Abstract
In this article we extend the Iterative Protein Redesign and Optimization (IPRO) framework for the design of protein libraries with targeted ligand specificity. Mutations that minimize the binding energy with the desired ligand are identified. At the same time explicit constraints are introduced that maintain the binding energy for all decoy ligands above a threshold necessary for successful binding. The proposed framework is demonstrated by computationally altering the effector binding specificity of the bacterial transcriptional regulatory protein AraC, belonging to the AraC/XylS family of transcriptional regulators for different unnatural ligands. The obtained results demonstrate the importance of systematically suppressing the binding energy for competing ligands. Pinpointing a small set of mutations within the binding pocket greatly improves the difference in binding energies between targeted and decoy ligands, even when they are very similar.
INTRODUCTION AND OBJECTIVES
As we enter the postgenomic era we have in our hands a plethora of protein designs, experimental techniques, and computational methods. Recent developments (1–3) have made it clear that given sufficient resources and screening capabilities, directed evolution can be used to drastically improve protein function. A variety of protocols are available for performing targeted mutagenesis or constructing combinatorial libraries with customized statistics of mutations and/or parental fragments (see Moore and Maranas (4) for a review), raising the question of what type of mutations and/or recombination events are likely to yield functionally enriched protein libraries. This underlines the task of constructing protein libraries enriched with desired functions compared to a random sampling of protein sequence space. The challenge here is in effectively searching sequence space to find improved variants containing multiple mutations and particularly to identify interacting mutations whose effects on fitness are nonadditive.
Computer simulations play an increasingly significant role in understanding the underlying physical principles that dictate protein folding, stability, and function, leading to greatly improved protein design predictions (4). Although it is not yet feasible to consistently predict structure and function de novo, it is possible to assess the impact of mutations on existing, well-characterized proteins (5–8). The goal of this study is to modify the Iterative Protein Redesign and Optimization (IPRO) computational protein library design framework (1) to enable the systematic redesign of proteins for desired ligand specificity while suppressing the affinity toward competing molecules. The approach is demonstrated through a comprehensive computational study involving the redesign of the l-arabinose-responsive bacterial transcriptional regulatory protein AraC to accept targeted unnatural ligands as transcriptional activating “effector” molecules (9). This is an important endeavor because the precise control of gene transcription in response to specific stimuli has wide implications ranging from synthetic biology and metabolic engineering to the development of customized genetic selections for use in subsequent protein engineering projects. Furthermore, the regulatory properties of AraC make it a good candidate for protein engineering because of the natural coupling of molecular recognition to gene transcription, enabling the use of a genetic selection and/or high-throughput screening procedure to rapidly identify mutants with improved binding specificity. Finally, the availability of high-resolution atomic-level x-ray crystal structures of the effector-binding/dimerization domain of AraC in the presence and absence of l-arabinose (10) allows for computationally modeling novel effector recognition.
We describe the use of simulation and optimization methods to accurately reflect the relative strengths with which wild-type AraC binds various compounds. IPRO is subsequently used to predict mutagenesis strategies resulting in altered binding selectivity. Specifically, we explore the design of AraC variants responding to novel effector molecules that increasingly resemble l-arabinose (e.g., cis-verbenol, followed by d-arabinose). Our interest in binding target molecules such as cis-verbenol stems from a need to develop biocatalysts capable of converting renewable and abundant natural resources (including plant oils such as those containing α-pinene) into value-added products such as antibiotics, pharmaceutical intermediates, and chemicals for the flavor and fragrance industry.
AraC SYSTEM
The AraC monomer is a 292–amino acid polypeptide composed of an N-terminal effector binding/dimerization domain (residues 1–170) followed by a C-terminal DNA-binding domain. Under physiological conditions, the AraC protein exists primarily as a dimer that tightly regulates transcription from the PBAD promoter by acting as a repressor in the absence of inducer (by forming a DNA loop in the promoter region) and as an activator in response to inducer (l-arabinose) (10,11). The l-arabinose binding “signal” is a conformational change in the AraC dimer that consequently disrupts the DNA loop and activates transcription. This signal is transmitted from the N-terminus to the DNA binding domain via movement of the N-terminal arm. In the absence of the inducer, this arm is believed to make contacts with the C-terminal domain (12), whereas in the presence of l-arabinose, this arm closes over the N-terminal binding pocket.
Induction of the ara operon is specific to l-arabinose: Structurally and chemically similar sugars such d-xylose and d-arabinose fail to act as wild-type AraC effectors (13). d-Fucose, which is identical to l-arabinose at all positions except C5 (where fucose contains a methyl group instead of a hydrogen), acts as a competitive inhibitor that binds AraC (in the same position as l-arabinose) but fails to induce gene expression in vivo or in vitro (13,14). AraC mutants have been isolated that are induced by fucose (13,15). Thus, as in the case of other receptors (16,17), very similar small molecules can have drastically different binding affinities and stimulatory effects.
COMPUTATIONAL PROCEDURE
Modification of IPRO framework
We use binding calculations to score the relative strengths with which AraC binds various compounds and subsequently deploy mathematical optimization to suggest mutagenesis strategies resulting in the desired altered binding selectivity. Binding energy, which is computationally approximated using the CHARMM (18,19) energy function, accessed through the IPRO optimization framework, serves as a surrogate of molecular recognition (i.e., binding affinity) (1). The optimization step identifies mutations that lead to stronger binding scores for the desired ligand while at the same time depressing binding scores for competing molecules.
The protein redesign framework IPRO provides the backbone of the computational environment for the redesign of AraC binding specificity (1). Briefly, it involves iterative optimal protein redesign of residues/rotamers (near the binding pocket) followed by backbone relaxation and ligand(s) redocking. Specifically, during each iteration a local backbone perturbation window (i.e., one to five residues) is randomly selected, and a perturbation of the backbone is imposed. New residues (i.e., mutations) and corresponding rotamers are identified by globally optimizing the binding score within the redesign window and readjusting rotamers within a wider window (11–15 residues) around the region of perturbation. This optimization step is followed by backbone relaxation and ligand(s) redocking (20). If the redesign and corresponding structural modifications lead to an improved binding score, then the perturbation is accepted. If the redesign leads to a worse binding score, then it is accepted or rejected based on the Metropolis criterion (21). This iterative cycle forms the basic working paradigm of IPRO.
Improving binding affinity of a regulatory protein must also take into account the competitive nature of the process. Specifically, at the same time that binding affinity for the targeted ligand is improved, the affinity for competing molecules must be depressed. This new design paradigm warrants a number of modifications in the general IPRO procedure. We address this challenge in this article by putting forth and solving a two-level optimization problem. In the outer level, new designs (i.e., residue choices) are made, while in the inner level separate rotamer sets are identified that optimize the binding with respect to the desired and undesired substrates. A constraint ensures that the binding score for even the best conformation (i.e., rotamer choices) for the undesirable ligand(s) remains greater than what is needed for successful binding of the desired ligand. When this threshold is exceeded, the corresponding design choice is deemed infeasible. The structure of the proposed two-stage optimization formulation is as follows:
 |
The inner minimization problems identify separate rotamer combinations that minimize the binding energy E with respect to the desired L1 and competing (L2, L3, …, Ln) ligands. For all competing ligands this minimum binding energy is constrained to be above a high enough threshold M preventing effective binding (see Fig. 1). We use known good/poor binders for a given protein system to arrive at appropriate values for M. Note that the inner part of the optimization problem is decomposable into n separable minimization problems that can be run on separate processors. Similar to the original IPRO procedure, the outer optimization problem is solved using the Metropolis criterion to update amino acid choices after each iteration. Backbone relaxation and ligand-redocking steps can also be used after each time the inner rotamer optimization problems are solved. Fig. 2 pictorially illustrates the computations workflow of the modified IPRO framework.
FIGURE 1.
Two-level optimization formulation of the modified IPRO. In the outer level new designs with respect to amino acids choices are generated, whereas the inner level identifies the rotamer choices that minimize the binding energy with respect to various ligands. By changing rotamers, the amino acids are chosen such that the binding energy with respect to all undesired ligands is above a cutoff value, preventing their binding while simultaneously ensuring sufficiently low binding energy with respect to the targeted ligand to enable binding.
FIGURE 2.
(A) Local region of the protein (1–5 consecutive residues around the targeted ligand) is randomly chosen for perturbation. The ϕ and ψ angles of the targeted position (as shown in the circle) are perturbed by up to 5°. (B) All amino acid rotamers consistent with these torsion angles are selected at each position from the Dunbrack and Cohen rotamer library (35,36). Rotamer-backbone and rotamer-rotamer energies are calculated for all the selected rotamers. (C) The binding energy is minimized using a MILP formulation to select the optimal rotamer at each of these positions. (D) After rotamer selection for the target molecule, the nontarget ligand is docked, and its binding energy is calculated using the CHARMM energy function (18,19). The best conformation (i.e., rotamer choices) is accepted if it ensures that the binding score for the undesirable ligand(s) remains greater than what is needed for successful binding. When this threshold is exceeded, the corresponding design choice is deemed infeasible. (E) In this step the backbone and the targeted ligand are allowed to relax to adjust to the changes in the side chains. This is achieved by allowing ϕ and ψ to vary freely and to be determined during energy minimization. (F) ZDOCK software is employed to readjust the targeted ligand position regarding the modified backbone and side chains (20). (G) Protein-ligand binding energy is computed using CHARMM energy functions. If the binding energy for the target ligand is lower than the previous best ligand structure then this move is accepted as best solution otherwise. (H) Metropolis criterion is used to decide whether to accept or reject the move.
COMPUTATIONAL PREDICTIONS
A key consideration for any successful AraC redesign is to retain the “light-switch” mechanism of the ara regulatory operon (22) that preserves the coupling between binding and transcriptional activation. Extensive mutagenesis analyses by Schleif and colleagues have identified a series of “hemiplegic” AraC mutations that specifically block either induction (I-) or repression (R-) at PBAD (22,23). Many other mutations in the N-terminal arm are reported to cause constitutivity or uninducibility (23). In our studies it is important for the N-terminal arm in engineered AraC variants to maintain contact with the C-terminal domain in the absence of inducer and to disfavor contact in the presence of an inducer (favoring instead arm-inducer interactions). Based on these requirements, we have computationally disallowed critical residue positions from being mutated. Sixteen residues of the 32 residues forming the binding pocket were selected as design positions. Mutations at these selected positions (located in the N-terminal domain) are presumed to weaken l-arabinose binding interactions while preserving the repression of the ara regulatory operon in the absence of the effector (22,23). Therefore, these positions were deemed to be viable candidates to be considered as design positions to confer novel specificity in AraC protein.
The structure of AraC complexed with l-arabinose shows an extensive network of water molecules within the ligand-binding pocket (10). This network of water molecules mediates hydrogen bonds between the ligand and AraC, thus affecting the binding and location of the ligand in the pocket. We computationally explored the effect of placing 16 structural water molecules in the binding pocket in the docking calculations. It has been acknowledged (24–26) that water-mediated interactions can affect the stability, dynamics, and the placement of the protein backbone. We find that adding water molecules improves protein docking and thus results in more accurate ligand positioning. Predicted ligand positions for l-arabinose and d-fucose more closely match the known crystal structures (calculated RMSD = 0.20 Å for the two sugars) when water molecules are included in the calculations compared to the predicted positions in the absence of water (RMSD = 3.53 Å). Therefore, in the following detailed studies we report on results in the presence of structural water molecules.
The validity of using computationally derived binding energy as surrogate for molecular recognition was first tested by calculating binding energies for different sugars (i.e., l-arabinose, d-fucose, d-arabinose, l-lyxose, d-lyxose, l-xylose, d-xylose, l-ribose, and d-ribose) using the CHARMM (18,19) energy function. The calculated values were subsequently contrasted against experimental data available in the literature (13,23,27–30). We find that the calculated binding energies qualitatively reflect the experimentally observed absence of transcriptional activation for the tested sugars (Fig. 3). Specifically, l-arabinose and d-fucose, two sugars known to bind to the AraC protein, have the two most negative binding energy scores. Several of the tested sugars including l,d-xylose and l-lyxose were also verified by Doyle et al. to not inhibit induction by l-arabinose, implying that these sugars are certainly not bound by AraC (13). These results bolster the assumption that binding energy is a reasonable surrogate for ligand binding by AraC, which is at least a requirement for transcriptional activation. Further experimental analysis is necessary to determine whether (or how readily) binding energy correlates with a ligand's ability to induce transcription.
FIGURE 3.
Binding energies of the various sugars with AraC protein. l-Arabinose and d-fucose, known to bind to AraC, have the most negative binding energies.
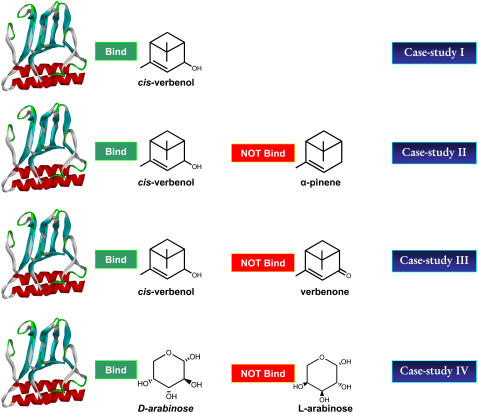
Four case studies are addressed here to demonstrate the proposed computational procedure. The first study involves engineering AraC variants that bind cis-verbenol, one of the oxidized forms of the bicyclic monoterpene α-pinene without proactively depressing affinity for competitive ligands. This study explores the ability of modified IPRO to redesign a transcription factor (i.e., AraC) to recognize an effector molecule very different in structure and chemistry from l-arabinose (Fig. 4). In the second study, we redesign AraC to recognize cis-verbenol but at the same time not bind its reduced form α-pinene. In the third case study, we again redesign AraC to selectively bind cis-verbenol but at the same time not bind verbenone, an alternative oxidized product of α-pinene that is chemically and structurally very similar to cis-verbenol. Finally, in the fourth case study we computationally redesign AraC protein to impart novel effector selectivity capable of distinguishing between different chiral forms of the arabinose sugars (i.e., l- and d-arabinose). The binding energy values for known poor binders for the AraC protein were used to choose appropriate binding energy cutoff values. For the second case study, this cutoff value was set at −20.0 kcal/mol. This value is higher than the binding energies of d-xylose (Fig. 3), which is known not to bind AraC. Furthermore, for the third and fourth case–studies, a tighter cutoff value of −30 kcal/mol was chosen to help elucidate mutations that sharpen specificity toward the target ligand from very similar competing ligands. In these four studies, several computational libraries were constructed using different sets of randomization seeds for the iterative backbone perturbation employed by IPRO during each design cycle. We found that in all cases although the amino acid design choices can vary between different randomization runs, the underlying properties of the selected amino acids are preserved. The modified IPRO procedure is run for all studies on a Linux PC cluster with 3.06-GHz Xeon CPU/4GB RAM, for a total of 4000 major iterations.
FIGURE 4.
Structures of the targeted and decoy molecules in the four case studies.
Binding of cis-verbenol
We have verified that neither α-pinene nor its oxidized forms cis-verbenol and verbenone induce transcription from the ara regulatory operon (H. Fazelinia, P. Cirino, and C. D. Maranas, The Pennsylvania State University, unpublished data). Meanwhile their calculated binding energies using CHARMM-based energy functions are significantly higher than those calculated for native inducers, indicating that these compounds are not bound by AraC.
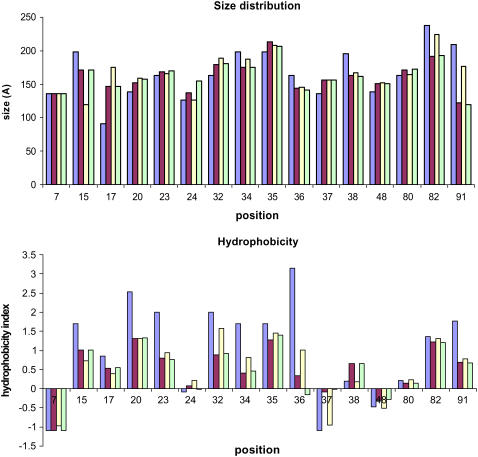
In the first case study, we address the engineering of AraC to bind cis-verbenol without considering the effect of the identified mutations on the binding of other competitive ligands. Computational results for redesigning the effector-binding site of AraC for cis-verbenol have revealed a number of important redesign trends. Of 16 design positions (i.e., positions allowed to mutate), positions Asp7, Phe35, Asn48, and His80 are always conserved as wild-type. Several mutations within the binding pocket are found that can significantly alter the calculated binding specificities of the receptor (see Table 1). Predicted mutations in positions Phe15, Phe34, Ile36, Arg38, Tyr82, and Trp91 are found to significantly lower the volume of the binding site, consistent with the fact that the new ligand (i.e., cis-verbenol) is 45% larger than l-arabinose (see Fig. 5). Also, hydrophilic amino acids tend to replace Ala17, Val20, and Leu23, which are located in a solvent-exposed area of AraC and do not directly affect the binding of the ligand.
TABLE 1.
Identified mutations for improving the binding of cis-verbenol in the presence of water molecules for the first three case studies
| Computational design
|
||||
|---|---|---|---|---|
| Design position | Wild-type residue | Binding of cis-verbenol (case study 1) | Binding of cis-verbenol but not α-pinene (case study 2) | Binding of cis-verbenol but not verbenone (case study 3) |
| 7 | Asp | WT* | WT | WT |
| 15 | Phe | Gly, Gln, His | Gly, His, Thr | Gly, Gln, His |
| 17 | Ala | Gln, Arg | Asn, Arg | Gln, Arg |
| 20 | Val | Arg, Thr, His | Trp, Gln | Tyr, Arg, Thr |
| 23 | Leu | Gln, Glu, Lys | Trp, Gln, Lys | Phe, Gln, Glu, Lys |
| 24 | Thr | Val, Asn, Gln | WT | His, Gln, Glu |
| 32 | Leu | Tyr, Asn, Arg | Gln, His, Phe | Ala, Ser, Tyr, Asn, Arg |
| 34 | Phe | Met, Glu, His | Tyr, Gln, Arg, His | Arg, Glu, Gln, His |
| 35 | Phe | WT | WT | WT |
| 36 | Ile | Thr, Asn | Asn | Asn, Asp |
| 37 | Asp | Val, Lys, Arg | Lys, Arg | Val, Lys, Arg |
| 38 | Arg | Ala, His, Trp | Ala, His | His, Ala, Trp |
| 48 | Asn | WT | WT | WT |
| 80 | His | WT | WT | WT |
| 82 | Tyr | Ala, Phe | WT | Gly, Ala, Phe |
| 91 | Trp | Gly, Ala, His | Phe, Arg, His | Ala, Gly, His |
WT refers to wild-type AraC.
FIGURE 5.
Size and hydrophobicity distribution of the wild-type amino acids (blue column), designs for case study 1 (red column), case study 2 (yellow column), and case study 3 (green column) for all design positions.
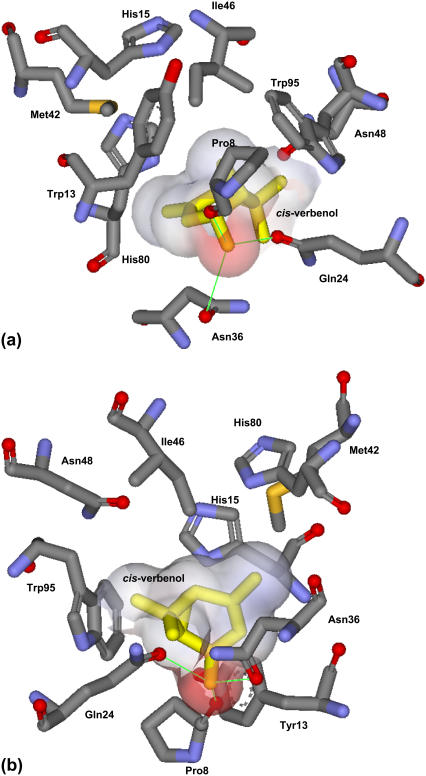
Similar to the position of l-arabinose bound to AraC, cis-verbenol is predicted to stack against the indole ring of Trp95 (see Fig. 6). Ligand binding is stabilized by hydrogen bonds and van der Waals (vdw) interactions between side chains of residues within the binding pocket and the hydroxyl and aliphatic groups of cis-verbenol, respectively. The N-terminal arm of AraC is predicted to form both direct and indirect contacts with the verbenol, resulting in complete burial of the new ligand. The hydrogen bond between the OH group of cis-verbenol and the main-chain carbonyl of Pro8 and vdw interactions between the cis-verbenol C5 and C7 methyl groups and the side chain of amino acids in positions 13 and 15 stabilize the position of the N-terminal arm. Although His93 plays an important role in binding l-arabinose, this residue is not involved in cis-verbenol binding. In contrast, amino acids in positions 36 and 42, not involved in binding and recognition of l-arabinose, are predicted to play significant roles in binding cis-verbenol. Hydrophilic amino acids predicted to replace Ile36 create a new hydrogen bond with the OH group of the new ligand and are in vdw contact with its C3 methyl group (see Fig. 6). Furthermore, wild-type Met42 is predicted to be in vdw contact with the C10 methyl group of cis-verbenol. The replacement of Thr24 (involved in binding l-arabinose) with larger amino acids such as Gln stabilizes the position of the new ligand in the binding pocket by creating a new hydrogen bond with the OH group of cis-verbenol.
FIGURE 6.
Best predicted orientation of cis-verbenol (shown with its vdw surface area) in the redesigned binding pocket of AraC from two different angles. Hydrogen bonds are shown with green lines; cis-verbenol is predicted to stack against the indole ring of Trp95 and networks of hydrogen bonds, and vdw interactions are responsible for placing the ligand in the binding pocket.
Overall, computational results (see Table 1) indicate that despite the structural and chemical difference between cis-verbenol and l-arabinose, IPRO does identify sets of mutations typically involving ∼12 mutated positions within the binding site that lower the binding energy from −16.82 kcal/mol to as low as −55.54 kcal/mol.
Binding of cis-verbenol but not α-pinene
Although the redesigns described above managed to lower the binding score substantially for cis-verbenol, this does not necessarily sharpen ligand specificity. Specifically, the binding energy of the redesigned AraC with α-pinene not only remains negative but also increases in absolute value (i.e., changes from −12.58 to −46.54 kcal/mole). This quantitatively demonstrates that when the binding energy for a new ligand is optimized with no regard to the binding energy for the competitive ligand(s), it typically leads to a redesign that appears to have broader specificity (31–33). This result motivates the need to proactively suppress the binding energy for α-pinene while optimizing the binding energy for cis-verbenol. We use the modified IPRO procedure (as described above) to accomplish this objective. With the modified version of IPRO, the binding energy of cis-verbenol is lowered from −16.82 kcal/mol to −50.19 kcal/mol while at the same time the binding energy of the redesigned AraC with α-pinene remains approximately the same (i.e., changes only from −12.58 kcal/mol to −10.03 kcal/mol). In addition to the same four positions that remain unmutated in the previous case (i.e., 7, 35, 48, and 80), residues Thr24 and Tyr82 are also conserved. The overall mutated amino acid size patterns between the two computed libraries are very similar (see Table 1 and Fig. 5), except for positions 15 and 91, where in the second case study predicted residues are more than 20% different in size. Smaller amino acids are preferred in the second library compared with the solutions found in the first library at position 15, whereas larger ones are favored at position 91. Having a smaller amino acid at position 15 reduces the magnitude of a vdw interaction implicated in the binding of α-pinene. The role of larger residues at position 91 is less clear. The hydrophobicity patterns of the mutated residues in the two libraries are also very similar. Only subtle differences can be discerned at positions 15 and 38, which are presumably implicated in the destruction of the hydrophobic interactions needed for the binding of α-pinene with AraC. Consistent with the previous case study, more hydrophilic amino acids are favored to replace the wild-type amino acids at positions Val20 and Leu23, which are located in the solvent-exposed area. Fig. 7 contrasts in a Venn diagram the mutations found in the two case studies along with the quantitative impact of each single-point mutation on the binding energy for the two ligands. We see that some mutations, when their impact on α-pinene binding is ignored, tend to improve both binding scores, whereas others only improve the binding score with cis-verbenol alone. Mutations found on systematically suppressing the binding score with α-pinene consistently favor binding only cis-verbenol. Among these mutations there is a subset common to both case studies. Notably, in both cases there seems to be a strong additive component in the action of the mutations. If the mutations in all three regions shown in Fig. 7 are combined, the binding score changes are almost additively amplified.
FIGURE 7.
AraC protein (PDB: 2ARC) was redesigned using IPRO to bind cis-verbenol without any regard to the corresponding binding score of competitive ligands (mutations are shown within the dark gray circle). The modified version of IPRO was also employed to proactively suppress the binding energy for α-pinene while optimizing the binding energy for cis-verbenol (mutations are shown within the white circle). Common mutations between the two are placed in the overlapping regions between circles.
Binding of cis-verbenol but not verbenone
Next we attempt to redesign AraC computationally to discriminate between different oxidized forms of the bicyclic monoterpene α-pinene (i.e., cis-verbenol and verbenone; see Fig. 4). These two molecules are identical at all positions except C4, where hydroxyl oxygen and carbonyl oxygen are present for cis-verbenol and verbenone, respectively. Therefore, the computational redesign simulation must identify suitable amino acid choices for the binding pocket residues based only on this small difference. Comparison between the computed libraries for this case study and the first one, where only cis-verbenol was considered as the target ligand, reveals, as expected, only subtle differences in size, hydrophobicity, and charge (see also Table 1 and Fig. 5). Notably, at positions 24 and 36, more hydrophilic amino acids are favored.
In verbenone the carbonyl oxygen acts only as a hydrogen-bond acceptor, whereas the hydroxyl oxygen in cis-verbenol is a hydrogen donor and also an acceptor. Different amino acids are selected to form hydrogen bonds with the two ligands. To discern what amino acids favor the binding of verbenone, results from the predicted library in the first case study are contrasted against results from the library in which only verbenone was considered as the target molecule. We find that wild-type Thr24 and Lys36 are favored, acting as hydrogen bond donors to interact with the nonbonding electron pairs in the carbonyl oxygen in verbenone. On the other hand, computed libraries in the third case study favor His, Gln, and Glu for position 24 and acidic amino acids Asn and Asp for position 36. These mutations allow the unprotonated imidazole nitrogen of histidine and carbonyl oxygen of the acidic amino acids to form hydrogen bonds with the hydroxyl group of cis-verbenol and thereby stabilize the position of the target molecule in the pocket.
Overall, in this case study, binding energy of cis-verbenol improves from −16.82 kcal/mol to −51.23 kcal/mol, while at the same time the binding energy of the redesigned AraC with verbenone also decreases from −15.85 kcal/mol to −34.03 kcal/mol. The inability to further suppress binding with verbenone compared with the second case study is presumably a consequence of the fact that the competing molecule here is extremely similar to the targeted ligand (see Fig. 8).
FIGURE 8.
AraC protein (PDB: 2ARC) was redesigned using IPRO to bind cis-verbenol without any regard to the corresponding binding score of competitive ligands (mutations are shown within the dark gray circle). The modified version of IPRO was also employed to proactively suppress the binding energy for verbenone while optimizing the binding energy for cis-verbenol (mutations are shown within the white circle). Common mutations between the two are placed in the overlapping regions between circles.
Specificity alteration from l- to d-arabinose
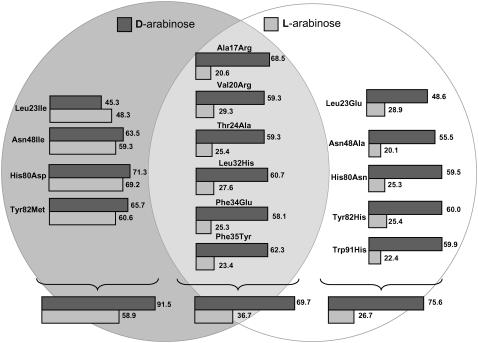
It has been shown that, in contrast to the stimulatory effect of l-arabinose, its enantiomer d-arabinose is unable to activate transcription in the ara regulatory operon (34). Here we address the computational redesign of AraC to enhance the binding score for d-arabinose as opposed to l-arabinose. This corresponds to a challenging task given that both enantiomers have exactly the same molecular groups. Therefore, the computational redesign procedure must identify appropriate residue choices for the active site based on only the differing stereogeometries between the two enantiomers. As before, first we identified mutations that improve the binding score of d-arabinose without any regard to the corresponding binding score for l-arabinose. We next accumulated (see Table 2) all the AraC redesigns that improve the d-arabinose binding score and disfavor the l-arabinose at the same time. The role of the identified mutations is more clearly elucidated by considering the underlying impact of these mutations on the volume, hydrophobicity, and charge at each position. Notably, the predicted mutations are few and involve only subtle changes in the size and hydrophobicity of the AraC binding pocket. Of 16 positions considered for redesign, we found that 10 positions are mutated away from wild-type (Table 2). As expected, because l-arabinose and its enantiomer d-arabinose have exactly the same size, we found that the average volumes at each position remain very close to those of the wild-type residues. The same holds, to a lesser extent, for the average hydrophobicity of the redesigned AraC pocket. For 11 design positions, the average hydrophobicity of residues in the redesigned AraC is very similar to the wild-type residues, although the hydrophobic characteristics of amino acids predicted to replace positions 20, 23, 36, 37, and 48 differ from the wild-type residues (Table 2).
TABLE 2.
Identified mutations for improving the binding of d-arabinose in the presence of water for the fourth case study
| Computational design
|
|||
|---|---|---|---|
| Design position | Wild-type residue | Binding of d-arabinose | Binding of d- but not l-arabinose |
| 7 | Asp | WT* | WT |
| 15 | Phe | WT | WT |
| 17 | Ala | Gly,Tyr, Arg | Gly, Tyr, Arg |
| 20 | Val | Gln, Glu, Arg | Thr, Gln, Arg |
| 23 | Leu | Met,Trp, Lys, Ile | Lys, Glu, Gln |
| 24 | Thr | Asn, Gly, Ala | Ala |
| 32 | Leu | Tyr, Phe, His | Ala, Gly, Phe, His |
| 34 | Phe | Met, Arg, Glu | His, Met, Glu |
| 35 | Phe | Gln, Tyr | Glu, Tyr |
| 36 | Ile | WT | WT |
| 37 | Asp | WT | WT |
| 38 | Arg | WT | WT |
| 48 | Asn | Lys, Gln, Ala, Ile | Gln, Ala |
| 80 | His | Gln, Asp, Ala | Gln, Phe, Asn |
| 82 | Tyr | Thr, Ala, Met | Phe, Glu, His |
| 91 | Trp | WT | His, Phe, Met |
WT refers to wild-type AraC.
Interestingly, despite the difference in topology between l- and d-arabinose, the sugar is stabilized similarly in the wild-type and all redesigned AraC variants (10). Specifically, in the redesigned AraC, Arg38 forms a bidentate interaction with two of the hydroxyl groups of the bound sugar, resembling the interactions that exist between Arg38 and l-arabinose in wild-type AraC. Meanwhile, as in the wild-type AraC, all predicted structures have d-arabinose stacking against the indole ring of Trp95, and the ligand position is stabilized by a network of hydrogen bonds between d-arabinose and binding pocket residue side chains (see Fig. 9).
FIGURE 9.
Location of l-arabinose (a) and d-fucose (b) in the binding pocket of the AraC (PDBs:2ARC, 2AAC). Model-predicted positions of d-arabinose in the presence (c) and in the absence (d) of water-mediated interactions. IPRO-predicted ligand positions more closely match those found from the crystal structures when water is included. Computational results indicate that in both cases the position of the N-terminal arm of AraC is stabilized by the main chain carbonyl of Pro8, which makes a hydrogen bond with one of the hydroxyl groups of sugars. In both cases, the sugar stacks against the indole ring of Trp95.
In all computed redesigns of AraC in this case study, the position of the N-terminal arm is stabilized by a hydrogen bond between the main chain carbonyl of Pro8 and one of the hydroxyl groups of the bound sugar (see Fig. 9). This interaction is very similar to that found in wild-type AraC, where the anomeric hydroxyl group (OH-1) of the bound sugar interacts with the main chain carbonyl of Pro8 (10). It is important to note that no specific information about the stabilizing interactions was a priori provided to the IPRO model. As in the previous case studies, the redesign of AraC substantially improves the binding energy for d-arabinose (from 15.43 kcal/mol to −107.41 kcal/mol), but the binding score for l-arabinose is also lowered (from −37.21 kcal/mol to −78.35 kcal/mol).
In the modified version of IPRO, mutation changes that suppress binding with l-arabinose were next identified (see Table 2). In 12 of 16 design positions, the mutations are very similar to the ones found when the binding score of d-arabinose was minimized. The binding score with d-arabinose is lowered from 15.43 kcal/mol to −89.698 kcal/mol, whereas the binding score for l-arabinose increases in this scenario from −37.21 kcal/mol to −18.03 kcal/mol. Comparisons between computed libraries for these two cases reveal only subtle differences in charge, hydrophobicity, and size distributions. One such difference is the replacement of Thr24 with aliphatic residues, partly destroying the hydrogen bond network involved in binding l-arabinose and thus diminishing the affinity of AraC for its natural effector. In contrast, mutating His80 to hydrophilic residues (Gln, Ser, Asn) creates a new hydrogen bond with d-arabinose (but not l-arabinose). The binding scores for individual mutations were calculated and are presented in Fig. 10 in the form of a Venn diagram for both cases.
FIGURE 10.
AraC protein (PDB: 2ARC) was redesigned using IPRO to bind d-arabinose without any regard to the corresponding binding score for l-arabinose (mutations are shown within the dark gray circle). The modified version of IPRO was also employed to proactively suppress the binding energy for l-arabinose while optimizing the binding energy for d-arabinose (mutations are shown within the white circle). Common mutation positions are placed in the overlapping region between the two circles.
DISCUSSION
In this article, we introduced a modified version of the IPRO protein design framework to enable the systematic redesign of proteins for improved binding affinity for a targeted ligand while the binding affinity for decoy ligands remains low. Computationally, this leads to a nested optimization structure where, in the inner stage, the rotamer optimization problem is solved separately for all ligands, whereas in the outer stage, residue redesign choices are made (see Fig. 1). This procedure was benchmarked using AraC as a model system by favoring the binding of targeted ligands (i.e., cis-verbenol or d-arabinose) while suppressing the binding energy of competing molecules (i.e., verbenone, α-pinene, and l-arabinose).
We found that failure to suppress the binding affinity for competing ligands leads to a universal improvement in the binding scores not only for the targeted but also for the decoy ligands. The modified IPRO procedure was shown to be capable of decoupling the two and identifying mutations that improve the binding only with the desired ligand. As expected, this decoupling was most difficult to achieve for very similar molecules (i.e., cis-verbenol and verbenone), which differ by only one group. Somewhat surprisingly, this decoupling was much easier for enantiomers (i.e., l- and d-arabinose), suggesting that proteins can be more readily modified to discern differences in ligand topology rather than ligand small group substitutions.
In all four case studies the ligand was stacked against the indole ring of Trp95, and networks of hydrogen bonds and vdw interactions were responsible for placing the respective ligand in the binding pocket. The position of the N-terminal arm, which plays a crucial role in the “light-switch” mechanism of the AraC protein, was universally stabilized by direct hydrogen bonding between the oxygen of the main chain carbonyl of Pro8 and one hydroxyl group of the target ligand. The average volume of the amino acids in the binding pocket was generally changed according to the size of the target ligand to improve the ligand-protein fit by compensating for differences in ligand structure.
Comparisons between the different computed libraries reveal that, in all case studies, all mutations found on systematically suppressing the binding score with the decoy consistently favor binding with only the target ligand. The number of common mutations predicted with and without a decoy strongly depends on the similarity of the chemistry and structure between the target and decoy molecules. Finally, in all cases, improvements in the binding scores are largely cumulative with respect to individual point mutations, alluding to a strongly additive mechanism of the effect of mutations.
Acknowledgments
We thank Dr. Manish Saraf for useful discussions and suggestions and Chuan Nain Kenny-Ou for his help with some of the docking calculations.
We gratefully acknowledge financial support from the National Science Foundation Awards BES0331047 (to C.D.M.) and BES0519516 (to P.C.C. and C.D.M.).
References
- 1.Saraf, M. C., G. L. Moore, N. M. Goodey, V. Y. Cao, S. J. Benkovic, and C. D. Maranas. 2006. IPRO: an iterative computational protein library redesign and optimization procedure. Biophys. J. 90:4167–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold, F. H., and G. Georgiou. 2003. Directed Evolution Library Creation. Methods in Molecular Biology, Vol. 231. Humana Press, Totowa, NJ.
- 3.Arnold, F. H., and G. Georgiou. 2003. Directed Enzyme Evolution: Screening and Selection Methods. Methods in Molecular Biology, Vol. 230. Humana Press, Totowa, NJ.
- 4.Moore, G. L., and C. D. Maranas. 2004. Computational challenges in combinatorial library design for protein engineering. AIChE J. 50:262–272. [Google Scholar]
- 5.Looger, L. L., M. A. Dwyer, J. J. Smith, and H. W. Hellinga. 2003. Computational design of receptor and sensor proteins with novel functions. Nature. 423:185–190. [DOI] [PubMed] [Google Scholar]
- 6.Allert, M., S. S. Rizk, L. L. Looger, and H. W. Hellinga. 2004. Computational design of receptors for an organophosphate surrogate of the nerve agent soman. Proc. Natl. Acad. Sci. USA. 101:7907–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korkegian, A., M. E. Black, D. Baker, and B. L. Stoddard. 2005. Computational thermostabilization of an enzyme. Science. 308:857–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dwyer, M. A., L. L. Looger, and H. W. Hellinga. 2004. Computational design of a biologically active enzyme. Science. 304:1967–1971. [DOI] [PubMed] [Google Scholar]
- 9.Schleif, R. 2003. AraC protein: a love-hate relationship. Bioessays. 25:274–282. [DOI] [PubMed] [Google Scholar]
- 10.Soisson, S. M., B. MacDougall-Shackleton, R. Schleif, and C. Wolberger. 1997. Structural basis for ligand-regulated oligomerization of AraC. Science. 276:421–425. [DOI] [PubMed] [Google Scholar]
- 11.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schleif, R., and C. Wolberger. 2004. Arm-domain interactions can provide high binding cooperativity. Protein Sci. 13:2829–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doyle, M. E., C. Brown, R. W. Hogg, and R. B. Helling. 1972. Induction of the ara operon of Escherichia coli B-r. J. Bacteriol. 110:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soisson, S. M., B. MacDougall-Shackleton, R. Schleif, and C. Wolberger. 1997. The 1.6 angstrom crystal structure of the AraC sugar-binding and dimerization domain complexed with d-fucose. J. Mol. Biol. 273:226–237. [DOI] [PubMed] [Google Scholar]
- 15.Wu, M., and R. Schleif. 2001. Mapping arm-DNA-binding domain interactions in AraC. J. Mol. Biol. 307:1001–1009. [DOI] [PubMed] [Google Scholar]
- 16.Chen, Z., B. S. Katzenellenbogen, J. A. Katzenellenbogen, and H. Zhao. 2004. Directed evolution of human estrogen receptor variants with significantly enhanced androgen specificity and affinity. J. Biol. Chem. 279:33855–33864. [DOI] [PubMed] [Google Scholar]
- 17.Krishnan, A. V., X. Y. Zhao, S. Swami, L. Brive, D. M. Peehl, K. R. Ely, and D. Feldman. 2002. A glucocorticoid-responsive mutant androgen receptor exhibits unique ligand specificity: therapeutic implications for androgen-independent prostate cancer. Endocrinology. 143:1889–1900. [DOI] [PubMed] [Google Scholar]
- 18.Wernisch, L., S. Hery, and S. J. Wodak. 2000. Automatic protein design with all atom force-fields by exact and heuristic optimization. J. Mol. Biol. 301:713–736. [DOI] [PubMed] [Google Scholar]
- 19.MacKerell, A. D., B. Brooks, C. L. Brooks, L. Nilsson, B. Roux, Y. Won, and M. Karplus. 1998. CHARMM: The energy function and its parameterization with an overview of the program. In The Encyclopedia of Computational Chemistry, R. Schleyer, editor. John Wiley & Sons, Chichester. 271–277.
- 20.Chen, R., L. Li, and Z. Weng. 2003. ZDOCK: an initial-stage protein-docking algorithm. Proteins. 52:80–87. [DOI] [PubMed] [Google Scholar]
- 21.Jiang, X., H. Farid, E. Pistor, and R. S. Farid. 2000. A new approach to the design of uniquely folded thermally stable proteins. Protein Sci. 9:403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reed, W. L., and R. F. Schleif. 1999. Hemiplegic mutations in AraC protein. J. Mol. Biol. 294:417–425. [DOI] [PubMed] [Google Scholar]
- 23.Ross, J. J., U. Gryczynski, and R. Schleif. 2003. Mutational analysis of residue roles in AraC function. J. Mol. Biol. 328:85–93. [DOI] [PubMed] [Google Scholar]
- 24.Levy, Y., and J. N. Onuchic. 2004. Water and proteins: a love-hate relationship. Proc. Natl. Acad. Sci. USA. 101:3325–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papoian, G. A., J. Ulander, M. P. Eastwood, Z. Luthey-Schulten, and P. G. Wolynes. 2004. Water in protein structure prediction. Proc. Natl. Acad. Sci. USA. 101:3352–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eisenberg, D., and A. D. McLachlan. 1986. Solvation energy in protein folding and binding. Nature. 319:199–203. [DOI] [PubMed] [Google Scholar]
- 27.Wilcox, G. 1974. The interaction of l-arabinose and d-fucose with AraC protein. J. Biol. Chem. 249:6892–6894. [PubMed] [Google Scholar]
- 28.Greenblatt, J., and R. Schleif. 1971. Arabinose C protein: regulation of the arabinose operon in vitro. Nat. New Biol. 233:166–170. [DOI] [PubMed] [Google Scholar]
- 29.Englesberg, E., J. Irr, J. Power, and N. Lee. 1965. Positive control of enzyme synthesis by gene C in the l-arabinose system. J. Bacteriol. 90:946–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soisson, S. M., B. MacDougall-Shackleton, R. Schleif, and C. Wolberger. 1997. The 1.6 A crystal structure of the AraC sugar-binding and dimerization domain complexed with d-fucose. J. Mol. Biol. 273:226–237. [DOI] [PubMed] [Google Scholar]
- 31.Varadarajan, N., J. Gam, M. J. Olsen, G. Georgiou, and B. L. Iverson. 2005. Engineering of protease variants exhibiting high catalytic activity and exquisite substrate selectivity. Proc. Natl. Acad. Sci. USA. 102:6855–6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hardt, M., and R. A. Laine. 2004. Mutation of active site residues in the chitin-binding domain ChBDChiA1 from chitinase A1 of Bacillus circulans alters substrate specificity: use of a green fluorescent protein binding assay. Arch. Biochem. Biophys. 426:286–297. [DOI] [PubMed] [Google Scholar]
- 33.Wilks, H. M., K. W. Hart, R. Feeney, C. R. Dunn, H. Muirhead, W. N. Chia, D. A. Barstow, T. Atkinson, A. R. Clarke, and J. J. Holbrook. 1988. A specific, highly active malate dehydrogenase by redesign of a lactate dehydrogenase framework. Science. 242:1541–1544. [DOI] [PubMed] [Google Scholar]
- 34.Schleif, R. 2000. Regulation of the l-arabinose operon of Escherichia coli. Trends Genet. 16:559–565. [DOI] [PubMed] [Google Scholar]
- 35.Dunbrack, R. L., Jr., and M. Karplus. 1993. Backbone-dependent rotamer library for proteins. Application to side-chain prediction. J. Mol. Biol. 230:543–574. [DOI] [PubMed] [Google Scholar]
- 36.Dunbrack, R. L., Jr., and M. Karplus. 1994. Conformational analysis of the backbone-dependent rotamer preferences of protein sidechains. Nat. Struct. Biol. 1:334–340. [DOI] [PubMed] [Google Scholar]