Abstract
The microscopic phenotype of cervical intraepithelial neoplasia (CIN) reflects a fine balance between factors that promote or reduce CIN development. A shortcoming of the current grading system is its reliance on static morphology and microscopic haematoxylin–eosin features of the epithelium alone. In reality, CIN is a dynamic process, and the epithelium may exhibit differing results over time. Functional biomarkers p16, Ki‐67, p53, retinoblastoma protein cytokeratin (CK)14 and CK13, help in the assessment of an individual CIN's lesion's potential for progression and regression. The aggregate information provided by these biomarkers exceeds the value of the classic grading system. Consequently, many more CINs that will either regress or progress can be accurately identified. These findings agree with known molecular interactions between HPV and the host. For accurate interpretation of a CIN, it is essential that these biomarkers be determined quantitatively and separately in the superficial, middle and deep layers of the epithelium. Such geography‐specific epithelial evaluations of quantitative biomarkers emphasise the dynamic nature of a particular CIN lesion, thereby changing the art of static morphology grading into dynamic interpretation of the diseased tissue, with a strong prognostic effect.
The microscopic phenotype of cervical intraepithelial neoplasia (CIN, also referred to as squamous intraepithelial lesion (SIL)) reflects a fine balance between factors that promote or accelerate the development of progressively more advanced disease and factors that reduce or decelerate its progression. Critical ingredients are genotypes of the human papillomavirus (HPV) and the patient's immune resistance. The severity of CIN or SIL is expressed by its microscopic grade, which is a standard part of the surgical pathology report, and greatly influences treatment of the patient (fig 1). This is understandable in view of the regression, persistence and progression figures of different CIN grades1 (table 1).
Figure 1 Treatment of cervical intraepithelial neoplasia (CIN) lesions depends heavily on CIN grade. HSIL, high‐grade squamous intraepithelial lesion; LSIL, low‐grade squamous intraepithelial lesion.
Table 1 Regression, persistence and progression rates of different CIN grades.
| Regression (%) | Persistence (%) | Progression to CIS (%) | Progression to invasive carcinoma (%) | |
|---|---|---|---|---|
| CIN1 | 57 | 32 | 11 | 1 |
| CIN2 | 44 | 35 | 22 | 5 |
| CIN3 | 32 | <56 | – | 12 |
Data from Osteer.1
CIN, cervical intraepithelial neoplasia; CIS, carcinoma in situ.
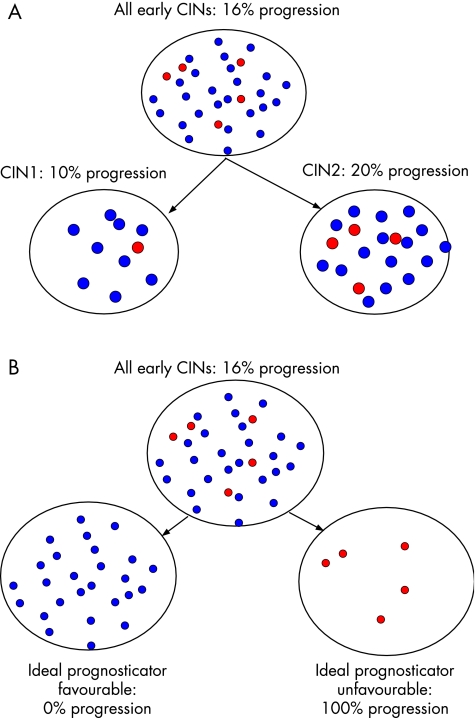
A shortcoming of the grading by microscopic pathology is that it assesses exclusively epithelial features and usually only those visible with the standard haematoxylin–eosin staining, thereby not taking into account other possibly valuable information. Another serious disadvantage is that the three distinct grades used in CIN (or two in SIL) can easily give a faulty static impression of a solidified sculpture, as if CIN or SIL were a static event, whereas in reality a CIN lesion is a dynamic process (a balance) that can progress and persist but also regress. Compounding the above are the well‐known issues of intraobserver and interobserver reproducibility, which, for grading of CIN, is far from perfect.2,3,4,5,6,7,8,9 It is also difficult to distinguish CIN reliably from non‐neoplastic lesions, resulting in either overtreatment or undertreatment.10,11 These points emphasise the need for adjuvant methods to interpret the actual morphological impression of a CIN lesion in dynamic terms rather than in static morphological grades. Such adjunctive methods are also important for better distinction of CIN from non‐neoplastic lesions and to predict accurately the risk for progression of low‐grade and regression of high‐grade CIN lesions. Even small improvements in prognostic accuracy will enormously reduce the number of patients erroneously or unnecessarily treated, as shown for CIN2 (table 2). Figure 2 shows the prognostic accuracy of CIN grade as opposed to the ideal prognosticator of a CIN lesion.
Table 2 CIN in the EU and annual estimates of the number of women with overtreatment.
| EU | |
|---|---|
| Number of inhabitants (2005) | 375 million |
| Number of cases of CIN1, CIN2 and CIN3 in the EU | 330 000 |
| Number of CIN2 cases in the EU | 82 500 |
| Annual number of overtreated patients with CIN2 in the EU by CIN grade | 66 000 |
| Annual number of overtreated patients with CIN2 in the EU by Q‐Ki‐67 | 57 750 |
| Annual number of overtreated patients with CIN2 in the EU by Q‐Ki‐67 and pRb | 47 025 |
| Improvement because of Q‐Ki‐67 per year | 8 250 |
| Improvement because of Q‐Ki‐67 and pRb per year | 18 975 |
CIN, cervical intraepithelial neoplasia; EU, European Union; pRb, retinoblastoma protein.
Data modified from Baak et al. Cell Oncol 2005;27:277–80.
Figure 2 The prognostic accuracy of cervical intraepithelial neoplasia (CIN) grade (A) and the ideal prognosticator (B).
Inasmuch as CIN causes progressive dysfunction in the proliferation and differentiation of cervical epithelial cells, many studies have focused on evaluating the merits of features related to proliferation and differentiation.12,13,14,15,16,17,18,19,20 Without doubt, p16 and Ki‐67 are the most widely available, robust, stable and strong predictive biomarkers currently available for handling CIN lesions. (The antibody MIB‐1 is often used in paraffin wax sections to assess Ki‐67 expression. In this article, for consistency, we use only the term Ki‐67, even if it relates to MIB‐1). Others, such as the retinoblastoma protein (pRb), p53, CK13 and CK14, add substantial insight.21,22,23,24,25 This paper summarises current knowledge and gives practical guidelines for using these most important biomarkers in daily surgical pathology practice.
CIN and HPV
Persistent infection with oncogenic HPV (high‐risk HPV or hrHPV) is necessary for cervical precursors to evolve into invasive carcinoma.26,27,28,29,30,31,32,33,34,35,36,37 Many believe that CIN does not exist without HPV and that earlier publications on HPV‐negative CINs are flawed by insensitive methods or materials. The viral load may be a predictor of the development of a CIN lesion, although this has been doubted.38,39,40,41,42 Large, worldwide vaccination programmes are in progress with promising results, clearly indicating the important role of the host's local immune response to HPV infection.43 HPV is clearly highly infectious, as in infected women it can be found not only in the cervix but also in the vagina, vulva, perianal region, urethra and even tampons.44 HPV is often also found in women without overt signs of infection.35 HPV enters only the parabasal cells of the cervical epithelium, not the superficial cells (in which the nuclear material for all intents and purposes is already largely dead), so infection requires small transepithelial microtraumas reaching to the basal cells of the cervical epithelium.
HPV infection often results in nothing more than a slightly increased proliferation of the epithelium. In fact, most (about 80%) of these women show no remarkable microscopic abnormalities of the epithelium or no more than a minimal glycogen loss from the epithelium, slightly enlarged nuclei, or sometimes parakeratosis or hyperkeratosis,45 but about 20% of those infected with hrHPV develop a real CIN lesion (fig 3).46,47,48,49,50,51,52,53,54,55 Clearance of the virus is followed by regression of cervical lesions. An altered transcriptional regulation of the viral oncogenes E6 and E7, especially in the oncogenic types, results in a topographic shift of E6 or E7 expression from the differentiated layers to the proliferating parabasal cell layers. When overexpressed in proliferating cells, E6 and E7 interfere with the cell cycle control regulated by p53 and pRb, respectively.56 These morphologically abnormal cells migrate towards the surface (just as normal cells do), but without normal maturation. The degree of abnormal maturation is expressed by the changes referred to as the poorly glycogenated epithelium or even CIN.45 The dysplastic cells are estimated to arrive at the surface in one to several weeks and then desquamate, so a full‐blown CIN3 may develop in up to a few weeks (fig 3B), although many CINs regress spontaneously, either slowly or after a prolonged period (fig 3C,D).
Figure 3 Morphological events in the cervical epithelium after being infected with high‐risk human papillomavirus (HPV). (A) 80% of the HPV infections occur without detectable morphological changes. (B) A full‐blown cervical intraepithelial neoplasia (CIN)3 may develop in 3 weeks. (C,D) Many CINs regress spontaneously, either slowly or after a prolonged period. nrm, normal ticology; react, reactive changes, non‐reoplastic.
Morphologically, a wide range of non‐neoplastic changes is associated with HPV infection. Features that characterise neoplastic change include decreased upward maturation and proliferation (mitoses above the level of the parabasal cells), variation in size, shape and polarity, and coarse chromatin of nuclei at all levels of the epithelium. In all cases of intraepithelial neoplasia, abnormal changes are visible throughout all layers of the epithelium, including the surface. However, when the bulk of the most abnormal changes are confined to the lower one third of the epithelium, the lesion is designated as CIN1. Similarly, CIN2 occurs when the most abnormal changes extend into the middle third. In CIN3, the changes are uniform and throughout the epithelium.
HPV and molecular mechanisms leading to CIN
Over the past years, several reviews have described the molecular biological mechanisms of HPV in detail.56,57,58 For the working surgical pathologists, we will give a summary here, as far as the mechanisms are relevant.
HPV is a double‐stranded circular (episomal) DNA virus whose genome can be divided into three regions: the upstream regulatory region, the early region and the late region. The upstream regulatory region is important in regulating viral replication and transcription of downstream sequences in the early region. The early region encodes predominantly viral proteins (E1–E8) that are important in viral replication. The late region encodes viral structural proteins (L1, L2) that are important in the formation of the capsid (a structural protein envelope that encapsulates each virion). Much research has been conducted over the past years on the functions of E1–E8, L1 and L2. E6 and E7 are the principal transforming proteins of HPV.59 The expression of these proteins in hrHPVs such as subtypes HPV16 and HPV18 (but not of those having low oncogenic risk such as HPV6 and HPV11) causes cells to become completely transformed,60 principally by inactivating the host's p53 and pRb, respectively. After getting access to the basal cell compartment of the cervical epithelium through microwounds, HPV infects these epithelial stem cells, and initially is a phenotypically latent passenger in the form of an episome (a DNA ring) in the cytoplasm of these basal layer cells. A few of the infected stem cells give rise to daughter cells capable of replicating the viral genome and synthesising capsid proteins in the terminally differentiated cell layers above the basal cell layer. The viral replicating cells display typical cytopathic changes, including, most prominently, koilocytosis or koilocytotic atypia—that is, the squamous cells with vacuolated appearance—as well as some minor changes in the nuclear morphology. Finally, mature viral particles are released along with the squamous debris that is exfoliated at the surface of the epithelium.
The transition from normal to dysplasia or invasive carcinoma is triggered by uncontrolled expression of E6 or E7 in proliferating basal and parabasal epithelial cells.61 This phenomenon distinguishes the process of cell transformation from productive viral infection and may be due to integration of the viral DNA in the host cell genome, which is observed in most invasive cancers and in a subset of high‐grade lesions.62 The integration of HPV DNA into the host's chromosomes is a critical event in HPV‐related oncogenesis, although little information is currently available on the effect of integration on the host genome. Conversely, viral integration may be the result of a genetically unstable environment in parabasal cells caused by p53 inactivation, the guardian of the human genome.56 Molecular studies have shown that HPV integration results in disruption of the open‐reading frames for E1 or E2 of the HPV genome, leading to increased expression of E6 and E7 in particular, favouring a growth advantage and neoplastic transformation.63,64,65 Both E6 and E7 proteins can bind to multiple cellular targets.66 Figure 4 shows a simplified scheme of the interactions that are thought to be most relevant for their transforming functions. A detailed discussion is beyond the scope of this article; useful information can be found at www.hpvtoday.com. E6 binds to the tumour suppressor gene product p53, which prevents cells from undergoing p53‐mediated apoptosis and p53‐independent activation of telomerase. E7 binds to the retinoblastoma tumour suppressor gene product pRb, resulting in hyperproliferation and inducing abnormal centrosome duplication independent of inactivation of pRb and its family members.67,68
Figure 4 Interaction of human papillomavirus (HPV) proteins E6 and E7 in the cell cycle. (A) The two conditions of a normal cell; (B) HPV stimulates proliferation through release of E6 and E7, interacting with p53 and retinoblastoma protein (pRb). Cdk, cyclin‐dependent kinase.
Viral and host factors in HPV persistence and progression
Viral and host factors play interdependent parts that foster the regression persistence and progression of HPV infections. Viral factors include viral variants, viral load and viral integration. Host factors include the host immune response and susceptibility genes.69
Viral factors
Several studies have now documented an association between HPV16 variants and the development of cervical cancer, with non‐European variants being associated with excess risk for cervical cancer.70 The limited data available for HPV types other than HPV16 suggest that non‐European variants of HPV18 and HPV58 are also associated with increased risk for cervical cancer.71,72 Cross‐sectional epidemiological studies showed an association between increasing HPV viral load and the risk for cervical cancer.37,38,39,40,41,42 However, the longitudinal data evaluating the pattern of viral load over time and the subsequent risk for progression of HPV infection to cervical neoplasms (CIN2 and CIN3) and cancer are insufficient to support this association.73,74 Although HPV is usually in the episomal form in cervical lesions, viral integration has generally been reported to be associated with oncogenesis. Unfortunately, the currently widely available tests determining HPV do not distinguish between episomal and integrated HPV. The frequency of HPV integration increases with the degree of disease severity, thus potentially correlating with progression to cervical cancer.62 Other viral events are epigenetic events in the HPV genome—that is, those events that alter gene expression (eg, phenotype) without a change in the DNA sequence (eg, genotype). Examples are hypermethylation or hypomethylation of viral oncogenes and the potential implications for suppression or activation, respectively, of viral oncogenic expression.
Host factors
A positive association exists between the detection of HPV antibodies (humoral immunity) and the risk for cervical neoplasia.75,76,77 Although these antibodies may effectively prevent infection, they seem to be unimportant effectors in causing regression in established HPV infections and related cervical lesions.78 In contrast to antibodies, T cell responses (cellular immunity) to HPV are likely to be an important effector mechanism for clearing established infections.79,80 Thus, adequate T cell responses generated after infection may help to protect against the progression of infection and against early lesions. Natural polymorphisms or genetic variations (genetic susceptibility) between people in immune‐related genes may also help to explain differences in the regulation of immune function. For example, human leucocyte antigen (HLA) DRB1*1301 seems to have a protective role in the pathogenesis of cervical cancer.70 However, this individual allele association is not likely to be explained by underlying haplotypes.81 Studies focusing on inherited susceptibility showed that having a sister or a mother with cervical cancer increases a woman's risk for cervical cancer twofold and that heredity may explain some of the variation in risk for cervical cancer.82,83 But the question is whether it is the presence of inherited susceptibility or the sharing of promiscuous sexual behaviour among family members that may cause cervical cancer. Shared familial environment was not found to be a major effect among mother–daughter relationships.83
Ki‐67 cell clusters and p16 distinguish CIN from reactive lesions
The development and behaviour of CIN is correlated with proliferation, an observation known for many decades. Ki‐67 is one of the most widespread biomarkers correlated with proliferation; one of the simplest applications therefore is to use Ki‐67 expression as a marker for CIN. In sections embedded in paraffin wax, the MIB‐1 equivalent of Ki‐67 is widely used, as its immunohistochemical staining pattern is stable, robust and gives rich contrast. Evaluation of Ki‐67‐positive cell clusters in the epithelium is a strong diagnostic adjunct in distinguishing CIN from normal or benign reactive cervical squamoepithelial lesions.18,25 However, to prevent overdiagnosis, Ki‐67‐positive, tangentially cut parabasal cells, inflammatory cells and immature metaplasia must be carefully excluded.
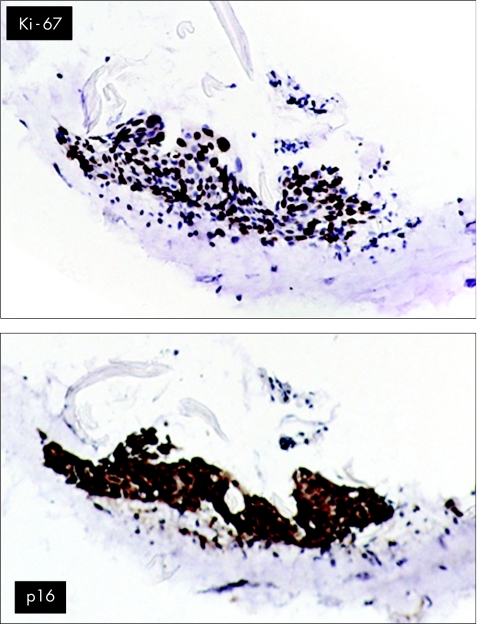
The cyclin‐dependent kinase inhibitor‐2A p16 is also useful in distinguishing CIN from reactive lesions.84 As p16 expression is regulated by a pRb‐dependent negative feedback loop, continuous inactivation of pRb by hrHPV E7 results in increased p16 levels. Hence, increased p16 levels may reflect HPV‐induced dysplasia with deregulated E7 expression.85 Information about the presence of HPV has no additional diagnostic value, as many non‐CIN epithelia can be HPV positive. Marked overexpression of p16—that is, diffuse and strong immunostaining—is seen in all cervical cancers and pre‐neoplastic lesions with infection by high‐risk and intermediate‐risk HPVs of subtypes HPV16, HPV18, HPV31, HPV33, HPV52 and HPV58 and weak or focal staining in lesions infected by HPV6 or HPV11.21 Others have confirmed the strong correlation between the presence of HPV DNA and p16 staining.23 At low magnification, p16 staining facilitates finding a dysplastic area, especially if the epithelium is heavily infested with leucocytes, as occurs often in CIN lesions.86 In addition, overexpression of p16 can be used to identify individual dyskaryotic cells in ThinPrep smears.87 Caution must be exercised, as occasionally inflammatory cells themselves may exhibit p16 reactivity in clearly non‐dysplastic lesions.24 However, during routine prospective evaluation of p16 over 12 months in a large gynaecopathology practice, no false‐positive p16 lesions have occurred (unpublished results), showing that false‐positive p16 is a very rare phenomenon and probably has a negligible role. The combination of Ki‐67 and p16 is useful to ascertain whether a suspicious yet uncertain cell group is indeed CIN and not atrophic epithelium (fig 5).
Figure 5 The use of Ki‐67 (top) and p16 (bottom) to ascertain that a suspicious yet uncertain cell group is indeed cervical intraepithelial neoplasia and not atrophic epithelium.
Ki‐67 and CIN grade
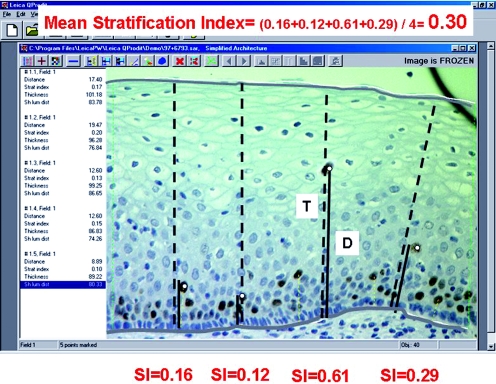
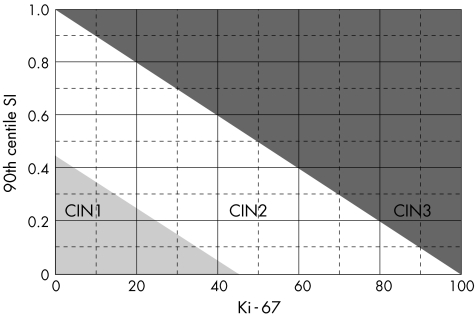
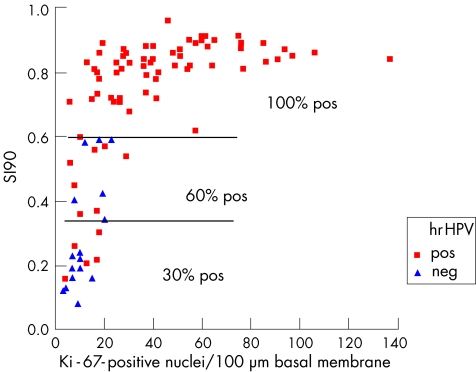
Immunoquantitation of Ki‐67 is an important diagnostic adjunct for the grading of CIN.19,20,88 Figure 6 shows the quantitative image analysis method. The combination of the Stratification Index (SI, which indicates, how high Ki‐67‐positive nuclei are located in the epithelium; the higher the SI, the higher the CIN grade) and the number of Ki‐67‐positive nuclei per 100 µm basal membrane (the more Ki‐67‐positive nuclei, the higher the grade) is the best discriminating set of features that distinguishes the three CIN grades at the same time (fig 7). Some CIN1 cases that were initially “misclassified” with these two quantitative Ki‐67 features showed a higher CIN grade on deeper levels from re‐cut of the paraffin wax blocks, whereas the other CIN1 cases that were correctly classified with Ki‐67 quantitation remained so in the deeper cuts. In a subsequent prospective evaluation of 121 routine CIN cases (test set), six independent pathologists, some of whom were general pathologists, achieved exact agreement in 78% of cases when they compared CIN grades with histology by staining with haematoxylin–eosin and quantitative Ki‐67. When the two reviewers were experts in gynaecological pathology, the agreement jumped to 97%, and the sensitivity, specificity and positive and negative predictive values were very high, further indicating the supportive role of Ki‐67 in general surgical pathology practice. Immunoquantitative parameters for Ki‐67 are also correlated with the presence of hrHPV in CIN lesions (fig 8).20
Figure 6 The quantitative image analysis method. The microscopic image of the epithelium is shown on the monitor of the image analysis system. With the mouse, the operator demarcates a diagnostic epithelium strip, carefully avoiding tangentially cut areas. The demarcation lines are shown as white lines (surface, basal membrane, left, right). The operator then clicks the mouse on all Ki‐67‐positive nuclei within the demarcated strip. After each click, the system automatically draws a perpendicular line from that point to the basal membrane and over the full thickness of the epithelium (these thin dotted lines are barely visible for all nuclei) and calculates several quantitative features such as thickness (T) of the epithelium at that point, distance (D) from the point to the basal membrane, the stratification index (SI = D/T) and others. These quantitative features are shown in the left panel after each click and are stored automatically. Lines D (dotted lines) and T (continuous lines) are emphasised for three nuclei at AA′, BB′ and CC′. The SIs are 0.16, 0.12, 0.61 and 0.29, respectively. The image analysis program automatically calculates many quantitative features per sample. For example, the mean SI of the four measurements used as an example is (0.16+0.12+0.61+0.29)/4 = 0.30. With multivariate analysis, the 90th centile (SI90) of all individual SI measurements in a case is the strongest factor (more relevant than the mean SI) to describe the grade and also to predict progression. The number of Ki‐67‐positive nuclei per 100 µm basal membrane is the next strongest discriminator for grade, whereas the percentage of Ki‐67‐positive nuclei in the middle third of the epithelium (MIDTHIRD) is the only factor that adds to the prognostic value of SI90.
Figure 7 Ki‐67 and grade. Scatter plot of the 90th centile of the stratification index (SI90) against the number of Ki‐67‐positive nuclei per 100 µm basal membrane for cases with cervical intraepithelial neoplasia (CIN)1, CIN2 and CIN3. The variables chosen were the two most discriminating ones in the multivariate stepwise regression analysis.
Figure 8 Ki‐67 correlates with hrHPV presence. Scatter plot of the 90th centile of the stratification index (SI90) against the number of Ki‐67‐positive nuclei per 100 µm basal membrane, for the oncogenic (high‐risk or hrHPV‐positive, pos) and hrHPV‐negative (neg) cases. Note the strong correlation between hrHPV and SI90.
Biomarkers and biological aggression of early CIN lesions
Koilocytosis
Pathologists often regard koilocytosis as an additional sign of CIN aggression (ie, is more likely to progress). In reality, the finding or absence of koilocytosis is of no value, and in fact may be misleading. Koilocytosis is the microscopic image of the productive phase of the HPV life cycle. In this state, the viral particles needed to form the koilocyte replicate and accumulate in the superficial differentiating layers of the epithelium, not in the basal proliferating part, which is the critical region for the developing dysplasia.59,61 The presence of koilocytosis is irrespective of HPV type. In a series where patients were evaluated only if the biopsy result was normal, atypical or CIN1, 35% of the patients with koilocytosis did not have HPV infection, 25% of patients with prominent koilocytes had a low‐risk HPV type and 40% had an hrHPV type.89 For a cell to show the presence of a well‐developed koilocyte, the cell should contain a considerably high viral load; thus, the absence of koilocytosis implies nothing about the presence of oncogenic organisms. In corroboration, progression‐or‐not analysis of original early CIN lesions showed that patients with koilocytosis had a considerably lower likelihood of progression than those without koilocytosis. Thus, CIN lesions with koilocytosis are productive viral infections, but are generally overdiagnosed when classified as high‐grade CIN. Instead, most should be grouped with low‐grade CIN1 at best. A further complicating evaluation of koilocytosis is that the presence of the features among observers is poorly reproducible.16
Prognostic value of Ki‐67 and other biomarkers in early CIN lesions
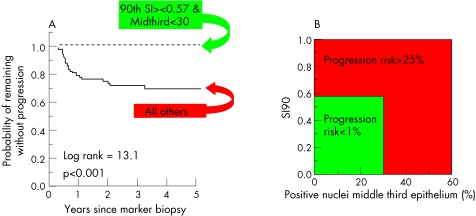
Ki‐67 features in small histological punch (marker) biopsy specimens predict progression to CIN3 in early CIN lesions more strongly than subjective CIN grade (both for routine and experts' review CIN grades).12,17 The prognostically strongest Ki‐67 features were the Ki‐67 SI (fig 6) and the percentage of the Ki‐67‐positive nuclei located in the middle third layer of the epithelium (MIDTHIRD; fig 9). The strongest prognostic thresholds of these features (SI = 0.57 and MIDTHIRD = 30%) were detected in the first study we conducted by multivariate regression analysis (Cox model). These features and thresholds are highly reproducible. High reproducibility is important but perhaps not enough to be used in another laboratory, as interlaboratory differences in tissue processing and staining of the sections undoubtedly occur and may perhaps influence the Ki‐67 features. The prognostic Ki‐67 test was therefore once more validated in two new patient sets from another country (Norway), first on historical archive material and then prospectively. Prospective validation is usually the most difficult phase when implementing a new laboratory test, as minor variations will occur despite the measurement and interpretation protocol being well defined. Although differences in the processing and staining procedures undoubtedly existed between the Dutch and Norwegian laboratories, the findings in the two Norwegian validation studies confirmed that the quantitative Ki‐67 test in CIN lesions was strongly predictive for CIN3 in the follow‐up (fig 9), whereas the routine CIN grade was not.12,14
Figure 9 (A) A quantitative progression model of early cervical intraepithelial neoplasia (CIN) lesions. (B) Survival curve of the Ki‐67 low‐risk and high‐risk groups. SI90, 90th centile of the Stratification Index.
The critical value of Ki‐67 SI = 0.57 needs some further explanation. In the normal epithelium, proliferation occurs in the parabasal cells. In high‐grade CIN, proliferation is abnormally high and is also found in the middle and superficial layers. As Ki‐67 SI = 0.57 and MIDTHIRD = 30%, the critical prognostic values, are in agreement with the idea of “upward proliferation” in high‐grade squamous intraepithelial lesion (HSIL), higher localisation of Ki‐67‐positive cells leads to an increase in Ki‐67 SI. Importantly, independent validation studies confirmed that these threshold values were essential to predict progression, making them strong and reliable prognostic factors.
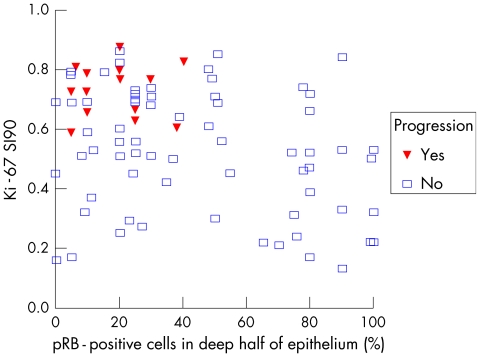
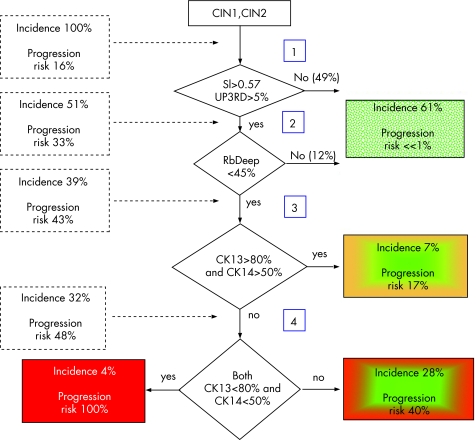
The prognostic value of the various cell cycle regulatory proteins and markers of squamous cell differentiation have been compared in various studies. Some found telomerase to be important in cytological material, but others had negative results90,91 and in one study on punch biopsy, telomerase was not a prognostic marker in early CIN.15 This is understandable as telomerase activity reflects a rather late step in the sequence from CIN3 to squamous cell carcinoma, explaining why it was not prognostic for early CIN progression.84 The results for pRb, p53, cyclins A, E and D, p16, p21, p27, telomerase, involucrin, CK13 and CK14 are not always in agreement with each other. However, this may be partly due to technical shortcomings, as an important biomarker function should also assess the dynamics of the cervical epithelium. Epithelial cells are born in the parabasal layer and then mature and migrate to the surface (where they are desquamated); therefore, an average assessment of biomarkers throughout the entire epithelial thickness may blur important dynamic information. All biomarker features were thus separately analysed in the basal, deeper and upper half of the epithelium with quantitative image analysis techniques to get the best reproducibility. Multivariate analysis showed that the combination of high Ki‐67 SI and reduced expression of pRb in the deeper half of the squamous epithelium predicted progression of CIN lesions (fig 10). Moreover, addition of reduced CK13 and CK14 expression identified a subgroup that had an even greater risk of progression, but this additional prognostic value of CK14 and CK13 was only in the prognostically high‐risk subgroup with high Ki‐67 and low pRb. Other studies found that loss of involucrin and CK13 expression occurred only in the high‐grade lesions and was therefore related to lesion grade. Loss of CK14 expression also occurred considerably more often in high‐grade than in low‐grade lesions.92 Quantitation of combined Ki‐67, pRb, CK13 and CK14 in early CIN lesions gives accurate information about their progression risk.15 These results are summarised in the prognostic decision scheme shown in fig 11.
Figure 10 Scatter plot of the 90th centile of the Stratification Index (SI90) of the Ki‐67‐positive nuclei (Ki‐67 SI90) and the percentage of retinoblastoma protein (pRb)‐positive nuclei in the deep half layer of the epithelium (RbDeep). None of the cases with Ki‐67 SI90<0.57 progresses. In these cases, pRb has no additional prognostic value. However, in the subgroup with Ki‐67 SI90>0.57 (which have a high risk of progression), many pRb‐positive nuclei (>40%) in the deep epithelial layer indicate no progression, in to contrast those with low pRb values (which have a very high progression risk).
Figure 11 Prognostic biomarker‐based decision scheme of an early cervical intraepithelial neoplasia (CIN) lesion. CK, cytokeratin; SI, Stratification Index; UP3RD, upper third.
On the basis of the above findings, a model for the development of an early CIN lesion has been developed (fig 12). Central to this hypothesis is that hrHPV E7, when expressed during HPV infection, impairs pRb. pRb normally acts to reduce growth. When impaired by E7, the growth‐reducing effect of pRb diminishes as shown by increased and upward proliferation (as a result, the Ki‐67 SI increases and finally exceeds a critical level of 0.57). When hrHPV is cleared by the host, the CIN lesion heals. The first sign (before disappearance of morphological haematonylin–eosin CIN features) is increased pRb levels in the deep layers of the epithelium. Shortly after that, proliferation reduces, and the subsequent upward spread of proliferating cells is also reduced, as manifested by lowering Ki‐67 reactivity. Naturally, p53 is also impaired (by E6), but the prognostic role of pRb is much stronger and overshadows the importance of p53 in predicting early CIN progression. However, in HSIL lesions, p53 has more value in identifying CIN3 lesions that will regress.
Figure 12 Human papillomavirus (HPV) and the development and course of an early cervical intraepithelial neoplasia (CIN) lesion. CK, cytokeratin; Cyc, cyclin; pRb, retinoblastoma protein.
Prediction of behaviour of CIN2 and CIN3 (HSIL) with biomarker patterns
Biomarker patterns are also prognostic in HSIL, which has important implications. When left untreated, many patients continue to show persistent disease (as shown by follow‐up biopsies in cases of HSIL or CIN2 and CIN3). Consequently, many doctors now ablate all HSILs; nonetheless, without treatment 15–45% will naturally regress (ie, no HSIL detected on follow‐up).1 The average age of patients with HSIL is around 29 years, whereas patients with (micro)invasive cancer are on average 9–10 years older. Complicating the decision of whether to treat HSIL is the reality that excision, with either cold‐knife cone or diathermic loop method, is a considerable medical procedure, with potentially serious complications, most commonly cervical insufficiency. As many patients treated for HSIL will at some later time become pregnant, cervical insufficiency becomes a major concern and commonly leads to immature birth,93 demanding preventive cervical cerclage under general anaesthesia at 16–20 weeks' gestation. Obviously, today's clinical goal is to prevent unnecessary treatment by cone or loop electrosurgical excision procedure. The remedy, naturally, is to identify those HSILs that would regress spontaneously.
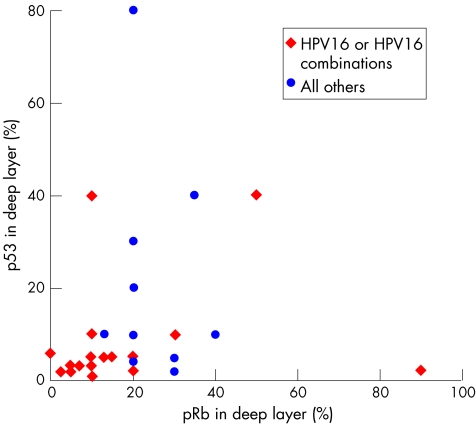
To identify factors related to histologically proven persistence or regression, small histological colposcopically directed (punch) biopsy specimens were analysed for HPV genotypes and different immunoquantitative proliferation, cell cycle regulation and differentiation markers.94 Special attention was paid to p53 and pRb in biopsy specimens as potential markers for hrHPV E6 and E7 function.95 All cases had a biopsy interval of at least 100 days between the marker and first follow‐up biopsy. This interval is important to mitigate the effects of the initial punch biopsy procedure, which causes considerable damage to the cervix and, consequently, a local inflammatory and repair response. Healing is generally complete after 3 months. Within this interval, there is a serious risk of overdiagnosing dysplastic remnant lesions because of the superimposed reactive changes in the epithelium. To minimise this risk, we and others excluded biopsy specimens taken shortly after the initial biopsy, and included only those with an interval of at least 100 days between the marker and follow‐up biopsy.94,96 On the basis of the microscopic findings in the diagnoses by follow‐up cone or loop electrosurgical excision procedure, each initial marker CIN lesion was classified as regressive or persistent.97 All lesions had hrHPV infection and were diffusely positive for p16: 63% were positive for HPV16 or HPV16 mixed with other hrHPV genotypes and 37% had other hrHPV types. The rate of spontaneous histological regression was 43%. Others have found similar rates of spontaneous regression.96,98,99,100 The initial marker (punch) biopsy specimens of the persistent HSILs had considerably lower levels of pRb and p53 in the deep half of the epithelium than those having non‐persistent (regressive) HSILs. The degrees of positivity of p16, Ki‐67, cyclin D1 reactivity, lesion extent in the punch (marker) biopsy specimen and patient age were all unrelated to persistence or regression, as found later in the follow‐up cone biopsies. HPV16‐positive HSIL had a lower regression percentage of pRb and p53 than those with other HPV types (fig 13), and we found a (not significant) trend for HPV16‐containing lesions to persist more often than non‐HPV16 lesions (in agreement with others94). However, the percentages of pRb–positive and p53‐positive nuclei in the deep half of the epithelium of a histological punch cervical biopsy specimen were much stronger predictors of CIN3 regression than HPV genotype (fig 14).
Figure 13 Scatter plot of retinoblastoma protein (pRb) and p53 content of human papillomavirus (HPV)16‐positive (red dots) versus other oncogenic HPV genotypes (blue dots) in the deep epithelium layer. A (not significant) trend that HPV16‐positive high‐grade squamous intraepithelial lesions have lower pRb and p53 values can be seen.
Figure 14 Scatter plot of retinoblastoma protein (Rb) and p53 content of HSIL lesions that regress (blue dots) and persist (red triangles). HSIL, with either Rb>55% or p53>15% have a high likelihood of regression, contrasting HSIC, with Rb<35% and p53<15% in the deep half of the epithelium.
In a prospective observational cohort study on prognostic variables associated with clinical behaviour, 100 women with HPV‐infected HSIL were followed up from about 4 months before the entire lesion was resected.96 Of them, 28% had spontaneous histological regression (CIN1 or less at resection). In the study, lesions in women with only HPV16 infection were less likely to regress than those in women with infection of HPV16 mixed with other HPV types, compared with lesions in women with infection of HPV types other than HPV16 (p = 0.049). In the cohort with only HPV16 infection, the presence or absence of the HLA*A201 allele had no effect on outcome, in contrast with patients having other HPV types in which persistence of the lesions was associated with the presence of the HLA*A201 allele. These results suggest that interactions among HPV type and HLA type as expressed by regression rates support a role for HLA‐restricted HPV‐specific immune responses in determining disease outcome.
It seems that the interaction (balance) between a patient's immune response and HPV factors results in a prognostically important epithelial cell reaction, measured by epithelial levels of pRb and p53. Future studies comparing the different markers will help to clarify the intriguing correlations among these biomarkers, hrHPV subtype, the host immune response and HLA allele interaction.
Why are the deep epithelial layer biomarkers especially prognostic in CIN?
We must realise that the microscopic image of a cervical epithelium is a snapshot (a photograph) of a dynamic process (a film) that normally would consist of many images taken over time (fig 15). The question of why measurement of biomarkers in the deep epithelium is especially predictive of CIN behaviour pulls together the different dynamic biological aspects already discussed earlier in this article.
Figure 15 The microscopic image of a cervical epithelium is a snapshot (a photograph) of a dynamic process (a film) consisting of many images.
The cervical epithelium is a dynamic tissue of socially and orderly moving cells with tightly balanced and controlled proliferation and differentiation. The HPV infection disturbs this process. Through oncoproteins E6 and E7, HPV immobilises or degrades and thereby reduces the amount of pRb and p53 available for normal function. This results in basal cells that do not enter into the normal maturation process, resulting after a short time in high (abnormal) proliferation of the cells of the superficial layers (mitoses or Ki‐67‐positive cells and abnormal chromatin patterns) and altered differentiation markers. Once the virus infection clears, p53 and pRb function in the deep layers restarts. This is so far the earliest sign of cure and normalisation of cell metabolism. Hereafter (perhaps after some weeks), proliferation and hence the number of Ki‐67‐positive cells falls, initially in the deep layer. These normal cells (now with mostly Ki‐67‐negative nuclei) will move up, mature and reach the epithelial surface in several weeks. Some time after the pRb and p53 have normalised, signs of abnormal proliferation and differentiation in the superficial layers (diagnostic for CIN3) will also disappear, and the condition becomes normal. Thus, the assessment of levels of p53 and pRb in the deep layer predicts the future events that will take place in the superficial epithelium, some weeks after the biopsy was carried out. The clearance and cure process expressed by the biomarkers is not always such that the CIN lesion just slowly disappears. The CIN3 lesion may regress to CIN2, hang there for some time, increase back to CIN3, and only then slide down to CIN1, totally regress or even persist. Quantifying biomarkers therefore can change the pathologist's role from reporting static morphology to the much more exciting possibility of dynamically interpreting and forecasting future events that will take place in the tissue. In this context, it is of utmost importance that biomarkers are analysed in certain specific geographical areas of the epithelium. An average measurement taken randomly in the epithelium, not considering the basal, deep and superficial layers, ignores the dynamic nature of the cervical epithelium, obscuring important prognostic information. The familiarity of pathologists with the biology and dynamics of tissues allows them to extract the biologically relevant information correctly. When this important facet of tissue—that is, geographical location—is not considered, important information is lost.
Handling and interpreting biomarker patterns in CIN lesions
On the basis of the discussion earlier, we now recommend the following routine when handling a cervical punch biopsy specimen in the surgical pathology laboratory, especially when an early CIN lesion is contemplated:
Analyse the diagnostic section stained with haematoxylin–eosin, for routine evaluation.
Scan a serial section, stained for p16, to identify diffusely reactive squamous areas. These are nearly always dysplastic (false‐positive p16 reactivity is rare and easily recognised).
Evaluate the next serial section with Ki‐67. Ki‐67‐positive cell clusters further support a diagnosis of CIN.
Carry out quantitative Ki‐67 image analysis for objective grading support and indication of progression risk in case of CIN1 and CIN2. If the Ki‐67 SI90>0.57 or the percentage of Ki‐67‐positive nuclei in the middle third layer of the epithelium exceeds 30%, the likelihood of progression to CIN3 in the follow‐up is high (about 30%).
In the subsequent section, analyse the percentage of pRb‐positive nuclei in the lower half of the epithelium.
Interpret the results as follows. If the combination of Ki‐67 SI90>0.57 occurs together with pRb<40%, progression risk in early CIN to CIN3 is high (about 50%). The progression risk in the remaining patients is almost zero. Moreover, in the subgroup at high risk according to Ki‐67 and pRb, a combination of CK13‐positive cells<80% and CK14‐positive cells<50% identifies patients with an excessively high progression risk. In other patients (with Ki‐67 SI90<0.57 or pRb>40%), the cytokeratins are not informative (fig 11).
The results for HSIL (CIN2 and CIN3) are promising; they also are in agreement with current molecular biological knowledge but have not been validated to the same degree as for low‐grade CIN. The following therefore should be used with care.
Evaluate the percentage of pRb‐positive and p53‐positive nuclei in the deep half of the epithelium. If either the percentage of pRb‐positive nuclei >40% or that of p53‐positive nuclei >15%, the likelihood of regression is high. All other HSIL lesions will probably persist or even progress.
Future developments
Biomarker‐mediated dynamic clinical behaviour prediction of CIN has been adequately validated. Routine biomarker applications can considerably change the manner in which afflicted women are followed up and treated. Definitive treatment in patients at high risk will be instituted earlier, whereas women whose biomarkers suggest that the process may resolve of its own accord will not receive treatment or will be treated only at a later time. Other important aspects of a dynamic CIN lesion—for example, the local immunoresponse factors—are relevant variables that may in the future affect prognosis and treatment. It is not yet certain whether the host's essential immune reactions can be adequately extracted from the usual paraffin‐wax‐embedded tissue alone, and independent validation studies are required. Moreover, other immunological and protein parameters must also be evaluated. Such proteomic analyses may go well beyond the analysis of classic formaldehyde‐fixed biopsy specimens.101
Acknowledgements
We thank Mrs Susan Allen‐de Jong for critically reading the text.
Abbreviations
CIN - cervical intraepithelial neoplasia
CK - cytokeratin
HLA - human leucocyte antigen
HPV - human papillomavirus
hrHPV - high‐risk human papillomavirus
HSIL - high‐grade squamous intraepithelial lesion
pRb - retinoblastoma protein
SI - Stratification Index
SIL - squamous intraepithelial lesion
Footnotes
Competing interests: None.
References
- 1.Ostor A G. Natural history of cervical intraepithelial neoplasia: a critical review. Int J Gynecol Pathol 199312186–192. [PubMed] [Google Scholar]
- 2.Heatly M K. How should we grade CIN? Histopathology 200240377–390. [DOI] [PubMed] [Google Scholar]
- 3.Keenan S J, Diamond J, McCluggage W G.et al An automated machine vision system for the histological grading of cervical intraepithelial neoplasia (CIN). J Pathol 2000192351–362. [DOI] [PubMed] [Google Scholar]
- 4.McCluggage W G, Bharucha H, Caughley L M.et al Interobserver variation in the reporting of cervical colposcopic biopsy specimens: comparison of grading systems. J Clin Pathol 199649833–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ismail S M, Colclough A B, Dinnen J S.et al Reporting cervical intra‐epithelial neoplasia (CIN): intra and inter‐pathologist variation and factors associated with disagreement. Histopathology 199016371–376. [DOI] [PubMed] [Google Scholar]
- 6.Ismail S M, Colclough A B, Dinnen J S.et al Observer variation in histopathological diagnosis and grading of cervical intraepithelial neoplasia. BMJ 1989298707–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robertson A J, Anderson J M, Swanson Beck J.et al Observer variability in histopathological reporting of cervical biopsy specimens. J Clin Pathol 198942231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stoler M H, Schiffman M. Atypical Squamous Cells of Undetermined Significance—Low‐grade Squamous Intraepithelial Lesion Triage Study (ALTS) Group. Interobserver reproducibility of cervical cytologic and histologic interpretations: realistic estimates from the ASCUS‐LSIL Triage Study, JAMA 20012851500–1505. [DOI] [PubMed] [Google Scholar]
- 9.Grenko R T, Abendroth C S, Frauenhoffer E E.et al Variance in the interpretation of cervical biopsy specimens obtained for atypical squamous cells of undetermined significance. Am J Clin Pathol 2000114735–740. [DOI] [PubMed] [Google Scholar]
- 10.AI Nafussi AI, Colquhoun MK Mild cervical intraepithelial neoplasia (CIN‐1)—a histological overdiagnosis. Histopathology 199017557–561. [DOI] [PubMed] [Google Scholar]
- 11.Creagh T, Bridger J E, Kupek E.et al Pathologist variation in reporting cervical borderline epithelial abnormalities and cervical intraepithelial neoplasia. J Clin Pathol 19954859–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kruse A J, Baak J P, Janssen E A.et al Ki‐67 predicts progression in early CIN: validation of a multivariate progression‐risk model. Cell Oncol 20042613–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kruse A J, Buhr‐Wildhagen S, Janssen E A.et al The relationship between syntactic structure analysis features, histological grade and high‐risk HPV DNA in cervical intraepithelial neoplasia. Cell Oncol 200426135–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kruse A J, Gudlaugsson E, Helliesen T.et al Evaluation of prospective, routine application of Ki‐67 immunoquantitation in early CIN for assessment of short‐term progression risk. Anal Quant Cytol Histol 200426134–140. [PubMed] [Google Scholar]
- 15.Kruse A J, Skaland I, Janssen E A.et al Quantitative molecular parameters to identify low‐risk and high‐risk early CIN lesions: role of markers of proliferative activity and differentiation and Rb availability. Int J Gynecol Pathol 200423100–109. [DOI] [PubMed] [Google Scholar]
- 16.Kruse A J, Baak J P, Helliesen T.et al Prognostic value and reproducibility of koilocytosis in cervical intraepithelial neoplasia. Int J Gynecol Pathol 200322236–239. [DOI] [PubMed] [Google Scholar]
- 17.Kruse A J, Baak J P, Janssen E A.et al Low‐ and high‐risk CIN‐1 and 2 lesions: prospective predictive value of grade, HPV, and Ki‐67 immuno‐quantitative variables. J Pathol 2003199462–470. [DOI] [PubMed] [Google Scholar]
- 18.Kruse A J, Baak J P, Helliesen T.et al Evaluation of KI‐67‐positive cell clusters as a diagnostic marker for cervical intraepithelial neoplasia. Am J Surg Pathol 2002261501–1507. [DOI] [PubMed] [Google Scholar]
- 19.Kruse A J, Baak J P, de Bruin P C.et al Relationship between the presence of oncogenic HPV DNA assessed by polymerase chain reaction and Ki‐67 immunoquantitative features in cervical intraepithelial neoplasia. J Pathol 2001195557–562. [DOI] [PubMed] [Google Scholar]
- 20.Kruse A J, Baak J P, de Bruin P C.et al Ki‐67 immunoquantitation in cervical intraepithelial neoplasia (CIN): a sensitive marker for grading. J Pathol 200119348–54. [DOI] [PubMed] [Google Scholar]
- 21.Sano T, Oyama T, Kashiwabara K.et al Expression status of p16 protein is associated with human papillomavirus oncogenic potential in cervical and genital lesions. Am J Pathol 19981531741–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klaes R, Friedrich T, Spitkovsky D.et al Overexpression of p16(INK4A) as a specific marker for dysplastic and neoplastic epithelial cells of the cervix uteri. Int J Cancer 200292276–284. [DOI] [PubMed] [Google Scholar]
- 23.Keating J T, Cviko A, Riethdorf S.et al Ki‐67, cyclin E, and p16INK4A are complimentary surrogate biomarkers for human papilloma virus‐related cervical neoplasia. Am J Surg Pathol 200125884–891. [DOI] [PubMed] [Google Scholar]
- 24.Nucci M R, Castrillon O H, Bai H.et al Biomarkers in diagnostic obstetric and gynecologic pathology: a review. Adv Anat Pathol 20031055–68. [DOI] [PubMed] [Google Scholar]
- 25.Pirog E C, Baergen R N, Soslow RA et a l. Diagnostic accuracy of cervical low‐grade squamous intraepithelial lesions is improved with Ki‐67 immunostaining. Am J Surg Pathol 20022670–75. [DOI] [PubMed] [Google Scholar]
- 26.Kiviat N. Natural history of cervical neoplasia: overview and update. Am J Obstet Gynecol 19961751099–1104. [DOI] [PubMed] [Google Scholar]
- 27.Koutsky L A, Holmes K K, Critchlow C W.et al A cohort study of the risk of cervical intraepithelial neoplasia grade 2 or 3 in relation to papillomavirus infection. N Engl J Med 19923271271–1278. [DOI] [PubMed] [Google Scholar]
- 28.Woodman C B J, Collins S, Winter H.et al Natural history of cervical human papillomavirus infection in young women: a longitudinal cohort study. Lancet 20013571831–1836. [DOI] [PubMed] [Google Scholar]
- 29.Saito K, Saito A, Fu Y S.et al Topographic study of cervical condyloma and intraepithelial neoplasia. Cancer 1987592064–2070. [DOI] [PubMed] [Google Scholar]
- 30.Schneider V. CIN prognostication: will molecular techniques do the trick? Acta Cytol 200347115–116. [DOI] [PubMed] [Google Scholar]
- 31.EI Hamidi A, Kocjan G, Du M‐Q Clonality analysis of archival cervical smears. Correlation of monoclonality with grade and clinical behavior of cervical intraepithelial neoplasia. Acta Cytol 200347117–123. [DOI] [PubMed] [Google Scholar]
- 32.Crum C P, Abbott D W, Quade B J. Cervical cancer screening: from the papanicolaou smear to the vaccine era. J Clin Oncol 200321224–230. [DOI] [PubMed] [Google Scholar]
- 33.Sun C A, Lai H C, Chang C C.et al The significance of human papillomavirus viral load in prediction of histologic severity and size of squamous intraepithelial lesions of uterine cervix. Gynecol Oncol 20018395–99. [DOI] [PubMed] [Google Scholar]
- 34.Clavel C, Masure M, Levert M.et al Human papillomavirus detection by the hybrid capture II assay: a reliable test to select women with normal cervical smears at risk for developing cervical lesions. Diagn Mol Pathol 20009145–150. [DOI] [PubMed] [Google Scholar]
- 35.Nobbenhuis M A E, Walboomers J M M, Helmerhorst Th JM.et al Relation of human papillomavirus status to cervical lesions and consequences for cervical‐cancer screening: a prospective study. Lancet 199935420–25. [DOI] [PubMed] [Google Scholar]
- 36.Bosch F X, Lorincz A, Munoz N.et al The causal relation between human papillomavirus and cervical cancer. J Clin Pathol 200255244–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cuschieri K S, Cubie H A, Whitley M W.et al Persistent high risk HPV infection associated with development of cervical neoplasia in a prospective population study. J Clin Pathol 200558946–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schiffman M, Herrero R, Hildesheim A.et al HPV DNA testing in cervical cancer screening: results from women in a high‐risk province of Costa Rica. JAMA 200028387–93. [DOI] [PubMed] [Google Scholar]
- 39.Healey S M, Aronson K J, Mao Y.et al Oncogenic human papillomavirus infection and cervical lesions in aboriginal women of Nunavut, Canada. Sex Transm Dis 200128694–700. [DOI] [PubMed] [Google Scholar]
- 40.Sherman M E, Schiffman M, Cox J T. Effects of age and HPV viral load on colposcopy triage: data from the randomized atypical squamous cells of undetermined significance/low‐grade squamous intraepithelial lesion triage study (ALTS). J Natl Cancer Inst 200294102–107. [DOI] [PubMed] [Google Scholar]
- 41.Van Duin M, Snijders P J, Schrijnemakers H F.et al Human papillomavirus 16 load in normal and abnormal cervical scrapes: an indicator of CIN II/III and viral clearance. Int J Cancer 200298590–595. [DOI] [PubMed] [Google Scholar]
- 42.Dalstein V, Riethmuller D, Pretet J L.et al Persistence and load of high‐risk HPV are predictors for development of high‐grade cervical lesions: a longitudinal French cohort study. Int J Cancer 2003106396–403. [DOI] [PubMed] [Google Scholar]
- 43.Koutsky L A, Ault K A, Wheeler C M.et al A controlled trial of a human papillomavirus type 16 vaccine. N Engl J Med 20023471645–1651. [DOI] [PubMed] [Google Scholar]
- 44.Fairley C K, Robinson P M, Chen S.et al The detection of HPV DNA, the size of tampon specimens and the menstrual cycle. Genitourin Med 199470171–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robboy S J, Anderson M C, Morse A.et al Cervical precancer (intraepithelial neoplasia). In: Robboy SJ, Anderson MC, Russell P, eds. Pathology of the female reproductive tract. London: Churchill Livingstone, 2002165–193.
- 46.Hildesheim A, Schiffman M H, Gravitt P E.et al Persistence of type‐specific human papillomavirus infection among cytologically normal women. J Infect Dis 1994169235–240. [DOI] [PubMed] [Google Scholar]
- 47.Hinchliffe S A, van Velzen D, Korporaalet al Transience of cerical HPV infection in sexually active, young women with normal cervicavaginal cytology. Br J Cancer 199572943–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wheeler C, Greer C E, Becker T M.et al Short‐term fluctuations in the detection of cervical human papillomvirus DNA. Obstet Gynecol 199688261–268. [DOI] [PubMed] [Google Scholar]
- 49.Moscicki A, Shiboski S, Broering J.et al The natural history of human papillomavirus infection as measured by repeated DNA testing in adolescent and young women. J Pediatr 1998132277–284. [DOI] [PubMed] [Google Scholar]
- 50.Ho G Y F, Bierman R, Beardsly L.et al Natural history of cervicovaginal papillomavirus infection in young women. N Engl J Med 1998338423–428. [DOI] [PubMed] [Google Scholar]
- 51.Rozendaal L, Walboomers J M M, van der Linden J C.et al PCR‐based high risk HPV test in cervical cancer screening gives objective risk assessment of women with cytomorphologically normal cervical smears. Int J Cancer 199668766–769. [DOI] [PubMed] [Google Scholar]
- 52.Ho G Y F, Burk R D, Klein S.et al Persistent genital human papillomavirus infection as a risk factor for persistent cervical dysplasia. J Natl Cancer Inst 1995871365–1371. [DOI] [PubMed] [Google Scholar]
- 53.Koutsky L A, Holmes K K, Critchlow C W.et al A cohort study of the risk of cervical intraepithelial neoplasia grade 2 or 3 in relation to papillomavirus infection. N Engl J Med 19923271272–1278. [DOI] [PubMed] [Google Scholar]
- 54.Cuschieri K S, Cubie H A, Whitley M W.et al Multiple high risk HPV infections are common in cervical neoplasia and young women in a cervical screening population. J Clin Pathol 20045768–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baseman J G, Koutsky L A. The epidemiology of human papillomavirus infections. J Clin Virol 200532S16–S24. [DOI] [PubMed] [Google Scholar]
- 56.Steenbergen R D M, de Wilde J, Wilting S M.et al HPV‐mediated transformation of the anogenital tract. J Clin Virol 200532SS25–S33. [DOI] [PubMed] [Google Scholar]
- 57.Milde‐Langosch K, Riethdorf S. Role of cell‐cycle regulatory proteins in gynecological cancer. J Cell Physiol 2003196224–244. [DOI] [PubMed] [Google Scholar]
- 58.Trunk M J, Wentzensen N, von Knebel Doeberitz M. Molecular pathogenesis of cervical cancer and its first steps. Pathologie 200526283–290. [DOI] [PubMed] [Google Scholar]
- 59.Wright T C, Kurman R J, Ferenczy A. Precancerous lesions of the cervix. In: Kurman RJ, eds. Blaustein's pathology of the female genital tract. 5th edn. New York: Springer, 2002278–279.
- 60.Zur Hausen H. Papillomavirus causing cancer: evasion from host‐cell control in early events in carcinogenesis. J Natl Cancer Inst 200092690–698. [DOI] [PubMed] [Google Scholar]
- 61.von Knebel Doeberitz M. New markers for cervical dysplasia to visualise the genomic chaos created by aberrant oncogenic papillomavirus infections. Eur J Cancer 2002382229–2242. [DOI] [PubMed] [Google Scholar]
- 62.Klaes R, Woerner S M, Ridder R.et al Detection of high‐risk cervical intraepithelial neoplasia and cervical cancer by amplification of transcripts derived from integrated papillomavirus oncogenes. Cancer Res 1999596132–6136. [PubMed] [Google Scholar]
- 63.Nishimura A, Ono T, Ishimoto A.et al Mechanisms of human papillomavirus E2‐mediated repression of viral oncogene expression and cervical cancer cell growth inhibition. J Virol 2000743752–3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Riley R R, Duensing S, Brake T.et al Dissection of human papillomavirus E6 and E7 function in transgenic mouse models of cervical carcinogenesis. Cancer Res 2003634862–4871. [PubMed] [Google Scholar]
- 65.Jeon S, Allen‐Hoffmann B L, Lambert P F. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J Virol 1995692989–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zur Hausen H. Papillomavirus and cancer: from basic studies to clinical application. Nat Rev Cancer 20022342–350. [DOI] [PubMed] [Google Scholar]
- 67.Munger K, Howley P M. Human papillomavirus immortalization and transformation functions. Virus Res 200289213–228. [DOI] [PubMed] [Google Scholar]
- 68.Duensing S, Duensing A, Crum C P.et al Human papillomavirus type 16 E7 oncoprotein‐induced abnormal centrosome synthesis is an early event in the evolving malignant phenotype. Cancer Res 2001612356–2360. [PubMed] [Google Scholar]
- 69.Wang S S, Hildesheim A. Viral and host factors in human papillomavirus persistence and progression. J Natl Cancer Inst 20033135–40. [DOI] [PubMed] [Google Scholar]
- 70.Hildesheim A, Wang S S. Host and viral genetics and risk of cervical cancer: a review. Virus Res 200289229–240. [DOI] [PubMed] [Google Scholar]
- 71.Villa L L, Sichero L, Rahal P.et al Molecular variants of human papillomavirus types 16 and 18 preferentially associated with cervical neoplasia. J Gen Virol 2000812959–2968. [DOI] [PubMed] [Google Scholar]
- 72.Chan P K, Lam C W, Cheung T H.et al Association of human papillomavirus type 58 variant with the risk of cervical cancer. J Natl Cancer Inst 2002941249–1253. [DOI] [PubMed] [Google Scholar]
- 73.Ylitalo N, Sorensen P, Josefson A M.et al Consistent high viral load of human papillomavirus 16 and risk of cervical carcinoma in situ: a nested case‐control study. Lancet 20003552194–2198. [DOI] [PubMed] [Google Scholar]
- 74.Lorincz A T, Castle P E, Sherman M E. Viral load of human papillomavirus and risk of CIN3 or cervical cancer. Lancet 2002360228–229. [DOI] [PubMed] [Google Scholar]
- 75.Schiller J T, Hildesheim A. Developing HPV virus‐like particle vaccines to prevent cervical cancer: a progress report. J Clin Virol 20001967–74. [DOI] [PubMed] [Google Scholar]
- 76.Lehtinen M, Dillner J, Knekt P.et al Serologically diagnosed infection with human papillomavirus type 16 and risk of subsequent development of cervical carcinoma: nested case‐control study. BMJ 1996312537–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.De Sanjose S, Hamsikova E, Munoz N.et al Serological response to HPV16 in CIN‐III and cervical cancer patients. Case‐control studies in Spain and Colombia. Int J Cancer 19966670–74. [DOI] [PubMed] [Google Scholar]
- 78.Sun Y, Eluf‐Neto J, Bosch F X.et al Serum antibodies to HPV16 proteins in women from Brazil with invasive cervical carcinoma. Cancer Epidemiol Biomarkers Prev 19998935–940. [PubMed] [Google Scholar]
- 79.Tsukui T, Hildesheim A, Schiffman M H.et al Il‐2 production by peripheral lymphocytes in response to human papillomavirus‐derived peptides: correlation with cervical pathology. Cancer Res 1996563967–3974. [PubMed] [Google Scholar]
- 80.Kadish A S, Timmins P, Wang Y.et al Regression of CIN and loss of HPV infection is associated with cell‐mediated immune responses to an HPV type 16 E7 peptide. Cancer Epidemiol Biomarkers Prev 200211483–488. [PubMed] [Google Scholar]
- 81.Carreon J D, Martin M P, Hildesheimet al Human leukocyte antigen class I and II haplotypes and risk of cervical cancer. Tissue Antigens 200566321–324. [DOI] [PubMed] [Google Scholar]
- 82.Hemminki K, Li X, Mutanen P. Familial risk in invasive and in situ cervical cancer by histological type. Eur J Cancer Prev 20011083–89. [DOI] [PubMed] [Google Scholar]
- 83.Magnusson P K, Lichtenstein P, Gyllensten U B. Heritability of cervical tumours. Int J Cancer 200088698–701. [DOI] [PubMed] [Google Scholar]
- 84.Klaes R, Benner A, Friedrich T.et al p16INK4a immunohistochemistry improves interobserver agreement in the diagnosis of cervical intraepithelial neoplasia. Am J Surg Pathol 2002261389–1399. [DOI] [PubMed] [Google Scholar]
- 85.Snijders P J, Steenbergen R D M, Heideman D A.et al HPV‐mediated cervical carcinogenesis: concepts and clinical implications. J Pathol 2006208152–164. [DOI] [PubMed] [Google Scholar]
- 86.Kruse A J. Quantitative biomarkers to identify low‐ and high‐risk early CIN lesions [dissertation]. Vrije Universiteit Amsterdam 2003
- 87.Murphy N, Ring M, Killalea A G.et al P16ink4a as a marker for cervical dyskaryosis: CIN and cGIN in cervical biopsies and ThinPrep smears. J Clin Pathol 20035656–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bulten J, Van der Laak J A, Gemmink J H.et al Ki‐67, a promising marker for the classification of cervical intraepithelial neoplasia. J Pathol 1996178268–273. [DOI] [PubMed] [Google Scholar]
- 89.Abadi M A, Ho G Y F, Burk R D.et al Stringent criteria for histological diagnosis of koilocytosis fail to eliminate overdiagnosis of human papillomavirus infection and cervical intraepithelial neoplasia grade 1. Hum Pathol 19982954–59. [DOI] [PubMed] [Google Scholar]
- 90.Reesink‐Peters N, Helder M N, Wisman G B.et al Detection of telomerase, its components, and human papillomavirus in cervical scrapings as a tool for triage in women with cervical dysplasia. J Clin Pathol 20035631–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bravaccini S, Sanchini M A, Amadori A.et al Potential of telomerase expression and activity in cervical specimens as a diagnostic tool. J Clin Pathol 200558911–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Southern S A, McDicken I W, Herrington C S. Loss of cytokeratin 14 expression is related to human papillomavirus type and lesion grade in squamous intraepithelial lesions of the cervix. Hum Pathol 2001321351–1355. [DOI] [PubMed] [Google Scholar]
- 93.Sadlar L, Saftlas A, Wang W.et al Treatment for cervical intraepithelial neoplasia and risk of preterm delivery. JAMA 20042912100–2106. [DOI] [PubMed] [Google Scholar]
- 94.Baak J P A, Kruse A ‐ J, Garland S M.et al Combined p53 and retinoblastoma protein detection identify persistent and regressive cervical high‐grade squamous intraepithelial lesions. Am J Surg Pathol 2005291062–1066. [PubMed] [Google Scholar]
- 95.Zielinsky G D, Snijders P J F, Rozendaal L.et al The presence of high‐risk HPV combined with specific p53 and p16ink4a expression patterns points to high‐risk HPV as the main causative agent for adenocarcinoma in situ and adenocarcinoma of the cervix. J Pathol 2003201535–543. [DOI] [PubMed] [Google Scholar]
- 96.Trimble C L, Piantadosi S, Gravitt P.et al Spontaneous regression of high‐grade cervical dysplasia: effects of human papillomavirus type and HLA phenotype. Clin Cancer Res 2005114717–4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hagen B, Skjeldestad F E.A guide in gynecological oncology 2002. Oslo: The Norwegian Medical Association, 2002
- 98.Follen M, Atkinson E N, Schottenfeld D. A randomized clinical trial of 4‐hydroxyphenylretinamide for high‐grade squamous intraepithelial lesions of the cervix. Clin Cancer Res 200173356–3365. [PubMed] [Google Scholar]
- 99.Meyskens F L, Jr, Surwit E, Moon T E. Enhancement of regression of cervical intraepithelial neoplasia II (moderate dysplasia) with topically applied all‐trans‐retinoic acid: a randomized trial. J Natl Cancer Inst 199486539–543. [DOI] [PubMed] [Google Scholar]
- 100.Keefe K A, Schell M J, Brewer C.et al A randomized, double blind, phase III trial using oral beta‐carotene supplementation for women with high‐grade cervical intraepithelial neoplasia. Cancer Epidemiol Biomarkers Prev 2001101029–1035. [PubMed] [Google Scholar]
- 101.Baak J P, Janssen E A, Soreide K.et al Genomics and proteomics—the way forward. Ann Oncol 20051630–44. [DOI] [PubMed] [Google Scholar]