Abstract
A panel of five quasimonomorphic mononucleotide repeats that dispenses with the need to analyse corresponding germline DNA was proposed by Suraweera et al for the detection of high‐frequency microsatellite instability (MSI) in colorectal cancer. Using this panel, a simplified and a more sensitive (compared with the original) algorithm (p<0.05) was developed to define the instability of each repeat by assessing the morphological shape of its plot and not its absolute length. 103 cases of colorectal tumours were investigated and the results compared with those obtained by the analysis of five consensus microsatellites (Bethesda reference panel). By the proposed method, a higher specificity, but no loss of sensitivity, was found. Thus, the use of the five mononucleotide repeats in combination with the modified assessment technique simplifies the assessment of MSI, while retaining the sensitivity of the Bethesda panel for the detection of high‐frequency MSI.
The microsatellite instability (MSI) phenotype is characteristic of the hereditary non‐polyposis colorectal cancer syndrome and is also found in about 15% of sporadic colorectal cancers.1,2,3 A National Cancer Institute consensus meeting proposed the analysis of two mononucleotide and three dinucleotide repeats for the assessment of MSI (Bethesda reference panel; BAT‐25 (GenBank accession no. 9834508), BAT‐26 (GenBank accession no. 9834505), D5S346 (GenBank accession no. 181171), D2S123 (GenBank accession no. 187953) and D17S250 (GenBank accession no. 177030)).4 The evaluation of these markers requires the analysis of tumour DNA and corresponding germline DNA. Bacher et al5 and Suraweera et al6 have since described multiplexed, fluorescence‐based assays. Suraweera uses a panel of five mononucleotide repeats (mononucleotide repeats pentaplex (MRP) and polymerase chain reaction (PCR)) that are quasi‐monomorphic in most Caucasian populations. With these five repeats, the MSI status can be assessed without analysing the corresponding germline DNA. We used this method to analyse 103 samples of colorectal tumours, whereas Suraweera et al6 defined instability as deviation from a predetermined size window for each repeat. We therefore applied a simplified algorithm to detect MSI.
Methods
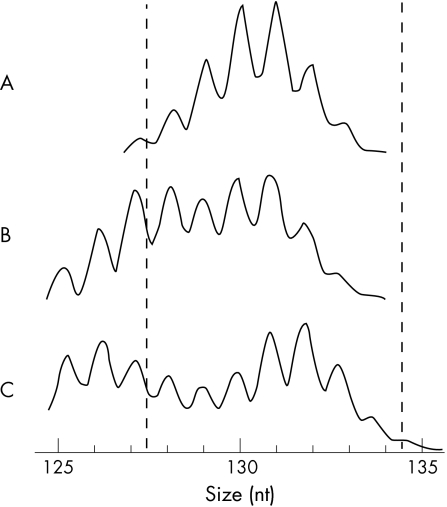
In all, 103 specimens of colorectal tumours were investigated by the MRP method described by Suraweera et al6 and re‐evaluated with the Bethesda panel of markers. In our study, a repeat was classified as instable if the plot of the PCR product showed a double‐peaked pattern (fig 1A: one peak, stable; fig 1B,C: biphasic, instable—one peak preceded and followed by two higher peaks, respectively) rather than referring to its absolute length as proposed by Suraweera et al.6 To determine the specificity of the proposed assessment algorithm, 78 control samples of haematological malignancies that were mismatch repair competent and microsatellite stable (MSS: 72 cases of acute lymphoblastic leukaemia; 6 cases of acute myeloblastic leukaemia; containing 10–90% malignant cells) were also screened by MRP. All the samples originated from a Caucasian population and were anonymised and examined retrospectively.
Figure 1 Assessment of microsatellite instability in a mononucleotide repeat. (A) Microsatellite stable colorectal carcinoma. (B) Tumour no 674‐61: biphasic shape of the polymerase chain reaction product. Both peaks lie in the window (vertically plotted lines) defining microsatellite stability according to Suraweera et al.6 (C) Microsatellite instable colorectal carcinoma (repeat NR‐24; medium length of NR‐24 in our study was 131 bp).
Results
We analysed 103 samples of colorectal tumours by MRP to evaluate the MSI assessment method described by Suraweera et al.6 The simplified algorithm “biphasic shape” for the detection of MSI showed 29 cases of MSI and 74 cases of MSS. The two different definitions of MSI yielded discrepant results in two tumours. Even though one of the repeats showed a biphasic shape in these tumours, both peaks lay in the window, defining stability according to Suraweera (eg, tumour no 674‐61, vertically plotted lines, fig 1B). With only two unstable mononucleotide repeats, these two tumours would be classified as MSS by the length assessment technique. When analysed by the Bethesda panel, the two tumours displayed instability at three and four of five repeats, verifying the classification as MSI on the basis of the assessment algorithm biphasic shape. The suggested algorithm seems to be more sensitive than the original proposal (p<0.05; χ2 test).6
The use of the Bethesda panel confirmed the results of the MRP analysis in 101 of the 103 colorectal cancer samples. The two discrepant tumours were MSS (stability at all five repeats) by MRP, but showed instability at two dinucleotide repeats with the Bethesda panel. Both cases exhibited instability in two dinucleotide repeats. Interestingly, for the Bethesda reference panel, the finding of two unstable dinucleotide markers in the absence of BAT‐26 mutations has been described as a source of misclassification of tumours as MSI.5,7,8 After evaluation with the extended Bethesda panel (10 markers), the two tumours were classified as low‐frequency MSI in concordance with the clinical and histological picture (age at diagnosis, 63 and 65 years; immunohistochemical expression of the mismatch repair genes MLH1, MSH2 and MSH6).
To further clarify the specificity of the MRP assay, we screened a group of 78 samples of mismatch repair‐competent malignancies. MSI instability was not found in any of these control samples.
Discussion
We showed that the mononucleotide repeat panel proposed by Suraweera et al6 is appropriate for the detection of MSI: the results of assessment by this panel were confirmed by the Bethesda panel in 101 of the 103 cases with colorectal cancer that were investigated. The two discrepant tumours were identified as MSS by MRP, but as low‐frequency MSI by the extended Bethesda reference panel. As described by Suraweera, MRP subsumes microsatellite stable and low‐frequency unstable tumours as MSS and separates this group from MSI tumours. This distinction is sufficient for clinical practice.6
Suraweera et al6 propose evaluation of the absolute length of a repeat, which requires exact calibration of the detection system depending on the kind of polymerase and the type of dyes used.6 We introduced an algorithm assessing the shape of a repeat's plot, instability being indicated by a biphasic shape (fig 1B,C). This algorithm facilitated the analysis and proved to be more sensitive than the technique proposed by Suraweera et al.6
The MRP assay identified no false‐positive cases of MSI among the colorectal tumour specimens. Additionally, no cases of MSI were detected in the control group of 78 samples of mismatch repair‐competent malignancies. These results confirm the high specificity of MRP.
To summarise, in a Caucasian study population, the mononucleotide repeat panel proved to be as specific as the Bethesda panel for the detection of MSI. It determines the MSI status of DNA extracted from complete tissue sections in situations in which no matching normal DNA would be available without microdissection. Our investigations also show that the biphasic shape of a mononucleotide repeat can be used to define instability, instead of the absolute repeat size. This facilitates analysis while preserving the high sensitivity associated with the Bethesda panel.
Acknowledgements
We thank M Ruck for helpful advice and for revising the manuscript. This work was supported by the German José Carreras Leukemia Foundation, grant no DJCLS F05/02.
Abbreviations
MRP - mononucleotide repeat pentaplex
MSI - microsatellite instability
MSS - microsatellite stable
PCR polymerase chain reaction -
Footnotes
Ethical approval: The samples were anonymised and examined retrospectively according to the requirements of the local ethics committee (Ethikkommission der Medizinischen Fakultät der Universität Tübingen).
Competing interests: None.
References
- 1.Aaltonen L A, Peltomaki P, Leach F S.et al Clues to the pathogenesis of familial colorectal cancer. Science 1993260812–816. [DOI] [PubMed] [Google Scholar]
- 2.Ionov Y, Peinado M A, Malkhosyan S.et al Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature 1993363558–561. [DOI] [PubMed] [Google Scholar]
- 3.Thibodeau S N, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science 1993260816–819. [DOI] [PubMed] [Google Scholar]
- 4.Boland C R, Thibodeau S N, Hamilton S R.et al A National Cancer Institute Workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998585248–5257. [PubMed] [Google Scholar]
- 5.Bacher J W, Flanagan L A, Smalley R L.et al Development of a fluorescent multiplex assay for detection of MSI‐high tumors. Dis Markers 200420237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suraweera N, Duval A, Reperant M.et al Evaluation of tumour microsatellite instability using five quasimonomorphic mono‐nucleotide repeats and pentaplex PCR. Gastroenterology 20021231804–1811. [DOI] [PubMed] [Google Scholar]
- 7.Loukola A, Eklin K, Laiho P.et al Microsatellite marker analysis in screening for hereditary nonpolyposis colorectal cancer (HNPCC). Cancer Res 2001614545–4549. [PubMed] [Google Scholar]
- 8.Buhard O, Suraweera N, Lectard A.et al Quasimonomorphic mononucleotide repeats for high‐level microsatellite instability analysis. Dis Markers 200420251–257. [DOI] [PMC free article] [PubMed] [Google Scholar]