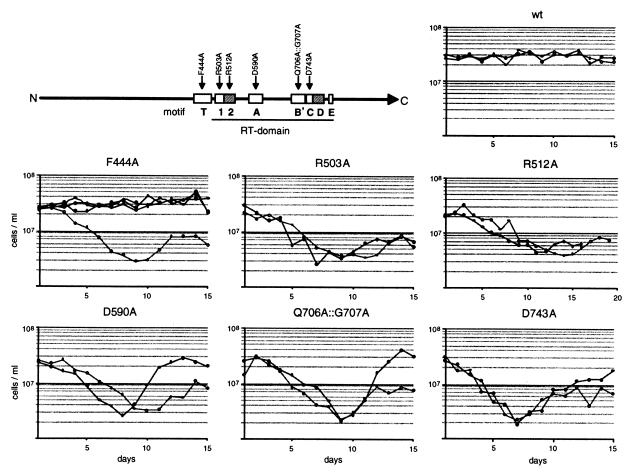
Figure 1.
Growth curves of Trt1p mutants. Precultures inoculated with colonies after plasmid shuffle were grown in YES liquid media containing geneticin at 32°C for 24 h. Each day thereafter, liquid cultures were inoculated at a density of 5 × 104 cells/ml, grown at 32°C for 24 h, and the cells were counted. Those cells not used for inoculation were harvested by centrifugation and were stored frozen for DNA preparations. For every mutant, at least two growth curves from independent colonies were recorded. In the case of F444A, the one curve showing a growth defect was subsequently found to be caused by a second-site two-amino acid deletion in Trt1p, whereas those cells carrying only the F444A mutation grew normally. The positions of the mutations in the T-motif and in the RT-domain of Trt1p are indicated in the diagram.
Effect of trt1 Mutations on Cell Growth.
The ability of mutated trt1 genes to complement the genomic trt1−∷his3+ deletion was tested by recording growth curves (Fig. 1). The growth of mutants R503A, D590A, Q706A∷G707A, and D743A was indistinguishable from that of trt1− cells (12), with a point of lowest viability varying between days 8 and 10 followed by the generation of survivors. Strains containing the R512A mutation in motif 2 of the RT-domain also went through a phase of low viability, but the point of lowest viability was reached later (days 11–14). Three of four cultures of the T-motif mutant F444A showed no growth defect for at least 15 days, as seen when the wild-type trt1+ gene was introduced by plasmid shuffle. The fourth culture of this mutant went through a phase of low viability; however, when the plasmid was reisolated and resequenced, it was found to contain an in-frame deletion of 6 bp in front of the RT-domain in addition to the desired T-motif mutation. We conclude that this additional mutation led to the loss of Trt1p function in vivo whereas the mutation F444A alone did not.
Telomere Length.
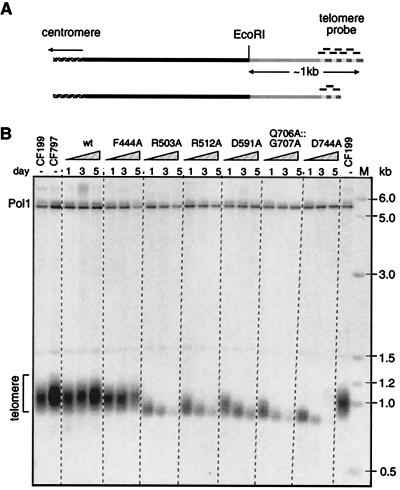
To examine telomere length during the course of growth curves, total DNA was prepared from cultures at days 1, 3, and 5. DNA was digested with EcoRI, a restriction enzyme that cuts approximately 1 kb from wild-type chromosome ends in S. pombe (16). With the loss of telomeric sequences, these EcoRI fragments were expected to decrease in size and give weaker hybridization signals (Fig. 2A). When telomere length for cultures from two growth curves per mutant was examined, a loss of telomeric sequences could be observed for all mutants that displayed a growth defect: R512A, D591A, Q706A∷G707A, and D743A (Fig. 2B; data not shown). For DNA preparations from the growth curve culture of T-motif mutant F444A that had the additional deletion and went through a phase of low viability, a decrease in telomere length was also observed (data not shown) whereas telomeres were maintained at wild-type length in a F444A mutant that did not show any growth defect (Fig. 2B).
Figure 2.
Telomere shortening caused by Trt1p mutations. (A) Diagram of hybridization with telomeric probe. In wild-type S. pombe telomeres, EcoRI cuts ≈1 kb from the telomere end. With shortening telomeres, shorter EcoRI-fragments and weaker hybridization signals are expected up to the point at which the whole telomeric repeat sequence is lost. (B) Telomere blot of EcoRI-cut DNA from S. pombe strains CF199 (wild-type), CF797 (starting strain for plasmid shuffle), and frozen cultures of days 1, 3, and 5 from growth curves of strains carrying pKAN1-trt1+ (wt) or pKAN1-trt1mut. To test for equal loading, the blot was also hybridized with a probe to the single-copy gene encoding DNA polymerase α (pol1+), which lies on a 5.6-kb EcoRI-fragment.
Protein Levels of Mutant Trt1p.
A loss of function of any mutant Trt1p protein could be attributable to a loss of its catalytic activity or to reduced protein levels caused by lower expression of the trt1mut gene or destabilization and degradation of the mutant protein. To examine the protein levels of mutant Trt1p in the cell, nine repeats of amino acids 410–419 of the human transcription factor c-myc (myc9) were placed at the C terminus of Trt1p, and the gene encoding this fusion protein was transformed on the pKAN1 plasmid into the genomic trt1−∷his3+ strain. The presence of the myc9 tag led to ≈50 bp shorter telomeres in vivo, when compared with telomeres of the wild-type strain CF199 (data not shown). The telomere blot showed that telomeres were stably maintained at this shorter length for at least 75 generations after plasmid shuffle, and cells did not display any growth defects.
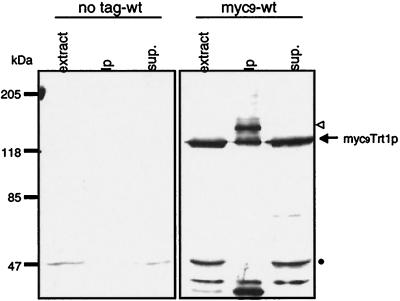
A strong band at a molecular weight of ≈128 kDa was observed when crude cell extract was probed with a polyclonal antibody against the c-myc epitope tag (Fig. 3). In immunoprecipitations with monoclonal anti-myc antibody conjugated to agarose beads, a second band at ≈147 kDa was observed whereas the 128-kDa band was not depleted from the supernatant. The calculated molecular weight of the myc9Trt1p fusion protein is 134 kDa. Both the 128- and 147-kDa bands were specific to extracts from cells expressing the fusion protein (compare with “no tag-wt” lanes in Fig. 3). Thus, it appears that there are two versions of the protein, both capable of being immunoblotted with anti-c-myc antibodies but one with more myc tags being much more efficiently immunopurified on antibody beads; we hypothesize that this heterogeneity arises by recombination within the repeated myc codons of the gene or proteolytic degradation of some of the myc tags.
Figure 3.
Detection of Cmyc9Trt1p fusion protein in cell extracts. Immunoblot of extracts prepared from a wild-type S. pombe strain with untagged Trt1p (CF199) and a strain expressing C-terminal myc9-tagged Trt1p (CF830). Crude extract (50 μg of total protein), immunoprecipitation with monoclonal anti-c-myc-antibody bound to agarose beads, and the supernatant thereof were separated by SDS/PAGE. Tagged proteins were detected by immunoblotting using a rabbit polyclonal anti-c-myc primary antibody (Santa Cruz Biotechnology) and peroxidase-conjugated goat anti-rabbit IgG secondary antibody (Boehringer Mannheim). Putative bands for the myc9Trt1p fusion protein are indicated by an arrow and open triangle (see text). The band at ≈50 kDa indicated by a dot is unrelated to Trt1p.
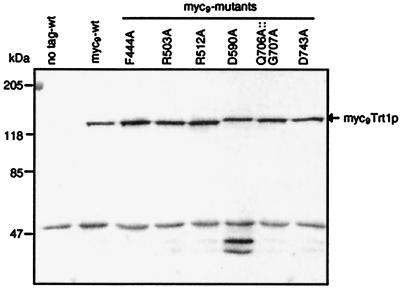
Plasmids were constructed bearing genes encoding mutant myc9Trt1p fusion proteins on the pKAN1 vector and introduced into CF797 by plasmid shuffle. Extracts were prepared from transformants and assayed on immunoblots. All mutant myc9Trt1p proteins were expressed at levels similar to that of the wild-type protein (Fig. 4). Two additional bands at low molecular weight detected for mutant D590A appear to be C-terminal degradation fragments, perhaps caused by a higher protease susceptibility of the mutant protein.
Figure 4.
Trt1p mutants are expressed at wild-type level. Cellular extracts prepared from S. pombe strains expressing untagged Trt1p and wild-type or mutant myc9Trt1p were separated on an SDS polyacrylamide minigel, were transferred to a nylon membrane, and were probed with polyclonal anti-c-myc antibody. The putative band for the myc9Trt1p fusion protein is indicated by an arrow. To confirm equal loading, identical samples were run on a second minigel, and proteins were stained with Coomassie blue (data not shown).
In Vitro Telomerase Activity Assay.
We next examined the DNA polymerization activity of the mutant myc9Trt1p fusion proteins in vitro. The myc9 epitope tag allowed efficient immunopurification of the fusion proteins by using monoclonal anti-c-myc antibodies bound to agarose beads. Using an adaptation of a method developed with epitope-tagged Est2p in S. cerevisiae [ref. 10; R. Weilbaecher and V. Lundblad, personal communication], we were able to assay the activity of bead-bound telomerase. The observed activity closely resembled that described earlier by Lue and Peng (17), except their higher molecular weight products were not observed. The bead method proved to be very reliable and more sensitive than determining S. pombe telomerase activity after partial purification of the enzyme by chromatography as described (17).
After immunoprecipitation, aliquots of beads were incubated with several different primer oligonucleotides and all four deoxynucleotides (dATP, dTTP, dCTP, and [α-32P]dGTP). Six different primer oligonucleotides composed of S. pombe telomeric sequence repeats GGTTAC(A) and two primers consisting of S. cerevisiae telomeric sequences, containing only G- and T-nucleotides, were used. It was reported previously that S. cerevisiae telomeric primers were efficiently elongated by partially purified S. pombe telomerase (17). For all primer oligonucleotides used, elongation products were observed (data not shown). Surprisingly, the most intense elongation bands were seen when S. cerevisiae telomeric primer PBoli 14 was used. Because we wanted to measure in vitro telomerase activity of the Trt1p mutants with the highest sensitivity possible, PBoli 14 was used for subsequent experiments.
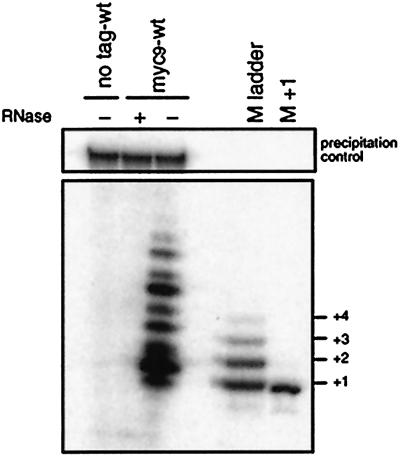
Nine nucleotides were added to PBoli 14 in vitro, with the most intense bands at positions primer+2 and primer+6 (Fig. 5). No elongation products could be seen when the immunoprecipitation beads were preincubated with RNase or in immunoprecipitations from a wild-type strain (CF199), which does not express myc9-tagged Trt1p (Fig. 5). Using primers ending in different positions of the telomeric repeat and dideoxynucleotides, we determined the sequence being added to PBoli14 as well as to a primer made of S. pombe telomeric repeats in vitro. With both sets of primers, the sequence CGGTKAV was added (K indicates that the results were consistent with G or T; V indicates A, G, or C). The band observed at the +1 position in Fig. 5 may be caused by misincorporation of dG or by primer degradation. Our sequence results are in agreement with the predominant S. pombe telomeric repeat sequence VGGTTAC (data not shown).
Figure 5.
Validation of the in vitro telomerase assay. Cell extracts prepared from wild-type cells with untagged Trt1p (CF199) and cells expressing myc9Trt1p fusion protein (CF830) were subject to immunopurification on monoclonal anti-c-myc antibodies conjugated to agarose beads. After incubation with (+) and without (−) RNase A, telomerase activity was assayed. A 32P-5′-end labeled 100-mer oligonucleotide (Upper) was added to test that no reaction products were lost during phenol-chloroform extraction or ethanol precipitation. Products were separated by sequencing gel electrophoresis. As length markers, PBoli 14 oligonucleotide was elongated by using [α-32P]dGTP (M ladder) or [α-32P]ddGTP (M+1), respectively, by terminal deoxynucleotidyltransferase. Because electrophoretic mobility depends on nucleotide composition, the M ladder does not align with the telomerase extension products.
In Vitro Activity of Telomerase Mutants.
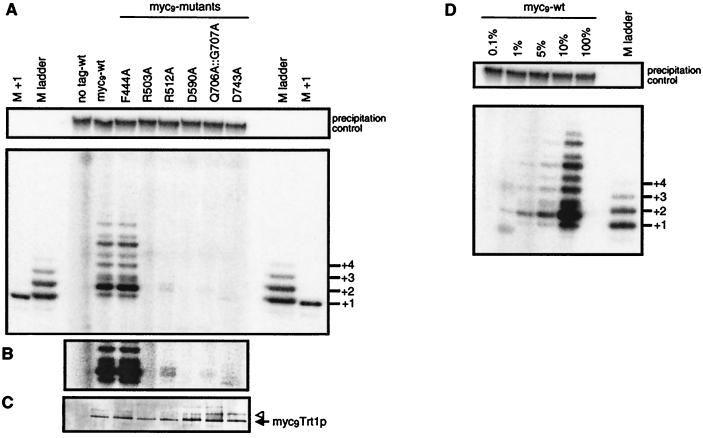
Telomerase activity of S. pombe expressing mutant myc9Trt1p fusion proteins was assayed after immunopurification from extracts of two independent strains per mutant (Fig. 6A; data not shown). For the mutant in the T-motif, F444A, in vitro primer elongation activity could be detected at the wild-type level. Mutant R512A showed only ≈5% of wild-type activity (Fig. 6B) whereas no convincing elongation products could be detected for the other mutants. Immunoblot analysis confirmed that similar amounts of myc9Trt1p were bound in each immunopurification (Fig. 6C). To determine the sensitivity of the in vitro assay, dilutions of wild-type myc9Trt1p were tested. The primer+2 extension band could still be detected with as little as 1% of wild-type extract (Fig. 6D). Therefore, the in vitro activities of mutants R503A, D590A, Q706A∷G707A, and D743A are less than 1% of wild-type activity.
Figure 6.
In vitro telomerase activity of Trt1p mutants. (A) Extracts from S. pombe cells expressing untagged Trt1p, or wild-type or mutant myc9Trt1p fusion protein were immunopurified on monoclonal anti-c-myc antibodies conjugated to agarose beads. Extension of primer oligonucleotide PBoli 14 was assayed as in Fig. 5, where the precipitation control and length marker are also described. (B) A darker exposure of a section better illustrates the primer+2 reaction product for mutant R512A. (C) To test whether equal amounts of myc9Trt1p were bound to the immunoprecipitation beads, the other half of the beads used for the telomerase assay was boiled in Laemmli loading buffer, proteins were resolved on a 6% SDS polyacrylamide minigel, and tagged proteins were revealed by immunoblotting. The arrow and triangle indicate the putative myc-tagged Trt1p species described in the text. (D) Dilution assay to determine the sensitivity of the in vitro telomerase assay. After immunoprecipitation from cells expressing wild-type myc9Trt1p fusion protein, immunoprecipitation beads suspended in TMG(200) were successively diluted and were used in in vitro activity assays as described in A.
Discussion
Telomerase adds telomeric repeats to the termini of chromosomes by reverse transcription of a small part of its RNA subunit. This model is supported by the homology of the RT-domain of the TERT subunit to other reverse transcriptases and by mutational analysis of the RT-domain of budding yeast and human TERTs. We have now extended the study of structure-function relationships in TERT to the evolutionarily distant fission yeast.
Mutations of Conserved Residues in the RT-Motifs Lead to a Loss of Telomerase Activity in Vivo and in Vitro.
We showed that, when some residues in the RT-motifs of Trt1p, that are identical in all eight known members of the TERT polymerase subclass, are changed to alanine, S. pombe cells expressing these mutant TERTs show a growth defect and loss of telomeric sequences over time. When the primer extension activities of the mutant proteins were tested in an in vitro assay, polymerization activity was reduced to less than 1% of wild-type level for mutants R503A, D590A, Q706A∷G707A, and D743A and to ≈5% of wild-type level for mutant R512A. As cellular Trt1p protein levels were similar to wild-type, the loss of activity is not caused by degradation of the mutant proteins. Mutagenesis of several conserved amino acid residues in TERTs from human (hTERT) and Tetrahymena thermophila (Tt_TERT) and testing their ability to catalyze a primer extension reaction in an in vitro reconstitution assay have been described (refs. 11 and 18; T. Bryan, K. Goodrich, and T.R.C., unpublished work). However, the only other system in which it has been possible to assess the activity of TERT mutants both in vitro and in vivo has been S. cerevisiae Est2p (4, 10, 19).
The complete loss of telomerase activity was expected for mutants D590A and D743A and is consistent with previous observations for alanine mutations of the analogous residues in Est2p (4, 19) and hTERT (11). Both residues are thought to be necessary for the coordination of two magnesium ions that are directly involved in the catalysis of the polymerization reaction (8).
In the case of the motif 2 mutant R512A, a growth defect was observed. However, the point of lowest viability was shifted from days 8–10 to days 11–14. Consistent with a more benign phenotype, some residual primer extension activity was measured for this mutant enzyme in vitro (≈5% wild-type activity). In contrast, no activity (less than 1% of wild-type activity) was observed for the analogous mutant R631A of hTERT in an in vitro reconstitution assay (11).
T-Motif Mutant F444A Is Functional in the Fission Yeast.
A short stretch of amino acid residues within the T-motif (FYxTE) is strictly conserved among all known TERT sequences. Surprisingly, mutating the phenylalanine residue in this sequence to alanine did not cause growth defects or telomere shortening in vivo. Moreover, the mutant enzyme showed wild-type-like activity in vitro. In contrast, no activity (less than 1% of wild-type activity) could be measured for the analogous F560A mutant in hTERT after in vitro reconstitution (11). This apparent species-specific difference does not necessarily imply that the T-motif has a different function in the TERTs from human and S. pombe; for example, the S. pombe RNA-protein complex may be more stable and therefore not sensitive to a single point mutation, or reconstitution of human telomerase in rabbit reticulocyte lysates may be more sensitive to structural perturbations than assembly in vivo.
Cmyc9Trt1p Fusion Proteins and in Vitro Telomerase Assay.
The availability of a myc9 epitope tag at the C terminus of Trt1p provides a useful tool to determine expression of low levels of Trt1p. This is the first report of any yeast TERT being detected in crude cell extracts; enrichment of the protein by immunoprecipitation was not necessary for its detection, although it is still useful, as described below.
Enrichment of Cmyc9Trt1p fusion proteins on antibody beads and assaying for primer extension activity proved to be an effective and reliable method to quantify S. pombe telomerase activity in vitro. Activity depended on the presence of the myc9 epitope tag at Trt1p, was RNase-sensitive, and was absent for catalytic aspartate mutants D590A and D743A. Furthermore, S. pombe telomeric sequence was added to the primers (also see ref. 17). Taken together, these experiments provide strong evidence that the activity observed is caused by the action of the telomerase enzyme. The availability of the Cmyc9Trtp fusion protein and the in vitro telomerase assay, together with the ease of genetic manipulations and the excellent cytology of S. pombe, make it an attractive system for future studies of telomerase.
Acknowledgments
We thank M. Galova, K. Nasmyth, N. Rhind, and P. Russell for providing plasmids, R. Weilbaecher and V. Lundblad for sharing their protocol for the in vitro telomerase assay, K. Goodrich, E. Podell, and A. Gooding for oligonucleotide synthesis, and J. Cooper for critique of the manuscript. P.B. is supported by a Wellcome Prize Traveling Research Fellowship (Grant 054549/Z/98/Z). This work was supported by grants from the National Institutes of Health (GM28039) and the Human Frontier Science Program.
Abbreviations
- RT
reverse transcriptase
- TERT
telomerase reverse transcriptase
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.130187397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.130187397
References
- 1.Blackburn E H, Greider C W. Telomeres. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. [Google Scholar]
- 2.Lingner J, Cooper J P, Cech T R. Science. 1995;269:1533–1534. doi: 10.1126/science.7545310. [DOI] [PubMed] [Google Scholar]
- 3.Greider C W, Blackburn E H. Nature (London) 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 4.Lingner J, Hughes T R, Shevchenko A, Mann M, Lundblad V, Cech T R. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 5.Bryan T M, Cech T R. Curr Opin Cell Biol. 1999;11:318–324. doi: 10.1016/S0955-0674(99)80043-X. [DOI] [PubMed] [Google Scholar]
- 6.Xiong Y, Eickbush T H. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 8.Joyce C M, Steitz T A. Annu Rev Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- 9.Nakamura T M, Morin G B, Chapman K B, Weinrich S L, Andrews W H, Lingner J, Harley C B, Cech T R. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 10.Friedman K L, Cech T R. Genes Dev. 1999;13:2863–2874. doi: 10.1101/gad.13.21.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinrich S L, Pruzan R, Ma L, Ouellette M, Tesmer V M, Holt S E, Bodnar A G, Lichtsteiner S, Kim N W, Trager J B, et al. Nat Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura T M, Cooper J P, Cech T R. Science. 1998;282:493–496. doi: 10.1126/science.282.5388.493. [DOI] [PubMed] [Google Scholar]
- 13.Lundblad V, Blackburn E H. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- 14.Naito T, Matsuura F, Ishikawa F. Nat Genet. 1998;20:203–206. doi: 10.1038/2517. [DOI] [PubMed] [Google Scholar]
- 15.Kiely J, Hasse S B, Russell P, Leatherwood J. Genetics. 2000;154:599–607. doi: 10.1093/genetics/154.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sugawara N. Cambridge, MA: Harvard Univ.; 1988. [Google Scholar]
- 17.Lue N F, Peng Y. Nucleic Acids Res. 1997;25:4331–4337. doi: 10.1093/nar/25.21.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collins K, Gandhi L. Proc Natl Acad Sci USA. 1998;95:8485–8490. doi: 10.1073/pnas.95.15.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Counter C M, Meyerson M, Eaton E N, Weinberg R A. Proc Natl Acad Sci USA. 1997;94:9202–9207. doi: 10.1073/pnas.94.17.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]