Abstract
Repair of DNA double-strand breaks in vertebrate cells occurs mainly by an end-joining process that often generates junctions with sequence homologies of a few nucleotides. Mre11 is critical for this mode of repair in budding yeast and has been implicated in the microhomology-based joining. Here, we show that Mre11 exonuclease activity is sensitive to the presence of heterologous DNA, and to the structure and sequence of its ends. Addition of mismatched DNA ends stimulates degradation of DNA by Mre11, whereas cohesive ends strongly inhibit it. Furthermore, if a sequence identity is revealed during the course of degradation, it causes Mre11 nuclease activity to pause, thus stabilizing the junction at a site of microhomology. A nuclease-deficient Mre11 mutant that still binds DNA can also stimulate degradation by wild-type Mre11, suggesting that Mre11-DNA complexes may interact to bridge DNA ends and facilitate DNA joining.
Damage to chromosomal DNA in the form of double-strand breaks is dangerous for cells because of the risk of deletions, translocations, and other types of misjoined products. Whereas an unrepaired break is lethal, a misrepaired break can potentially lead to cellular transformation and loss of growth control in multicellular organisms. Eukaryotic cells have two primary methods to handle double-strand breaks: homologous recombination and nonhomologous end joining (NHEJ), each of which uses a distinct set of protein factors. For instance, the Rad51/52/54/55/57 system functions in homologous recombination (1), whereas Ku, DNA-dependent protein kinase, Xrcc4, and ligase IV function in NHEJ (2, 3).
One protein complex that appears to play a role in both homologous and nonhomologous processes in yeast (but does not affect interhomolog exchange), is Mre11/Rad50/Xrs2. This complex is required for NHEJ, sister chromatid repair, and telomere maintenance, as well as for the initiation of meiotic recombination (4–11). Homologs of Mre11 and Rad50 are present in mammalian cells and associate with the Nbs1 protein, which appears to substitute for Xrs2 in higher organisms (12). Truncated forms of the NBS1 gene are responsible for the human autosomal recessive disorder Nijmegen Breakage Syndrome (NBS), which causes genomic instability, leading to cancer predisposition and immunodeficiency in affected patients (13). NBS is phenotypically very similar to ataxia telangiectasia (AT), a more common genetic disorder that also causes pleiotropic defects in DNA damage responses but is caused by mutations in the ATM gene (14). Recently, mutations in Mre11 were found to be responsible for an AT-like disorder, thus providing yet another link between the Mre11/Rad50/Nbs1 (M/R/N) complex and double-strand break repair in mammalian cells (15).
When a DNA break is repaired by NHEJ in vivo, there are often small deletions at the breakpoints, and microhomologies are found at the junctions. These small regions of sequence identity between the broken DNA ends range from one to several base pairs in length. Microhomology-mediated joining has been observed in yeast (9, 16–19) as well as in mammalian cells (20, 21), and can occur in the coding joints of V(D)J recombination in B and T cells (22–24).
Yeast strains deficient in any of the components of the Mre11 complex are 10- to 100-fold less efficient in NHEJ of DNA ends generated by restriction enzymes (9, 16, 25). On plasmid substrates, the small fraction of rejoined molecules formed in these strains does not contain microhomology-mediated junctions, suggesting that M/R/X may be required for use of microhomologies in DNA repair. A similar observation was made of chromosomal translocation and inversion products formed in Mre11, Rad50, and Xrs2 null cells, showing that use of microhomologies in joining of chromosomal DNA is also affected by these proteins (26).
Previous studies of the enzymatic activities of the human M/R/N complex in vitro have demonstrated that Mre11 by itself is a 3′ to 5′ exonuclease on DNA duplexes, as well as an endonuclease on single-stranded DNA and hairpin structures (27–29). We also showed that, in the presence of a DNA ligase, Mre11 can facilitate joining of mismatched ends in vitro, and the junctions of the products contain short homologies (28). In the work presented here, we show that the exonuclease activity of Mre11 bound to one DNA molecule can be stimulated by addition of another DNA end. In addition, the degree of homology between the two ends determines the degree of stimulation, thus providing a possible mechanistic explanation for the activity of the M/R/N complex in microhomology-mediated end joining.
Materials and Methods
DNA Substrates and Competitors.
In Figs. 1 and 2A, the substrate with a 4-nt 5′ overhang was composed of TP74(CTGCAGGGTTTTTGTTCCAGTCTGTAGCACTGTGTAAGACAGGCCA) annealed to TP124(CATCTGGCCTGTCTTACACAGTGCTACAGACTGGAACAAAAACCCTGCAG), with TP74 labeled with [32P] at the 5′ end. All of the double-stranded DNA substrates used were blunt on one end, unless stated otherwise. The poly(dI)⋅poly(dC) (Amersham Pharmacia) was an average of 600 bp in length. The 5′ overhang competitor DNA used in Fig. 1 and in lanes 3 and 4 in Fig. 2A was composed of TP127(AGGCTGCAGCCACTGCAGTTCTAGACCTCATCGAGGGATTACATG) annealed to TP128(CATGTAATCCCTCGATGAGGTCTAGAACTGCAGTGGCTGCA). The blunt competitor DNA used in Fig. 1 was composed of TP128 annealed to TP126(TGCAGCCACTGCAGTTCTAGACCTCATCGAGGGATTACATG). The 3′ overhang competitor DNA used in Figs. 1 and 2A was composed of TP128 annealed to TP130(GCCACTGCAGTTCTAGACCTCATCGAGGGATTACATG). The cohesive 5′ overhang DNA competitor used in Fig. 2A, lanes 6 and 7, was composed of TP128 annealed to TP139(GATGTGCAGCCACTGCAGTTCTAGACCTCATCGAGGGATTACATG). The 5′ overhang competitor used in Fig. 2A, lanes 9 and 10, was composed of TP128 annealed to TP140(AGACATGCAGCCACTGCAGTTCTAGACCTCATCGAGGGATTACATG). The substrate used in Fig. 2B was composed of DG50(GGCCTGTCTTACACAGTGCTACAGACTGGAACAAAAACCCTGCAG) annealed to DG9 (30), with DG9 labeled with [32P] at the 5′ end. The 5′ overhang competitor used in Fig. 2B, lanes 3 and 4, was composed of TP152(CGTACTGCTGATGCAGCCACTGCAGTTCTAGACCTCATCGAGGGATTACATG) annealed to TP128. The competitor used in lanes 6 and 7 was composed of TP154(TAAGACAGGCCTGCAGCCACTGCAGTTCTAGACCTCATCGAGGGATTACATG) annealed to TP128. The DNA used in the gel mobility shift assay was composed of DAR40 annealed to DG113 (29), with DAR40 labeled with [32P] at the 5′ end.
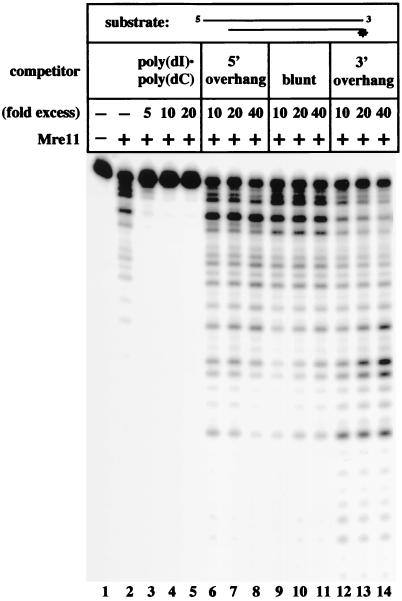
Figure 1.
Heterologous DNA molecules increase Mre11 exonuclease activity. A total of 25 ng (0.3 pmol) of human recombinant Mre11 was incubated with 0.04 pmol of double-stranded DNA containing a 4-nt 5′ overhang, labeled at the 5′ end of the recessed strand. Immediately after combining Mre11 with the labeled substrate, various unlabeled DNA molecules were added, as indicated. The fold excess of competitor was calculated by using moles of DNA molecules. The reactions were stopped after 30 min and then run on a denaturing polyacrylamide gel.
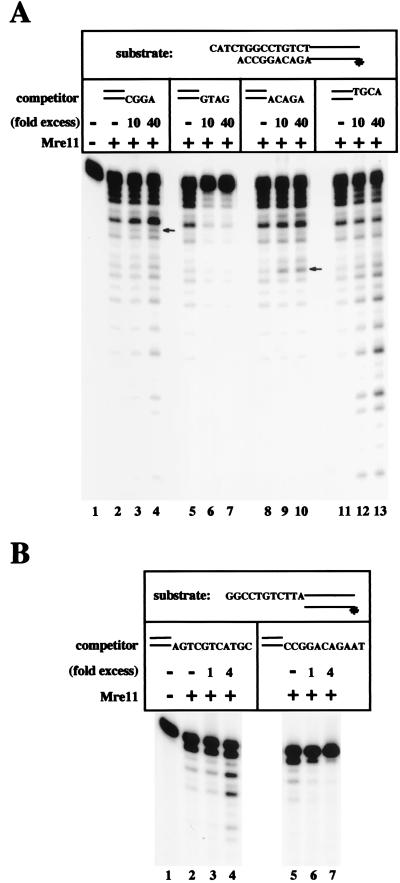
Figure 2.
The end sequence of heterologous DNA molecules determines the effect on Mre11 exonuclease activity. (A) 0.3 pmol Mre11 was incubated with 0.04 pmol of double-stranded DNA containing a 4-nt 5′ overhang and labeled at the 5′ end of the recessed strand, as shown. Various unlabeled DNAs were added, as in Fig. 1. The arrows indicate positions in the substrate DNA that correspond to the 5′ edge of sequence homologies present in both the substrate and the competitor. The presence of the specific competitor DNAs induces an increase in accumulation of the band indicated because of microhomology-induced pausing of Mre11 exonuclease activity (see model in Fig. 5). (B) 0.3 pmol Mre11 was incubated with 0.04 pmol of double-stranded DNA containing an 11-nt 5′ overhang and labeled at the 5′ end of the recessed strand, as shown. Various unlabeled DNAs containing 5′ overhangs were added as in Fig. 1, except that 10-fold lower amounts were used.
Proteins.
Wild-type Mre11 and H217Y Mre11 proteins were expressed by using a baculovirus system (BRL) and purified as described previously (28). The H217Y mutation was made in pTP17 by the Quickchange method (Stratagene), generating H217Y transfer vector pTP22. Mre11 H217Y virus was made from the corresponding bacmid, pTP25.
Exonuclease Reactions.
Reactions contained 25 mM Mops, pH 7.0, 150 mM KCl, 10% polyethylene glycol, 2 mM magnesium chloride, 2 mM manganese chloride, and 2 mM DTT. The reaction mix was prewarmed to 37° before addition to the proteins, in a volume of 10 μl each. The competitor DNA was added immediately after the substrate DNA, and incubations continued at 37° for 30 min. SDS, EDTA, and proteinase K were then added to final concentrations of 0.2%, 5 mM, and 0.1 mg/ml, respectively, and incubated for another 15 min. Four microliters of each reaction were lyophilized, resuspended in 6 μl formamide loading buffer, and run on a sequencing gel containing 15% acrylamide and 7 M urea. After the run, each gel was analyzed by using a phosphorimaging system (Molecular Dynamics).
Gel Shifts and Joining Reactions.
Gel mobility shift and plasmid joining assays were performed as described previously (29), except that the gel shift reactions were run on a 1/2× 90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3 (TBE) 6% acrylamide gel.
Results
Heterologous DNA Can Increase Mre11 Exonuclease Activity.
Because of the known involvement of Mre11 in DNA end joining in yeast and its ability to facilitate repair of mismatched ends in vitro, we investigated whether addition of heterologous DNA molecules would affect exonucleolytic degradation of a labeled DNA substrate by Mre11. We first tested a labeled substrate containing a 4-nt 5′ overhang that was labeled on the bottom, recessed strand. Incubation of recombinant human Mre11 protein with this substrate caused degradation of the labeled strand from 3′ to 5′ (Fig. 1, lane 2). When a 5- to 20-fold excess of poly(dI)⋅poly(dC) was added to this reaction directly after addition of the labeled substrate, degradation by Mre11 was completely abolished, as one would expect with unlabeled competitor DNA (Fig. 1, lanes 3–5). However, addition of a 10- to 40-fold excess of unlabeled, double-stranded DNA did not inhibit Mre11 exonuclease activity, but instead increased its activity by 2- to 10-fold (Fig. 1, lanes 6–14).
We then asked what properties of the added DNA were responsible for this stimulation and found that it depended on the structure of the DNA ends. For instance, in the presence of a 10- to 40-fold excess of unlabeled DNA containing a 4-nt 5′ overhang (not cohesive with or identical to the 5′ overhang on the labeled substrate), the amount of full-length substrate left undegraded was 30% less than without competitor, even though the competitor was itself a substrate for digestion by Mre11 (lanes 6–8). The length of DNA degraded also was increased: quantification of the bands in each lane showed that the amount of labeled DNA with more than 5 nt removed from the 3′ end was increased 3- to 4-fold in the presence of the unlabeled 5′ overhang DNA. This effect was also observed with unlabeled blunt DNA (Fig. 1, lanes 9–11), although the increase in degradation was less pronounced. Addition of homopolymer single-stranded DNA had no effect on the extent or efficiency of degradation by Mre11 (data not shown).
The competitor DNA with the greatest stimulatory effect was one containing a 4-nt 3′ overhang (Fig. 1, lanes 12–14). At the highest concentration, the amount of substrate left undegraded was 40% less than in the absence of heterologous DNA, and the amount of DNA shortened by more than 5 nt was increased by 5- to 6-fold. The 5′ overhang, blunt, and 3′ overhang unlabeled DNAs used in this experiment are all approximately 40–50 bp in length (similar to the labeled substrate), identical in sequence except for the overhangs, and blunt on the other end. The amounts of unlabeled DNA used here are the optimal concentrations; smaller amounts have little effect, and levels higher than those shown are increasingly inhibitory (data not shown).
The End Sequence of Competitor DNA Determines the Effect on Mre11 Exonuclease Activity.
To investigate this phenomenon further, we asked whether the sequence of the DNA overhangs affects the stimulation of Mre11 exonuclease activity. The labeled substrate used in this experiment was the same as in Fig. 1, and the sequence of the 5′ overhang end is shown in Fig. 2A. The first competitor is identical to the unlabeled DNA with a 5′ overhang used in Fig. 1, and the same stimulatory effect was seen (Fig. 2A, lanes 2–4). In the next set of reactions (Fig. 2A, lanes 5–7), the 5′ overhang sequence was changed to be complementary to the 5′ overhang present in the labeled substrate. In this case, addition of the cold competitor immediately after the labeled substrate was not stimulatory but inhibited the exonuclease activity of Mre11 almost completely.
This result suggested that the exonuclease activity of Mre11 could be regulated by the presence of single-stranded regions on the ends of duplex DNA, depending on the degree of complementarity between the overhang on the competitor and the overhang on the substrate. If so, then this effect might also occur when the region complementary to the single-stranded overhang on the competitor was not initially available for base-pairing; that is, if it had to first be revealed by exonuclease digestion of the labeled DNA. An example of this is shown in Fig. 2A, lanes 8–10, in which the sequence of the overhang on the competitor DNA, “ACAGA”, is identical to part of the sequence on the labeled strand of the substrate but is buried 10 nt from the 3′ end of that strand. Addition of this unlabeled competitor to the reaction with Mre11 was stimulatory but primarily increased the intensity of just one band (next to the arrow in Fig. 2A, lane 10), generated by degradation of the substrate to the exact position where the overhang sequence begins. With this combination of competitor and substrate, the increase in the extent of degradation was not seen, particularly when compared with the effect of the other 5′ overhang or the 3′ overhang shown in lanes 11–13. A similar but less pronounced effect was seen with the competitor used in lanes 3 and 4, in which the “CGGA” motif is present in the 5′ overhang of the competitor, as well as in the bottom, recessed strand of the labeled substrate (arrow next to lane 4 indicates the position of the sequence identity in the labeled substrate).
To verify that the effect of competitor DNA on Mre11 is not specific to any particular DNA sequence, we tested other substrate/competitor pairs containing completely different sequences and lengths of 5′ overhang (Fig. 2B). In this case, the overhangs were 11 nt on both the labeled substrate and the unlabeled competitors. Titration of the reaction components revealed that the optimal concentration of these competitors is approximately 10-fold lower than the DNAs with 4 nt overhangs used in Figs. 1 and 2A. As in the previous case, addition of unlabeled DNA with a nonmatching overhang increased the amount of labeled substrate consumed as well as the overall extent of degradation (Fig. 2B, lanes 1–4). An analysis of the initial rates of exonuclease digestion showed that the heterologous DNA increased exonuclease activity of Mre11 on this substrate by 1.6-fold (data not shown). In contrast to mismatched ends, the presence of a cohesive end on the competitor DNA molecule completely inhibited degradation by Mre11 (Fig. 2B, lanes 5–7).
Mre11S Mutant Protein Is Catalytically Inactive yet Affects Wild-Type Protein.
During the course of our studies, we have attempted to correlate in vitro activities of purified human Mre11 protein to phenotypes of yeast mre11 mutants, which have been described in the literature. mre11S yeast mutants have been isolated (31) that are similar to the set of rad50S mutants (32) in that double-strand breaks are introduced but not resected during meiosis. Although the rad50S mutants are all essentially wild type with respect to mitotic function, one of the mre11S strains, mre11–58S, exhibits sensitivity to ionizing radiation, a high spontaneous mutation rate, and delayed processing of double-strand breaks induced by HO endonuclease, similar to mre11 null strains. We made mutations in the human Mre11 gene equivalent to the critical mutation in the mre11–58S strain (H217Y in the human amino acid sequence) and expressed and purified this mutant protein.
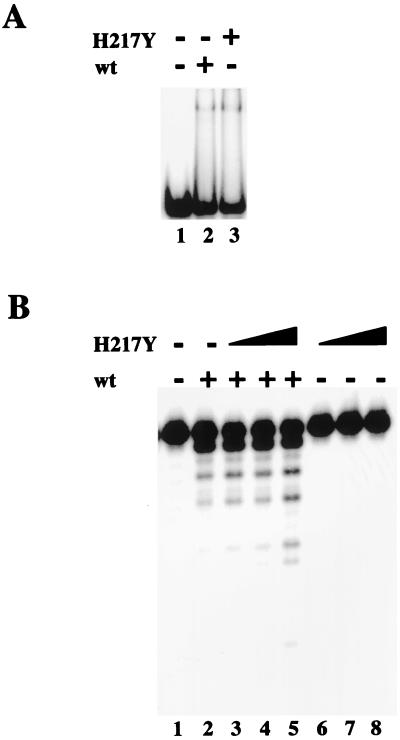
In a gel mobility shift assay for DNA-binding, Mre11 H217Y formed protein-DNA complexes at least as efficiently as the wild-type protein (Fig. 3A). The mutant protein was completely deficient in nuclease activity (Fig. 3B, lanes 6–8), as expected from observations of yeast Mre11–58 (33). Interestingly, however, addition of H217Y to reactions containing wild-type protein and a labeled DNA substrate caused an increase in the amount of full-length substrate degraded by wild-type Mre11 as well as the number of base pairs removed, similar to the effect of adding exogenous DNA (Fig. 3B, lanes 3–5). In the presence of wild-type Mre11 and a 4-fold excess of H217Y protein, 30% less of the full-length substrate was left intact, and the amount of DNA shortened by more than 3 nt was increased by 3- to 10-fold, compared with the reaction without H217Y protein.
Figure 3.
A nuclease-deficient but DNA-binding-proficient Mre11 mutant increases the activity of wild-type Mre11. (A) Gel mobility shift assay: 200 ng of wild-type or H217Y Mre11, as indicated, were incubated with a labeled double-stranded DNA containing 3′ overhangs on each end and run on a 6% acrylamide gel. (B) 30 ng of wild-type Mre11 was incubated with the same 5′ overhang substrate as in Fig. 2B. Increasing amounts (30 ng, lanes 3 and 6; 60 ng, lanes 4 and 7; 120 ng, lanes 5 and 8) of H217Y Mre11 protein were included in the reactions as indicated.
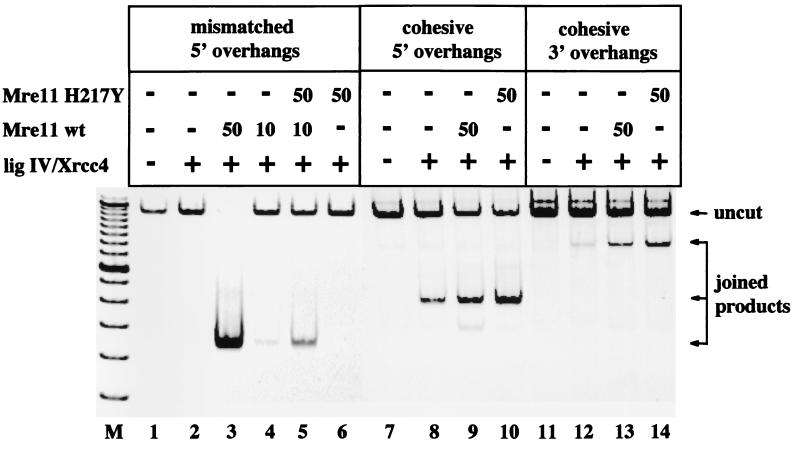
From these results, it seemed possible that H217Y would also increase end joining, so we used a PCR-based plasmid rejoining assay to investigate this question. In the experiment shown in Fig. 4, lanes 1–6, a plasmid was cut with two different restriction enzymes, generating mismatched 5′ overhangs. Incubation of this substrate with a ligase (in this case, a complex of human DNA ligase IV and Xrcc4) did not generate any joined products because the DNA ends were not cohesive (lane 2). Addition of wild-type Mre11 to this reaction yielded a large amount of joined product (lane 3), similar to the results shown previously (28). The amount of product obtained was very sensitive to the amount of wild-type Mre11 added (lanes 3 and 4). Addition of the catalytically inactive H217Y mutant to a reaction with a suboptimal level of wild-type Mre11 increased the yield of product 5- to 10-fold (lane 5), similar to the effect of H217Y on the wild-type protein in the exonuclease assays (Fig. 3B). By itself, H217Y did not facilitate joining of mismatched ends because it is lacking exonuclease activity (lane 6).
Figure 4.
Wild-type and H217Y Mre11 increase joining of nonhomologous and homologous ends. Plasmid DNA was cut with restriction enzymes to generate mismatched overhangs (XhoI/BamHI), cohesive 5′ overhangs (BamHI/BglII), or cohesive 3′ overhangs (SphI). The plasmids were incubated with varying amounts of wild-type or H217Y Mre11, as well as with ligase IV/Xrcc4 complex, as indicated. The products were amplified by PCR and run on a native polyacrylamide gel. Arrows indicate the positions of the uncut plasmid DNA and the joining products.
The ability of the H217Y mutant Mre11 to facilitate joining directed by the wild-type protein suggested that Mre11 might be bridging DNA ends. We tested this hypothesis by performing the joining assay using a substrate with cohesive 5′ or 3′ ends, as shown in Fig. 4, lanes 7–14. The presence of Mre11 in these reactions with ligase IV/Xrcc4 did, in fact, increase the yield of joined products 1.5- to 3-fold on both 5′ and 3′ cohesive ends, with the H217Y protein consistently yielding a higher level of joining than wild-type Mre11.
Discussion
Microhomologies have been observed very frequently at junctions of NHEJ (illegitimate recombination) products in both yeast and mammalian cells. A number of observations, both in vitro and in vivo, link the Mre11/Rad50/Xrs2(Nbs1) complex to the use of microhomology in NHEJ. In Saccharomyces cerevisiae strains lacking any one of the components of this complex, junctions made by NHEJ in plasmids lack the microhomologies that are usually present in strains deficient in other components of the NHEJ system (1, 9). In addition, Mre11, Rad50, and Xrs2 mutants show a 600-fold increase in the frequency of large-scale chromosomal rearrangements, and the junctions at these sites do not contain microhomologies, in contrast with other mutant strains (26). Finally, a very simple system containing only Mre11 and a DNA ligase can generate microhomology-dependent end joining in vitro (28). In this work we report activity of the Mre11 protein that may explain its role in this process.
Inhibition by Complementary Ends.
First, we find that the exonuclease activity of Mre11 is sensitive to the presence of sequence identities between a substrate DNA and other DNA ends present in the reaction. 3′ to 5′ resection of a labeled DNA molecule is strongly inhibited when a complementary single-stranded overhang is present on another DNA end. The strength of this inhibition depends on the length of the sequence homology, probably because of the greater stability of longer duplexes. Mechanistically, this effect of cohesive ends is probably due to the fact that the Mre11 exonuclease is at least 3- to 5-fold less active on a nick compared with an open DNA end (data not shown). Once two DNA ends are aligned, even with a small number of base-pairing interactions, they become essentially one DNA molecule containing two single-strand breaks.
Stimulation by Noncomplementary Ends.
In contrast to the inhibitory effect of cohesive overhangs, other kinds of ends actually stimulate Mre11 exonuclease activity. Mismatched overhangs, blunt ends, and overhangs of the opposite polarity increase exonuclease activity by as much as 10-fold. This effect is particularly striking considering that these molecules are also potential substrates for Mre11 and would therefore be expected under normal circumstances to reduce activity on the labeled substrate.
When a sequence homology is buried within the substrate DNA molecule, as in Fig. 2A, the addition of an unlabeled molecule containing the same sequence motif at its open end increases the amount of labeled substrate degraded, precisely to the edge of the homology. In the context of the joining reaction, this pause in the exonucleolytic degradation would allow a DNA ligase more time to bind to the short region of base-pairing at the DNA ends and to seal one (or both) strands. The stimulatory effect of exogenous DNA molecules combined with pausing at sites of sequence identity suggests that, in a joining reaction, Mre11 increases the rate of degradation on mismatched ends until a homology is uncovered (see also model in Fig. 5).
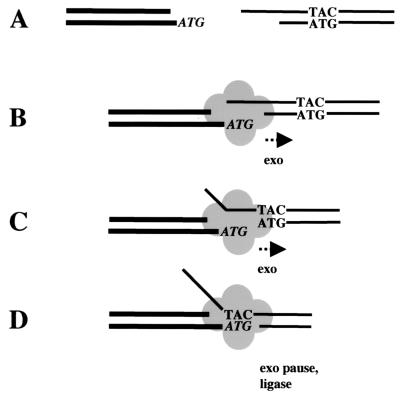
Figure 5.
A hypothetical model of microhomology searching by Mre11. (A) Broken DNA ends are generated that contain a sequence identity (“ATG”) in the single-stranded overhang of one end (thick lines) that is also present in the double-stranded region of the other end (thin lines). (B) A putative multimer of Mre11 binds both DNA ends simultaneously. The 5′ overhangs do not match, thus Mre11 exonuclease activity is stimulated on one end (here shown as the end on the right). (C) After excision of a few nucleotides, the overhangs still cannot base pair with one another, so Mre11 exonuclease activity proceeds further. (D) Degradation of the ATG present on the bottom strand of the right-hand DNA molecule uncovers the TAC on the top strand, thus creating cohesive ends and pausing exonuclease activity, which increases the probability of a DNA ligase sealing the nick on the bottom strand. Other enzymes would then be necessary to repair the top strand.
A DNA End-Bridging Model.
The observation that the end structure on a separate DNA molecule can influence Mre11 activity suggests that the two ends may be juxtaposed in contact with one another by the enzyme. Support for this hypothesis also comes from the observation that both wild-type Mre11 and an Mre11 mutant protein, catalytically inactive but able to bind DNA, have stimulatory effects on joining of cohesive ends by a DNA ligase. It is likely that Mre11 forms higher-order complexes in the presence of DNA that hold two ends in proximity with one another. We have shown previously that Mre11 by itself forms complexes that are larger than monomeric size (28), and we are currently investigating the stoichiometry of complexes in the presence of DNA. The finding that Mre11S alleles mutated in the 5′ end of the gene could complement alleles mutated at the 3′ end in S. cerevisiae also supports this hypothesis of a higher order Mre11 structure (34).
The presence of the H217Y mutant protein in minimal joining reactions also increased junction formation by the wild-type protein and DNA ligase on mismatched ends by nearly 10-fold. We suggest that both the wild-type Mre11 and the mutant proteins bind DNA ends, and that the interaction between these DNA-protein complexes increases the overall efficiency of the homology search by the wild-type protein as well as the efficiency of the ligation event. Support for this explanation comes from our finding that addition of excess H217Y protein to a reaction containing wild-type Mre11 stimulates degradation to a similar extent as addition of exogenous DNA. Furthermore, addition of H217Y protein to a reaction containing wild-type Mre11 and the maximum stimulatory amount of exogenous DNA does not stimulate the reaction further (data not shown), consistent with the idea that both molecules are contributing to the same effect.
The fact that the H217Y mutant lacks nuclease activity and yet is able to stimulate end processing by the wild-type protein suggests that the active, higher-order Mre11 structure can function with only one functional active site. This idea is consistent with our previous finding that junctions formed in vitro with a ligase and Mre11 only contain deletions on one of the DNA ends (28). Sequencing of junctions formed in vivo in mammalian cells has shown that, in some cases, deletions are made on one side of the double-strand break or the other, but not both (35). However, microhomology-mediated junctions with sequence deletions from both ends have also been found in mammalian cells (20), and we are currently investigating whether Mre11 complexes can perform a bidirectional homology search in vitro.
A speculative model for Mre11 DNA-binding and homology searching is shown in Fig. 5. In this model, two DNA ends are held in alignment by a multimer of Mre11 [or M/R/X(N)]. The presence of mismatched ends in this complex stimulates Mre11 exonuclease activity on one strand of one DNA until a sequence identity is revealed, allowing the single-stranded overhangs on the ends to base pair with one another. This cohesive interaction then inhibits further exonuclease activity, thus making the enzyme pause and allowing a DNA ligase to seal the bottom strand. Further processing by other factors would then be necessary to completely repair both strands.
The M/R/X(N) complex is, of course, not the only DNA end-binding factor that functions in double-strand break repair. The Ku protein, for instance, which also functions in NHEJ, binds DNA ends very tightly and can inhibit Mre11 nuclease activity (29). The results shown here provide a more detailed understanding of Mre11 function on repair intermediates, but more of the known factors will have to be studied in combination with each other and on a variety of substrates to address the complexities of double-strand break repair.
Acknowledgments
We thank our colleagues in the Laboratory of Molecular Biology for helpful comments. T.T.P. was supported by a Helen Hay Whitney Postdoctoral Fellowship.
Abbreviations
- NHEJ
nonhomologous end joining
- M/R/X(N)
Mre11/Rad50/Xrs2(Nbs1)
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.110144297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.110144297
References
- 1.Paques F, Haber J E. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Critchlow S E, Jackson S P. Trends Biochem Sci. 1998;23:394–398. doi: 10.1016/s0968-0004(98)01284-5. [DOI] [PubMed] [Google Scholar]
- 3.Tsukamoto Y, Ikeda H. Genes Cells. 1998;3:135–144. doi: 10.1046/j.1365-2443.1998.00180.x. [DOI] [PubMed] [Google Scholar]
- 4.Ivanov E L, Korolev V G, Fabre F. Genetics. 1992;132:651–664. doi: 10.1093/genetics/132.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ajimura M, Leem S H, Ogawa H. Genetics. 1993;133:51–66. doi: 10.1093/genetics/133.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Game J C. Semin Cancer Biol. 1993;4:73–83. [PubMed] [Google Scholar]
- 7.Johzuka K, Ogawa H. Genetics. 1995;139:1521–1532. doi: 10.1093/genetics/139.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kironmai K M, Muniyappa K. Genes Cells. 1997;2:443–455. doi: 10.1046/j.1365-2443.1997.1330331.x. [DOI] [PubMed] [Google Scholar]
- 9.Boulton S J, Jackson S P. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bressan D A, Baxter B K, Petrini J H. Mol Cell Biol. 1999;19:7681–7687. doi: 10.1128/mcb.19.11.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le S, Moore J K, Haber J E, Greider C W. Genetics. 1999;152:143–152. doi: 10.1093/genetics/152.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carney J P, Maser R S, Olivares H, Davis E M, Le Beau M, Yates J R, III, Hays L, Morgan W F, Petrini J H. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 13.Varon R, Vissinga C, Platzer M, Cerosaletti K M, Chrzanowska K H, Saar K, Beckmann G, Seemanova E, Cooper P R, et al. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- 14.Shiloh Y. Annu Rev Genet. 1997;31:635–662. doi: 10.1146/annurev.genet.31.1.635. [DOI] [PubMed] [Google Scholar]
- 15.Stewart G S, Maser R S, Stankovic T, Bressan D A, Kaplan M I, Jaspers N G, Raams A, Byrd P J, Petrini J H, Taylor A M. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- 16.Tsukamoto Y, Kato J, Ikeda H. Genetics. 1996;142:383–391. doi: 10.1093/genetics/142.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schiestl R H, Zhu J, Petes T D. Mol Cell Biol. 1994;14:4493–4500. doi: 10.1128/mcb.14.7.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kramer K M, Brock J A, Bloom K, Moore J K, Haber J E. Mol Cell Biol. 1994;14:1293–1301. doi: 10.1128/mcb.14.2.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mezard C, Nicolas A. Mol Cell Biol. 1994;14:1278–1292. doi: 10.1128/mcb.14.2.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roth D B, Wilson J H. Mol Cell Biol. 1986;6:4295–4304. doi: 10.1128/mcb.6.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rouet P, Smih F, Jasin M. Mol Cell Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feeney A J. J Immunol. 1992;149:222–229. [PubMed] [Google Scholar]
- 23.Gu H, Forster I, Rajewsky K. EMBO J. 1990;9:2133–2140. doi: 10.1002/j.1460-2075.1990.tb07382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerstein R M, Lieber M R. Nature (London) 1993;363:625–627. doi: 10.1038/363625a0. [DOI] [PubMed] [Google Scholar]
- 25.Lewis L K, Westmoreland J W, Resnick M A. Genetics. 1999;152:1513–1529. doi: 10.1093/genetics/152.4.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C, Kolodner R D. Nat Genet. 1999;23:81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- 27.Trujillo K M, Yuan S S, Lee E Y, Sung P. J Biol Chem. 1998;273:21447–21450. doi: 10.1074/jbc.273.34.21447. [DOI] [PubMed] [Google Scholar]
- 28.Paull T T, Gellert M. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 29.Paull T T, Gellert M. Genes Dev. 1999;13:1276–1288. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McBlane J F, van Gent D C, Ramsden D A, Romeo C, Cuomo C A, Gellert M, Oettinger M A. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 31.Tsubouchi H, Ogawa H. Mol Cell Biol. 1998;18:260–268. doi: 10.1128/mcb.18.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alani E, Padmore R, Kleckner N. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- 33.Usui T, Ohta T, Oshiumi H, Tomizawa J, Ogawa H, Ogawa T. Cell. 1998;95:705–716. doi: 10.1016/s0092-8674(00)81640-2. [DOI] [PubMed] [Google Scholar]
- 34.Nairz K, Klein F. Genes Dev. 1997;11:2272–2290. doi: 10.1101/gad.11.17.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roth D B, Wilson J H. Proc Natl Acad Sci USA. 1985;82:3355–3359. doi: 10.1073/pnas.82.10.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]