Abstract
The interaction between sensory rhodopsin II (SRII) and its transducer HtrII was studied by the time-resolved laser-induced transient grating method using the D75N mutant of SRII, which exhibits minimal visible light absorption changes during its photocycle, but mediates normal phototaxis responses. Flash-induced transient absorption spectra of transducer-free D75N and D75N joined to 120 amino-acid residues of the N-terminal part of the SRII transducer protein HtrII (ΔHtrII) showed only one spectrally distinct K-like intermediate in their photocycles, but the transient grating method resolved four intermediates (K1–K4) distinct in their volumes. D75N bound to HtrII exhibited one additional slower kinetic species, which persists after complete recovery of the initial state as assessed by absorption changes in the UV-visible region. The kinetics indicate a conformationally changed form of the transducer portion (designated Tr*), which persists after the photoreceptor returns to the unphotolyzed state. The largest conformational change in the ΔHtrII portion was found to cause a ΔHtrII-dependent increase in volume rising in 8 μs in the K4 state and a drastic decrease in the diffusion coefficient (D) of K4 relatively to those of the unphotolyzed state and Tr*. The magnitude of the decrease in D indicates a large structural change, presumably in the solvent-exposed HAMP domain of ΔHtrII, where rearrangement of interacting molecules in the solvent would substantially change friction between the protein and the solvent.
INTRODUCTION
Some species of archaea, such as Halobacterium salinarum and Natronomonas pharaonis, have several retinylidene photoreceptors (archaeal rhodopsins) in their cytoplasmic membrane. Four types of archaeal rhodopsins have been identified so far; bacteriorhodopsin (BR), halorhodopsin (HR), sensory rhodopsin I (SRI), and sensory rhodopsin II (SRII, also called phoborhodopsin) (1–5). All of these proteins show similar structural architectures, seven transmembrane α-helices enclosing a common all-trans retinal chromophore, which is bound to a lysine residue on the seventh helix via a protonated Schiff base linkage. They exhibit similar cyclic photochemical reaction cycles (photocycles) initiated by light-absorption. In BR, and with some variations in the others, photoisomerization of retinal from the all-trans to 13-cis configuration is followed by a proton transfer from the protonated Schiff base to its aspartate counterion, later reprotonation of the Schiff base from a different aspartic acid residue, and finally thermal-reisomerization of the chromophore and deprotonation of the counterion aspartic acid (recovery of the initial state). Each of these steps is accompanied by conversion of quasi-stable intermediates with distinct optical properties designated K (or K/L), M, N, O, and the initial (unphotolyzed) state.
Despite their similarities, the functions of these proteins are distinctly different. BR and HR are light-driven ion pumps, which transfer a proton or a chloride across the cytoplasmic membrane, respectively, and generate electrochemical gradients for ATP synthesis and other energy-requiring processes. In contrast, SRI and SRII are photosensory receptors mediating signal transduction controlling motility. SRI modulates the flagellar motor rotation pattern to achieve positive phototaxis toward orange light, and via two-photon photochemistry negative phototaxis away from harmful UV-violet light. SRII absorbs blue-green light and initiates negative phototaxis to prevent cell exposure to intense sunlight whose maximum intensity is located near 500 nm.
The photosensors send interprotein signals to bound transducers HtrI and HtrII. The transducers are homologous to the well-characterized eubacterial chemotaxis receptors, such as the aspartate and serine receptors in Escherichia coli (6–8). As in the homologous proteins, the transducers are anchored to the membrane with two N-terminal transmembrane helices TM1 and TM2, the second of which is linked through a histidine kinase-adenylyl cyclases-methyl-accepting chemotaxis protein-phosphatases (HAMP) domain consisting of two α-helical amphipathic sequences to a cytoplasmic domain in which each helical sequence assumes a hairpin structure and forms a four-helix coiled-coil bundle in transducer dimers. The contact sites of Htrs to SRs are largely within the membrane, are specific to the respective SRs (9), and for SRII-HtrII have been resolved by x-ray crystallography (10).
Complex formation of SRs and Htrs were first demonstrated for SRI by the loss of pH-dependence of the M-decay rate in the presence of HtrI, which was interpreted as blockage of SRI's cytoplasmic proton-conducting channel by HtrI (11). HtrII from H. salinarum was also found to alter the photochemistry of SRII by accelerating the photocycle, which was explained as HtrII facilitating proton circulation between the Schiff base, the counterion Asp73, and an unidentified proton release group (12,13).
Kinetic optical absorption spectroscopy has been effectively applied to monitor chemical reactions in the vicinity of the chromophore, but it is not suited for detecting conformational changes of the photoreceptors and transducers distant from the chromophore. To overcome this limitation, several photothermal techniques have been used to monitor the thermodynamic properties of transient species during reactions of photoresponse proteins (14,15). We previously studied the dynamics of SRII from N. pharaonis (NpSRII) and a fusion protein with the receptor-interacting N-terminal portion of the cognate transducer, ΔNpHtrII (residues 1–157) by monitoring volume change and enthalpy change in the time domain by the pulsed laser-induced transient grating (TG) method (16). Volume change should provide a more direct measure of the protein conformational change including regions of the protein complex far from the chromophore-binding pocket. We observed a relatively large volume change of the NpSRII-ΔNpHtrII complex compared with that of NpSRII during the M′′→O transition, which may be a key interprotein signal transfer step from NpSRII to ΔNpHtrII.
Although this TG technique has provided new information on the kinetics, there were experimental difficulties for the application to the wild-type NpSRII system. First, the TG signal measures not only the volume change and enthalpy change, but also is influenced by the absorption change that is known as the population grating component (population grating is explained in Principles and Analysis below). If the photoreaction induces a large spectral shift, the signal due to the absorption change may mask the other contributions, which contain valuable information on the conformational changes. In the case of NpSRII, the absorption due to the blue-shifted M intermediate generates a large TG signal due to its population grating contribution and it was difficult to monitor conformational changes in this transition. Furthermore, there were many dynamics accompanying the absorption spectrum changes in the photoreaction of NpSRII. Because of the multistep processes with large amplitudes of the population grating, the influence of the transducer on the volume change or molecular diffusion was difficult to assess.
The D75N mutant of NpSRII offers a possibility to overcome these difficulties. The Asp75 residue is the counterion of the protonated Schiff base and is the acceptor of the proton from the protonated Schiff base of 13-cis retinal. Deprotonation of the Schiff base upon the proton transfer causes the large blue shift of the absorption spectrum in the M state. Therefore, D75N lacks M in the photocycle (17), but does not abolish the photophobic response to green-blue light (18) excluding that M-formation is prerequisite for the molecular activation. Both HtrII-free and HtrII-complexed D75N SRII are converted upon light stimulation to more than one red-shifted intermediate, which were distinguished kinetically or by infrared spectroscopy (17), although silent in terms of the visible spectra (19).
The photocycle of D75N contains a single red-shifted intermediate in the visible region in its photocycle, which is very favorable for TG measurements. In this study, we measured the photoreaction dynamics of the D75N mutant and the fusion protein with the interacting N-terminal portion of NpHtrII by the TG method to clarify the signal transduction process.
Several spectroscopic methods have been applied to reveal aspects of the photoreaction dynamics of NpSRII and NpHtrII. Laser flash photolysis (20,21), FT-IR measurements (22–28), and site-directed spin-labeling and EPR (29,30), have provided valuable information regarding localized changes, but unlike TG measurements none of these methods focus on global conformational changes. The photoacoustic method (31–33) measures a global property but is largely limited to early times, unlike TG analysis, which is applicable in a wide time window.
The TG signal showed very prominent features in the photoreaction of D75N as well as that of the fusion protein of D75N with 120 amino-acid residues of the N-terminal part of NpHtrII (D75N-ΔNpHtrII). We identified four intermediates, which were not detected by the absorption measurements for D75N. For the fusion protein, D75N-ΔNpHtrII, the volume changes of intermediates were different from those of D75N, although the reaction rates were very similar. Furthermore, very interestingly, a large diffusion coefficient (D) change was detected for D75N-ΔNpHtrII. Such a D-change was absent in the case of D75N without the HtrII transducer. On the basis of previous reports showing that D carries information on the intermolecular interaction between protein and water molecules including global conformational changes of proteins (34–37), we interpret this result in terms of a large conformational change at the linker region (the HAMP domain). This process might be related to the physiological signal transferring process from the receptor binding region to the cytoplasmic domain and provide insights into several related types of receptors including bacterial chemotaxis receptors, which contain homologous HAMP domains.
MATERIALS AND METHODS
Measurements
The second harmonic light from an Nd:YAG laser (λ = 532 nm, Minilite Continuum, Santa Clara, CA) with a pulse width of ∼6 ns was used for excitation of sample solutions. For recording absorption changes over the near-UV to visible range at various times after excitation, light from a Xe-lamp was introduced to the sample by a counter-propagation geometry to the excitation light. The probe light was led to a monochrometer by an optical fiber, and detected by an ICCD camera system (PI-MAX/PI-MAX2 System, Roper Scientific, Trenton, NJ). The intensity at each wavelength was recorded by a computer. For measuring the time development of the absorption change in detail at a specific wavelength, a continuous wave probe beam (λ = 594 nm) from an He-Ne laser was used. The change of the probe light intensity was monitored by a photomultiplier tube and accumulated by a digital oscilloscope (model No. TDS-520, Tektronix, Beaverton, OR).
The experimental setup for the TG measurement was similar to that reported previously (38–43). Briefly, the excitation laser beam was split into two by a beam splitter, and crossed inside a sample cell. A continuous wave laser light from a diode laser (λ = 840 nm) was used for the probe light. A part of the probe beam diffracted by the refractive index modulation was detected by a photomultiplier tube and the temporal profile was recorded by a digital oscilloscope. Usually, 300 signals were averaged to improve the signal/noise ratio. The repetition rate of the pump beam was 0.5 Hz to avoid multiexcitation of the sample.
Behavioral analysis used H. salinarum Pho81Wr- cells, which lack expression of bop, hop, sopI, sopII, htrI, and htrII, and exhibit low level of carotenoid pigments. The cells were transformed with a H. salinarum expression plasmid containing NpsopII and NphtrII genes under control of the fdx promoter (pJS004) or an expression plasmid containing a gene encoding NpSRII with the D75N mutation joined by a flexible linker peptide (ASASNGAS) to full-length NpHtrII under the control of the fdx promoter (pJS004_FL3).
The cultures were grown to early stationary phase as described in McCain et al. (44). Motility responses by the transformed H. salinarum cells to light stimuli were assayed by computer-assisted cell tracking and motion analysis. The early stationary phase cultures were diluted 1:10 in fresh CM (complex media containing 250 g NaCl, 0.2 g MgCl2, 2 g KCl, 3 g Na3 citrate, 20g Mg2SO4, 10 g peptone per 1 liter) and incubated for 1 h at 37°C with agitation. Motility responses to green light photostimuli were recorded with infrared light (>700 nm). Cell motility was captured in real-time avi files using a Flashbus Spectrim Lite Video Capture PCI Card (Integral Technologies, Indianapolis, IN) on a Dell Dimension 8500 that was running VirtualDub 1.6.14 (www.virtualdub.org) AVI encoder software for video capture. VirtualDub was set to record 10 frames per second during 10 s of cell swimming. Swimming cells were subjected to a 100-ms pulse of 540 ± 20 nm light 2 s after initiation of video capture. The stimulus was delivered from a Nikon 100 W He/Xe short arc lamp (Nikon, Tokyo, Japan). The cell tracking and motion analysis software package Celltrak 1.2 Beta (Motion Analysis Corporation, Santa Rosa, CA), running on a Dell Dimension 9150, was then used to analyze the cell responses to the photostimuli. Reversal frequencies of the cells were assessed by monitoring the rate of change in direction (RCD) divided by the speed of the cells (SPD) as a function of time.
Plasmid construction and site-directed mutagenesis
The plasmid pCY9 encodes NpSRII with a six-histidine extension at the C-terminus (18), whereas construct FP120 encoded a fusion protein in which full-length SRII and the 120 N-terminal residues of HtrII containing an additional His6 tag at the C-terminus are joined by the flexible linker peptide. The fusion gene was placed in plasmid pET21d (Novagen, Madison, WI) under control of the T7 promoter. Asp75-to-Asn substitutions were as described in Bergo et al. (27). The plasmid was transformed into the Escherichia coli BL21 (DE3) strain.
Protein expression and purification
The BL21 (DE3) cells were grown in LB medium containing 50 μg/ml ampicillin to an absorbance at 600 nm of ∼0.3, and protein expression was induced by addition of 1 mM isopropyl β-D-thiogalactopyranoside and 5 μM all-trans-retinal. After 3 h of induction, the cells were harvested by centrifugation at 1000 × g, resuspended in 50 mM Tris/HCl, pH 7.0, 5 mM MgCl2, and 1 mM phenylmethylsulfonyl fluoride buffer, and disrupted by a microfluidizer (Microfluidics, Newton, MA). Cell debris was sedimented by centrifugation at 1000 × g and the membrane fraction in the supernatant was then harvested by ultracentrifugation. The membranes were solubilized in 300 mM NaCl, 10 mM imidazole, and 50 mM potassium phosphate, pH 7.6, 1.5% n-octyl-β-D-glucoside (OG). After centrifugation of the solubilized membranes, the supernatant was incubated with nickel-nitrilotriacetic acid-agarose (Qiagen, Venlo, The Netherlands), and the His-tagged protein was eluted with 250 mM imidazole in 50 mM Tris-HCl, pH 7.6, and 300 mM NaCl, 0.5% OG. To remove imidazole, the sample was dialyzed against 50 mM Tris-HCl, pH 7.0, and 300 mM NaCl for 1 h. Concentrated OG was added to the sample to the final concentration of 1%.
For a typical measurement, the concentration of sample protein was adjusted to 200 μM and solubilized in 1% OG, 50 mM Tris-HCl, pH 7.0, 300 mM NaCl. The sample solution was filtered by a cellulose acetate membrane filter (Cosmospin Filter G, pore size = 0.2 μm; Nacalai Tesque, Kyoto, Japan) and placed in a quartz cell (optical path length = 2 mm). Any scattered probe light from the sample solution was carefully removed to avoid a heterodyne contribution to the TG signal.
PRINCIPLES AND ANALYSIS
The principle of the TG measurement has been described previously (38,39,45–50). In a TG experiment, a photoinduced reaction is initiated by the spatially modulated light intensity that is produced by the interference of two excitation light waves. The sinusoidal modulations of the concentrations of the reactant and the product lead to the modulation with the same pattern in the refractive index (δn) at the probe wavelength. This modulation is monitored by the diffraction efficiency of a probe beam (TG signal). The intensity of the TG signal is proportional to the square of δn.
The refractive index change after photoexcitation comes mainly from the thermal energy released (thermal grating) and created (or depleted) chemical species by the photoreaction (species grating). The species grating signal consists of two contributions: the population grating, which comes from the absorption spectrum change; and the volume grating, which is due to the molecular volume change. If there is no absorption spectrum change, the population grating term vanishes and the volume change can be calculated from the amplitude of the species grating signal intensity as described before (16,44,51,52).
The temporal profile of the TG signal reflects kinetics of reaction including the volume change, energy change, and diffusion processes. If a reaction completes very quickly (Scheme 1),
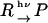 |
(Scheme1) |
where R and P denote the reactant and the product, respectively, the signal intensity decays by the smearing process of the spatial modulations of the refractive index by thermal diffusion or translational molecular diffusion. Solving diffusion equations, one finds that the decay rate constant of the thermal grating signal is given by Dthq2 (Dth, thermal diffusivity of the solution; q, grating wave number) (38,39,42). Similarly, the species grating signal decays with a rate constant of Dq2, where D is the diffusion coefficient of the chemical species. In such a case, the time development of the TG signal can be expressed by a sum of exponential functions (38,39),
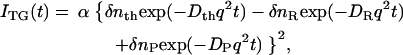 |
(1) |
where α is a constant, δnth (<0) is the refractive index change by the temperature rise, and δnR (>0) and δnP (>0) are the refractive index changes due to the presence of the reactant and the product, respectively. The sign of δnR is negative because the phase of the spatial concentration modulation is shifted 180° from that of the product.
If the back reaction from the product occurs,
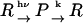 |
(Scheme 2) |
the rate constant of the back reaction (k) should be included in the rate constants and DPq2 should be replaced by DPq2+k. (40) The TG signal due to the species grating is expressed by
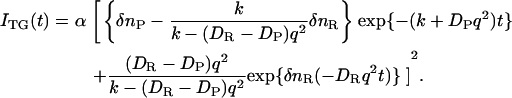 |
(2) |
In this research, we encountered a more complex reaction of the sensory rhodopsin. For example, let us consider the following reaction:
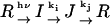 |
(Scheme 3) |
The diffusion equations for this reaction are expressed by
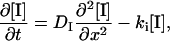 |
(3) |
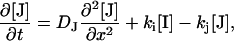 |
(4) |
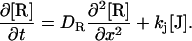 |
(5) |
Solving these equations, we obtain the following equation for describing the species grating signal:
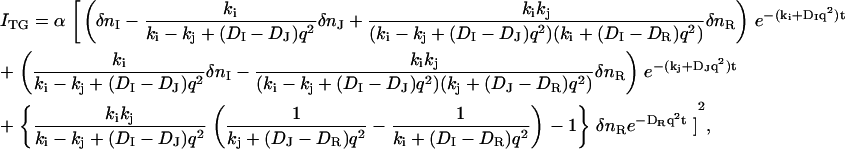 |
(6) |
where δn, k, and D denote the refractive index changes, decay rate constants, and diffusion coefficients, respectively. The subscripts I, J, and R indicate the I and J intermediates and the reactant (R), respectively.
RESULTS
Behavioral analysis of H. salinarum cells expressing D75N SRII-HtrII
It was previously shown that D75N of NpSRII coexpressed with NpHtrII mediates strong repellent responses, as does wild-type NpSRII, indicating that the D75N mutation does not interfere with molecular activation of the photoreceptor (18). Joining the C-terminus of NpSRII through a flexible peptide linker made up of ASASNGASA to the N-terminus of NpHtrII was also shown not to prevent signaling between the photoreceptor and the transducer (53).
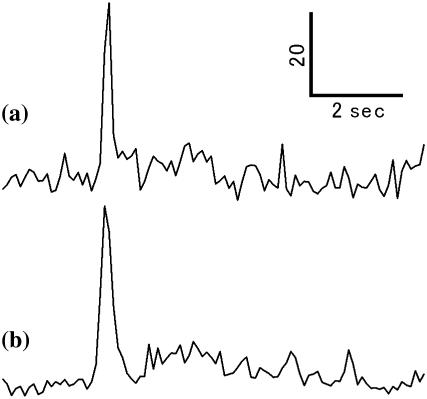
The effect of D75N mutation in the joined NpSRII and NpHtrII on phototaxis signaling was checked by measuring the behavior of H. salinarum transformants upon delivery of 540-nm light pulses (100 ms). As described in Materials and Methods, cell tracks were recorded and the reversal frequencies of the cells assessed by averaging RCD/SPD of ∼100 cells. Cells expressing NpSRII coexpressed with NpHtrII exhibit strong repellent responses to the light pulse, which are manifested as an abrupt transient increase in the reversal frequency of the cells in response to the light pulse stimulus (Fig. 1 a). A nearly identical reversal frequency response of the joined protein with the D75N mutation was observed (Fig. 1 b), proving that this molecule is also functional. Therefore, it is expected that conformational changes of the NpSRII D75N portion of the protein would be transmitted to the transducer portion of the joined protein in the same manner as in the wild-type complex. Below we analyzed kinetics of conformational changes of the joined protein with truncation at the position 120 of HtrII (ΔNpHtrII). ΔNpHtrII, which contains the two transmembrane helices and the cytoplasmic HAMP domain, was shown previously to confer binding affinity to SRII comparable to that of the longer version of HtrII (54).
FIGURE 1.
RCD/SPD transients showing reversal frequency responses of H. salinarum cells expressing NpSRII and NpHtrII (a) and D75N joined to full-length NpHtrII. (b) A pulse of 100-ms 540-nm light stimuli was delivered to the cells 2 s after initiation of the recording.
Reaction kinetics monitored by transient absorption changes
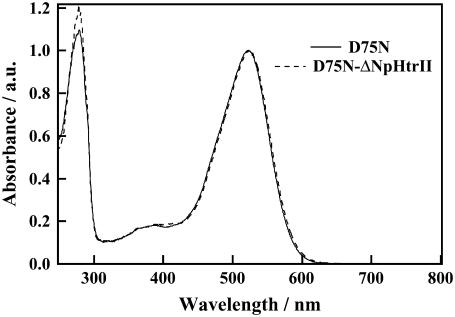
Fig. 2 shows absorption spectra of D75N and D75N-ΔNpHtrII. The peak wavelengths, λmax, in the visible region, were 522 nm and 524 nm for D75N and D75N-ΔNpHtrII, respectively. The nearly identical spectra show that the effect on the dark spectrum of the transducer protein attached to D75N is negligible.
FIGURE 2.
Absorption spectra of D75N and D75N-ΔNpHtrII in 1% OG, 50 mM Tris-HCl (pH = 7.0), 300 mM NaCl. Both spectra are normalized by the visible peak intensity.
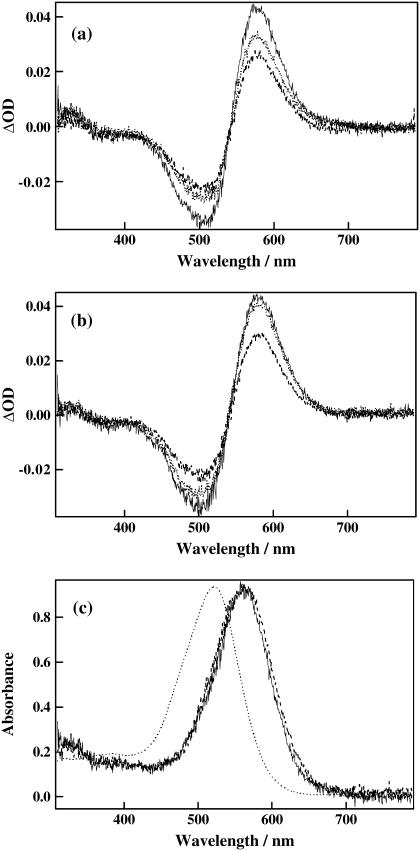
We examined the reaction kinetics of our samples by monitoring absorption spectrum changes after photoexcitation (Fig. 3). Both D75N and D75N-ΔNpHtrII showed similar red-shifts with an isosbestic point at 542 nm, indicating similar absorption spectra also for the intermediate species, although the decay rate of D75N-ΔNpHtrII is faster than that of D75N. The isosbestic points indicate presence of only one spectrally distinct intermediate species in the measured region of the spectrum in this time window (from microseconds to milliseconds).
FIGURE 3.
(a) Transient absorption spectra of D75N after 100 μs (solid line), 10 ms (dotted line), and 20 ms (dashed line) from photoexcitation. (b) Transient absorption spectra of D75N-ΔNpHtrII after 100 μs (solid line), 1 ms (dotted line), and 5 ms (dashed line) from photoexcitation. (c) Absorption spectra of the K-intermediate for D75N (solid line) and D75N-ΔNpHtrII (dashed line). For comparison, the absorption spectrum of the dark state of D75N is superposed (dotted line).
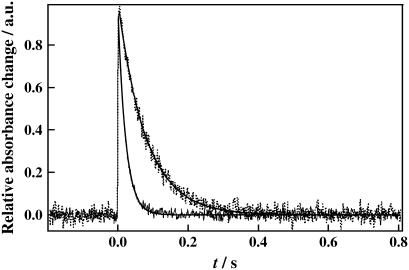
The fact that the kinetics of D75N and D75N-ΔNpHtrII, monitored at 594 nm (Fig. 4), can be fitted with a single exponential function with time constants of 81 ms and 12 ms, respectively, further supports existence of a single spectral species in their photocycles. The recovery time of D75N is more than ten times shorter than that of the wild-type NpSRII. Previously, it was shown that the reaction kinetics of D75N in purple membrane lipids showed two kinetic phases with lifetimes of 7 and 28 ms (17). The authors attributed the faster component to transformation from the K intermediate to the second intermediate, and the slower to the recovery of the unphotolyzed state. Sudo et al. (55) reported similar kinetics and designated the second intermediate as the Z-intermediate. The differences in the kinetics from our single exponential behavior might be due to the different solubilization conditions: we used OG detergent, whereas Schmies et al. (17) reconstituted the system in purple membrane lipids and Sudo et al. (55) solubilized in n-dodecyl-β-D-maltoside (DM).
FIGURE 4.
Time dependence of the transient absorption signal of D75N (dashed line) and D75N-ΔNpHtrII (solid line) monitored at 594 nm. The best fitted curves by a single exponential function are shown by the smooth lines.
Fig. 3 c shows the calculated spectra of this intermediate for D75N and D75N-ΔNpHtrII. Since both intermediates have a peak 45-nm red-shifted from the initial spectrum and this resembles the reported spectrum of K, we hereafter call this intermediate K. The shorter lifetime of K by interacting with the transducer is consistent with a previous report (55) and our observed lifetimes are close to those previously reported.
Transient grating signal of D75N
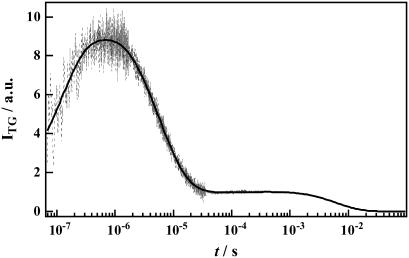
In contrast with the detection of only one intermediate in the photocycle of D75N and D75N-ΔNpHtrII by visible absorption changes, the TG signal resolved several kinetic components as shown in Fig. 5 for the free NpSRII D75N (q2 = 8.0 × 1011 m2/s). The signal rose quickly within the response time of our system (∼20 ns) and then more slowly in a submicrosecond time range. This slowly rising component was reproduced well by a biexponential function. After the rise, the signal decayed continuously in a wide time range up to tens of milliseconds. The decay was reproduced with three exponential functions. Hence, the temporal profile of the signal should be expressed by a sum of five exponential functions,
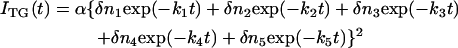 |
(7) |
where k1 > k2 > k3 > k4 > k5. The first and the second terms express the rising components of the signal, and the other terms represent the decaying components. The rising or decaying terms reflect the signs of the refractive index changes. By measuring the TG signal under various q2 conditions, it was found that the rate constants of k1, k2 and k4 do not depend on q2, and were determined to be (k1)−1 = 76 ± 19 ns, (k2)−1 = 460 ± 130 ns, and (k4)−1 = 8.0 ± 0.2 μs.
FIGURE 5.
A typical TG signal (dashed line) after photoexcitation of D75N in 1% OG, 50 mM Tris-HCl (pH = 7.0), 300 mM NaCl at q2 = 8.0 × 1011 m−2. The best fitted curve by Eq. 7 is shown by the solid line.
On the other hand, the rate constants k3 and k5 depended on q2. The rate constant k3 agreed well with that of the TG signal from the calorimetric reference sample (Evans Blue) under the same q2 condition, attributing this signal-to-refraction change to transient temperature changes by the released heat, the thermal grating: δn3 = δnth and k3 = Dthq2. From the profile of the signal and the fact that δnth is negative at this temperature, we can determine the signs of the preexponential factors as δn1 > 0, δn2 > 0, δn4 < 0 and δn5 < 0.
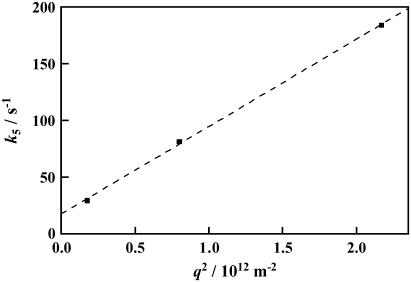
Another q2-dependent kinetic rate constant k5 in the millisecond time domain should be attributed to the molecular diffusion process. From the q2 dependence of k5 as shown in Fig. 6, the diffusion coefficient of D75N (DD75N) is calculated from the slope of this plot to be DD75N = 7.7 ± 0.2 × 10−11 m2/s (Eq. 2). The k5 value at q2 = 0 (18 ± 2 s−1 or 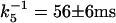 : intercept of the plot with the ordinate axis), represents an intrinsic reaction rate constant with no contribution of the molecular diffusion. The value obtained by the extrapolation is in good agreement with the lifetime of the photocycle in the visible (81 ms), attributing the refraction change in 56 ms either to population grating due to the absorbance change in the recovery process of the unphotolyzed state or to volume grating.
: intercept of the plot with the ordinate axis), represents an intrinsic reaction rate constant with no contribution of the molecular diffusion. The value obtained by the extrapolation is in good agreement with the lifetime of the photocycle in the visible (81 ms), attributing the refraction change in 56 ms either to population grating due to the absorbance change in the recovery process of the unphotolyzed state or to volume grating.
FIGURE 6.
Plot of the decay rate constant (k5) versus q2 for D75N (squares). The best fitted line by k5 = DN75Nq2 + 5 is plotted by the dashed line.
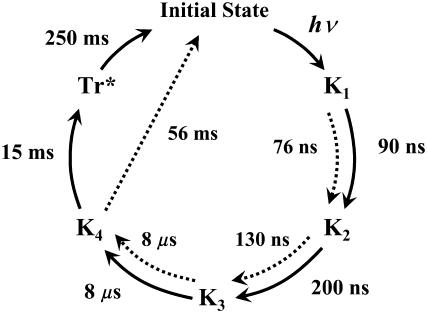
By use of the TG method, four spectrally silent intermediates states with distinct volume differences could be identified, denoted K1, K2, K3, and K4 in the following reaction scheme, since the absorption spectra of all intermediates resemble the reported spectrum of K:
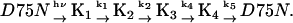 |
The spectrally silent nature of these processes indicates that the conformation around the chromophore is very similar for these species, but that the other parts of the protein that affect the volume are different.
In principle, the decay of the TG signal should be expressed by a biexponential function shown in Eq. 1 reflecting the diffusion of the reactant and the product. The presence of a single diffusing component in the millisecond range where K4 and unphotolyzed D75N coexist indicates that Ds of the reactant (D75N) and K4 are indistinguishable. Thus, conformational changes in the free photoreceptor in the K4 decay process are apparently not large enough to cause changes in the diffusion coefficient.
The change in the molecular volume of D75N up to the submicrosecond time region was calculated previously by the photoacoustic method (32) to be +16.7 ml/mol with a time constant of ∼360 ns at 20°C. Here we resolved two structural changes in the same time region. The amplitudes of the components reflect the volume change and the energetic change. The signs of the TG signal components imply that the molecular volume expands in the K1-to-K2 and K2-to-K3 conversions (48–50), which is consistent with the photoacoustic measurement. If we assume that there was no enthalpy change of the protein molecule after the K-like intermediate formation as concluded from the photoacoustic method, we calculate the volume changes to be +23 ml/mol and +6.7 ml/mol with 76 ns and the 460 ns kinetics, respectively. If energetic relaxation occurs, then the volume change would be smaller than the above estimated value. The difference between the two measurements could be due to the energetic contributions or more likely due to different detergents used (DM in their measurement versus OG in ours), because it was shown earlier that the volume change in this step is sensitive to the solvent environment (32). The molecular volume contraction in the K3-to-K4 conversion (8.0 μs) was calculated to be 4.8 ml/mol.
Transient grating signal of D75N-ΔNpHtrII
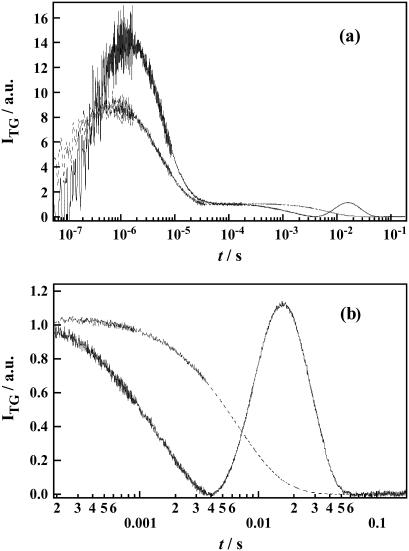
Fig. 7 depicts the TG signal of D75N-ΔNpHtrII at q2 = 7.6 × 1011 m−2. The TG signal in the submicrosecond-submillisecond time window is similar to that of free D75N: there are two rising phases (k1: 90 ± 35 ns, k2: 600 ± 200 ns) followed by the thermal grating signal k3. In the microsecond time region, one rising phase (k4: 8 ± 3 μs) was observed, indicating volume expansion for the complexed protein in this time domain, as opposed to the case of the free D75N, in which volume contraction was seen in a similar time domain.
FIGURE 7.
(a) A typical TG signal of D75N (dashed line) and D75N-ΔNpHtrII (solid line) in 1% OG, 50 mM Tris-HCl (pH = 7.0), 300 mM NaCl at q2 = 7.6 × 1011 m−2. (b) The TG signal of D75N-ΔNpHtrII (solid line) in the molecular diffusion time range is amplified to show the profile clearly. For comparison, D75N (dashed line) measured under the same condition is shown.
A remarkable difference in the TG signals of D75N-ΔNpHtrII compared to free D75N is appearance of biexponential kinetics in the millisecond time region (expanded in Fig. 7 b). The q2-dependence (Fig. 8 a) of the kinetics attributes these two kinetic components to two diffusing species with different D.
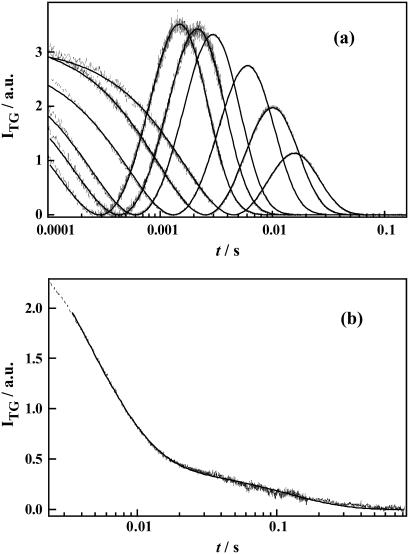
FIGURE 8.
(a) The TG signals (dashed lines) at various q2 in the molecular diffusion time range of D75N-ΔNpHtrII and the best fitted curves by Eq. 6 (solid lines) at q2 = 117, 78.7, 56.5, 26.1, 13.5, 6.78, and 2.17 × 1011 m−2 (from left to right). (b) A typical TG signal (dotted line) at a low q2 condition (q2 = 2.5 × 1010 m−2). The best fitted curve by a biexponential function is shown by the solid line.
Under a sufficiently small q2 condition (e.g., Dq2 ≪ k in the first term of Eq. 2) where we can ignore the contribution of protein diffusion on the decay, the remaining TG signal is ascribed solely to the kinetics of intrinsic reaction processes. The signal in Fig. 8 b (q2 = 2.5 × 1010 m−2) can be fitted by a biexponential function with lifetimes of 15 ms and 250 ms. The shorter lifetime is close to the lifetime of the absorption change of D75N-ΔNpHtrII (12 ms), attributing the 15-ms TG component to the species grating accompanying the relaxation process of the chromophore. Since there is no absorbance change in 250 ms, we attribute the longer-lived component (250 ms) of the TG signal to the volume grating. Therefore, we conclude that conformational change of the D75N-ΔNpHtrII molecular complex persists even after the photoreceptor spectral photocycle is completed. In free D75N, no delay of the TG signal is observed relative to the absorbance changes. Therefore, we attribute the delayed (250 ms) conformational change to the conformational changes in the NpHtrII portion. The negative sign of this TG component indicates that the partial volume of the protein decreases in this final step. We name this species Tr* to denote the different conformation of the transducer. Based on these results from the transient absorption and TG measurements, the reaction scheme of D75N-ΔNpHtrII may be described as in Fig. 9 and by the following reaction scheme:
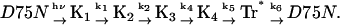 |
In the millisecond time region, three molecular species (K4, Tr*, and the unphotolyzed state) coexist, while only two Ds were detected. To identify the two species with the same D, we fitted the TG dynamics at various q2 in millisecond time range with Scheme 3 and Eq. 6, in which the molecular species appearing earlier than millisecond times were disregarded. Here, the parameters of DI, DJ, ki, and kj in Eq. 6 should be replaced by DK4, DTr*, k5, and k6, where DK4 and DTr* are the diffusion coefficients of K4 and Tr*, respectively. To obtain reliable parameters by the signal fitting, the number of adjustable parameters in Eq. 3 needs to be reduced. To accomplish this reduction, we first noted that the signal at a low q2, e.g., q2 = 2.5 × 1010 m−2 in Fig. 7 b, showed single exponential decay at >50 ms. This fact implies that there is only one diffusion coefficient at >50 ms, where only the reactant (unphotolyzed state) and the final intermediate Tr* coexist, hence, we concluded DTr* = DR. Second, the amplitudes of rise-decay signals around the 10-ms time region decrease with decreasing q2 (Fig. 8 a), which is a characteristic feature when D changes in the K4-to-Tr* process. Indeed, the TG signals at various q2 fit well with Eq. 6 by fixing k5 and k6 to the values determined at the low q2 measurement (12 ms and 250 ms) and by assuming DTr* = DR (Fig. 8 a). From this fitting, DR and DK4 were determined to be DR = 9.5 ± 0.1 × 10−11 m2/s, and DK4 = 4.7 ± 0.1 × 10−11 m2/s.
FIGURE 9.
Proposed reaction schemes of photocycles of D75N (dotted arrows) and D75N-ΔNpHtrII (solid arrows). The numbers indicates the lifetimes of each intermediates.
DISCUSSION
Phototaxis response of D75N-HtrII
Based on the close similarity of SRII to BR in structure and photochemistry, SRII was proposed (56) to open its cytoplasmic (CP) channel by a light-induced outward tilt of helix F, as occurs in the M and N states of BR (57–59). Evidence for such helix F movement in SRII has been reported from EPR spectroscopy of site-directed spin-labeled protein (30,60). Similar difference FTIR spectra of D75N and SRII in the amide band region when the red-shifted photoproducts are formed (19,61) strongly suggest that similar backbone conformational changes occur in D75N as in SRII.
In D85N of BR, a similar outward tilt of helix F was observed when the pH was changed from 6 to 9, which hydrates Asp96 in the CP channel, and also when Asp96 was mutated to Ala (62). D85N/F42C double mutation in BR was shown to stabilize an N-like state as characterized by its 13-cis chromophore, deprotonated Asp96, and the perturbed amide bonds at pH 8, which was photoconverted to a transient species with an all-trans chromophore, whereas, at pH 6, an O-like state with an all-trans chromophore is stabilized (63). These findings indicate that the conformational changes of D85N BR between the closed and open CP-channel structure are coupled to the chromophore isomerization state between all-trans and 13-cis. A similar coupling is likely to occur in the homologous D75N mutant of SRII.
Here we demonstrated that D75N-HtrII functions in phototaxis signaling despite the absence of a deprotonated Schiff base. Our interpretation is that retinal photoisomerization followed by proton transfer from the Schiff base to Asp75 may be necessary for signaling in the wild-type complex, but in D75N Schiff base deprotonation is not necessary, because Asp75 is replaced with an uncharged residue mimicking the protonated Asp75.
Comparison of volume change kinetics between D75N and D75N-ΔNpHtrII
The volume change dynamics are similar between D75N and D75N-ΔNpHtrII at <8.0 μs, indicating the early volume changes arise from the photoreceptor portion. Differences between them appear in the K3 →K4 transition in 8.0 μs, in which the sign of the volume changes were opposite. This difference is ascribable either to hydrogen-bonding alteration of the residues in the interface of the NpSRII-ΔNpHtrII binding region, which was shown to occur in the L to M process in wild-type NpSRII-ΔNpHtrII system (64) or to conformational changes of the transducer portion. The latter possibility is more likely, because only D75N-ΔNpHtrII but not free D75N undergoes substantial conformational changes in the K4 state, as demonstrated by the large decrease in D relatively to that of the unphotolyzed state. The conformational changes in the transducer portion contributing to the decrease in D return with the decay of K4 in 12 ms, and partial conformational changes in the transducer potion (Tr*) remain even after the decay of K4 and persist for ∼250 ms. Conformational changes in Tr* are relatively minor, since the D of Tr* is indistinguishable from that of the unphotolyzed state, although it has substantial difference in terms of the volume from the unphotolyzed state.
This volume expansion of ΔNpHtrII was not resolved in the case of the fusion protein of wild-type NpSRII-NpHtrII (16). We suggest that, in the case of wild-type NpSRII, the strong population grating signal due to M formation masked the volume grating signal. In other words, the absence of the M state for D75N-ΔNpHtrII allowed us to observe this structural change clearly.
Diffusion change of intermediate species of D75N-ΔNpHtrII
The D of the dark state D75N-ΔNpHtrII (39 kDa) (9.5 × 10−11 m2/s) is similar to D of many globular proteins with similar molecular weight (e.g., D of cytochrome peroxidase (35 kDa) is 9.4 × 10−11 m2/s) (54,65), suggesting that D75N-ΔNpHtrII exists in a 1:1 (not 2:2) complex under the conditions of our measurement, consistent with the expectation from Klare et al. (66). In our earlier observations, D of wild-type NpSRII with NpHtrII (157 residues) (NpSRII-ΔNpHtrII) was determined by the TG method to be 1.8 ± 0.2 × 10−11 m2/s (16). Since this value is considerably smaller than that expected from the molecular weight of NpSRII-ΔNpHtrII (43 kDa), it was concluded that NpSRII-ΔNpHtrII forms a 2:2 complex in the detergent solution, as in a lipid bilayer (67). The difference between the present result and that of wild-type NpSRII (16) may be due to the different detergents used (OG for the present case, DM for the previous study), which may alter the aggregation state.
The similar D of D75N-NpHtrII to that of a globular protein with a similar molecular weight suggests that D of D75N could be similar to that of a globular protein with a similar size. On the contrary, however, we found that D of D75N (DD75N = 7.7 × 10−11 m2/s) is rather small compared with D of globular proteins having a similar molecular weight (25 kDa) (e.g., D of chymotrypsin Aα (25kDa) is 10.2 × 10−11 m2/s) (65,68). This value is even smaller than that of D75N-ΔNpHtrII. We consider that the small D indicates that D75N exists as a dimer both in the initial state and the intermediate states. Both the two-dimensional and three-dimensional crystals of wild-type NpSRII made from OG-solubilized receptor show a dimeric arrangement of NpSRII with two dimers per unit cell (69,70). The present results indicate that the dimeric form of receptor protein observed in the two-dimensional and three-dimensional crystals may be maintained in 1% OG.
One of the most significant observations in this study is that D of the K4 intermediate is different from that of the unphotolyzed D75N-ΔNpHtrII protein, but there is no difference for transducer-free D75N. We will consider the cause of the transient D-change for the K4 intermediate. According to the Stokes-Einstein equation, D of a spherical molecule with a radius r is given by
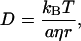 |
(8) |
where kB, T, η, and a are Boltzmann constant, temperature, viscosity, and a constant representing the boundary condition between the diffusing molecule and the solvent, respectively. To explain the large (approximately half) reduction in D for the creation of the K4 intermediate on the basis of the volume change alone, the molecular volume would need to expand almost eightfold compared to the unphotolyzed state. Such a large expansion seems unlikely. We examine several alternative possibilities to account for this change.
First, D depends on the molecular shape. Perrin theoretically derived the friction of the molecules with elongated shape compared to spherical (71). For example, if a molecule has an elongated shape, D will decrease compared to that of a spherical molecule with the same volume. However, to explain the observed reduction in D, the radius of the molecule along the semimajor axis should be at least 20-times longer than the one along the minor axis from the equation. This elongation is physically unreasonable. Therefore, the effect of molecular shape change is not expected to be a major factor of the D decrease.
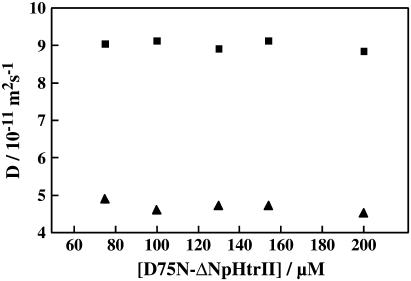
Second, the reduction in D might be explained by the photoexcitation of D75N-ΔNpHtrII inducing a transient dimerization or aggregation in the K4 state. If this is the case, D of the K4 state should be concentration-dependent. We examined this possibility by measurements of the concentration dependence of D. We did not observe a significant concentration dependence of the TG signal. Even at a low concentration (75 μM), we observed a rise-decay signal with no significant change in D for both states in a concentration range from 75 to 200 μM (Fig. 10). Therefore, we exclude the possibility of dimer formation or aggregation as a cause of the D change.
FIGURE 10.
Concentration dependence of the diffusion coefficients of initial state (squares) and K4 intermediate (triangles).
Finally, we consider an effect of intermolecular interaction. If the number of hydrogen bonds between the protein and water molecules increases, it will increase the friction for the translational diffusion to reduce D. Indeed, several studies have already shown that the formation of hydrogen bonds between protein and water molecules upon unfolding of α-helices decreases D (34,36,72). Therefore, one of the possibilities for the decrease in D upon K4 formation in D75N-ΔNpHtrII may be unfolding of α-helices, although other types of conformational changes that result in increases in the intermolecular interaction between the protein and water molecules cannot be excluded.
Which portion of the molecule is likely to be responsible for such large changes in D? Since the transmembrane domain of D75N-ΔNpHtrII is buried in the detergent micelle, it is unlikely that structural changes that occur in this domain result in increase of intermolecular hydrogen bonding with bulk water. Indeed, the free D75N, which is functional and is expected to undergo F-helix tilting as was reported in the case of wild-type NpSRII (29,30), did not elicit the D-change. Therefore, we may attribute the main origin of the D-change to the conformational change in the cytoplasmic domain of ΔNpHtrII, which in this case corresponds to its HAMP domain (85–120 of NpHtrII).
The HAMP domain consists of two helical amphipathic sequences linking the transmembrane domain and the cytoplasmic domain of the transducer protein and is believed to play an important role in connecting two subdomains of a protein and in transmitting conformational changes from one domain to the other. In the case of phototaxis transducer proteins, the HAMP domain connects the TM domain and the cytoplasmic domain, thus transmitting conformational changes from the membrane-embedded photoreceptor to the signaling domain in the distal end of the cytoplasmic domains, where activation of a histidine kinase, CheA, is regulated.
Conformational changes of a HAMP domain coupled to light activation of a photoreceptor were monitored earlier for the SRI-HtrI complex in which distances between two cysteine residues introduced at position 64 (contained in the HAMP domain at positions 55–100) of the two HtrI molecules in the dimer complex were monitored by their cross-linking rate. Formations of the attractant signaling state and of the repellent signaling state induced by light activation of wild-type and H166S SRI, respectively, elicit opposite effect on the crosslinking reactivity, indicating that the HAMP domain undergoes conformational changes upon light activation and that the direction of the movement of the HAMP domain defines the sign of the signal (CheA activation or CheA inhibition) (73).
Here we have obtained evidence of conformational changes of the HAMP domain of NpHtrII, which considerably increases friction with the aqueous solvent presumably resulted from alteration in hydrogen bonding with the solvent. A possible mechanism for the two amphipathic α-helices of HAMP to undergo such a large increase with interaction with the aqueous solvent is unfolding of the helices, although more detailed analysis is needed to verify this possibility.
In the full-length NpHtrII, the conformational changes of the HAMP domain that alter hydrogen bonding might provide a motive force for inducing concomitant conformational changes in the more distal coiled coil and CheA-interaction domains of NpHtrII, providing a insight for an on/off switch of CheA activity.
The conformational change of the HAMP domain takes place in 8 μs with formation of K4 and most of the change decay in 12 ms with the decay of K4, with delay of some portion of the structural changes of ΔHtrII, which decays in 250 ms. Despite the strong phototaxis signaling by D75N-HtrII comparable to that of wild-type, the lifetime of the spectral photocycle is considerably shorter (almost 10-fold) compared to the wild-type photocycle. One possible explanation is that the decay of the conformational changes in the cytoplasmic domain of HtrII is uncoupled from the decay of the receptor absorption changes and the HAMP domain alteration persists in D75N-HtrII for a similar time as in SRII-HtrII. Thus, both complexes would exhibit a similar extent of CheA-mediated phosphorylation of CheY and mediate comparable phototaxis responses.
CONCLUSION
The transient grating (TG) method detects the kinetics of molecular volume change, enthalpy change, and the diffusion coefficient as well as absorption changes. The technique enables us to detect global reaction steps of the photocycle, including those steps that are spectroscopically silent in the UV-visible region. In the D75N protein we identified four spectroscopically silent K-like intermediates (K1–K4) with distinct volume differences and with time constants for the interconversions at 76 ns, 460 ns, 130 ns, 8.0 μs, and 81 ms. Volume differences between the intermediates represent different conformations, but with the chromophore environment unchanged. The TG signal of the fusion protein of D75N with 120 residues of the N-terminal part of the transducer showed similar kinetics of these four intermediates and one additional transient: 90 ns and 600 ns, 8 μs, 12 ms, and 250 ms. The absorption spectrum of the final intermediate is the same as that of the original state, but the conformation of the transducer portion is altered, even after the conformation at the receptor recovered. Furthermore, it was found that the diffusion coefficient of K4 of this fusion protein is significantly different from that of the unphotolyzed state of the protein. We raise the possibility that this different D results from the unfolding of the α-helices in the linker region (HAMP domain) located between the transmembrane helices and cytoplasmic coiled-coil domain of NpHtrII and that this conformational change occurs with a time constant of 8 μs. If such unfolding occurs, our data indicates refolding would occur in 12 ms, but minor conformational changes in the transducer persist and decay completely only after 250 ms.
Acknowledgments
This work was supported by the Grant-in-Aid (No.13853002 and 15076204) from the Ministry of Education, Science, Sports and Culture in Japan (M.T.), National Institutes of Health grant R37GM27750 (J.L.S.), and the Robert A. Welch Foundation (J.L.S.).
K. Inoue and J. Sasaki contributed equally to the work.
References
- 1.Hoff, W. D., K. H. Jung, and J. L. Spudich. 1997. Molecular mechanism of photosignaling by archaeal sensory rhodopsins. Annu. Rev. Biophys. Biomol. Struct. 26:223–258. [DOI] [PubMed] [Google Scholar]
- 2.Kamo, N., K. Shimono, M. Iwamoto, and Y. Sudo. 2001. Photochemistry and photoinduced proton-transfer by pharaonis phoborhodopsin. Biochemistry (Mosc.). 66:1277–1282. [DOI] [PubMed] [Google Scholar]
- 3.Luecke, H., and J. K. Lanyi. 2003. Structural clues to the mechanism of ion pumping in bacteriorhodopsin. Adv. Protein Chem. 63:111–130. [DOI] [PubMed] [Google Scholar]
- 4.Klare, J. P., V. I. Gordeliy, J. Labahn, G. Büldt, H.-J. Steinhoff, and M. Engelhard. 2004. The archaeal sensory rhodopsin II/transducer complex: a model for transmembrane signal transfer. FEBS Lett. 564:219214. [DOI] [PubMed] [Google Scholar]
- 5.Spudich, J. L., and K.-H. Jung. 2005. Microbial rhodopsins: phylogenetic and functional diversity. In Handbook of Photosensory Receptors. Wiley-VCH, Weinheim, Germany.
- 6.Yao, V. J., E. N. Spudich, and J. L. Spudich. 1994. Identification of distinct domains for signaling and receptor interaction of the sensory rhodopsin I transducer, HtrI. J. Bacteriol. 176:6931–6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seidel, R., B. Scharf, M. Gautel, K. Kleine, D. Oesterhelt, and M. Engelhard. 1995. The primary structure of sensory rhodopsin II: a member of an additional retinal protein subgroup is coexpressed with its transducer, the halobacterial transducer of rhodopsin II. Proc. Natl. Acad. Sci. USA. 92:3036–3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim, S.-H., W. Wang, and K. K. Kim. 2002. Dynamic and clustering model of bacterial chemotaxis receptors: structural basis for signaling and high sensitivity. Proc. Natl. Acad. Sci. USA. 99:11611–11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang, X. N., and J. L. Spudich. 1998. HtrI is a dimer whose interface is sensitive to receptor photoactivation and His-166 replacements in sensory rhodopsin I. J. Biol. Chem. 273:19722–19728. [DOI] [PubMed] [Google Scholar]
- 10.Gordeliy, V. I., J. Labahn, R. Moukhametzianov, R. Efremov, J. Granzin, R. Schlesinger, G. Büldt, T. Savopol, A. J. Scheldig, J. P. Klare, and M. Engelhard. 2002. Molecular basis of transmembrane signaling by sensory rhodopsin II-transducer complex. Nature. 419:484–487. [DOI] [PubMed] [Google Scholar]
- 11.Spudich, E. N., and J. L. Spudich. 1993. The photochemical reactions of sensory rhodopsin I are altered by its transducer. J. Biol. Chem. 268:16095–16097. [PubMed] [Google Scholar]
- 12.Sasaki, J., and J. L. Spudich. 1998. The transducer protein HtrII modulates the lifetimes of sensory rhodopsin II photointermediates. Biophys. J. 75:2435–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sasaki, J., and J. L. Spudich. 1999. Proton circulation during the photocycle of sensory rhodopsin II. Biophys. J. 77:2145–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Terazima, M. 2002. Molecular volume and enthalpy changes associated with irreversible photo-reactions. J. Photochem. Photobiol. C. 3:81–108. [Google Scholar]
- 15.Losi, A., and S. E. Braslavsky. 2003. The time-resolved thermodynamics of the chromophore-protein interactions in biological photosensors as derived from photothermal measurements. Phys. Chem. Chem. Phys. 5:2739–2750. [Google Scholar]
- 16.Inoue, K., J. Sasaki, M. Morisaki, F. Tokunaga, and M. Terazima. 2004. Time-resolved detection of sensory rhodopsin II-transducer interaction. Biophys. J. 87:2587–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmies, G., B. Lüttenberg, I. Chizhov, M. Engelhard, A. Becker, and E. Bamberg. 2000. Sensory rhodopsin II from the haloalkaliphilic Natronobacterium pharaonis: light activated proton transfer reactions. Biophys. J. 78:967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang, C.-S., O. Sineshchekov, E. N. Spudich, and J. L. Spudich. 2004. The cytoplasmic membrane-proximal domain of the HtrII transducer interacts with the E-F loop of photoactivated Natronomonas pharaonis sensory rhodopsin II. J. Biol. Chem. 279:42970–42976. [DOI] [PubMed] [Google Scholar]
- 19.Hein, M., I. Radu, J. P. Klare, M. Engelhard, and F. Siebert. 2004. Consequences of counterion mutation in sensory rhodopsin II of Natronobacterium pharaonis for photoreaction and receptor activation: an FTIR study. Biochemistry. 43:995–1002. [DOI] [PubMed] [Google Scholar]
- 20.Chizhov, I., G. Schmies, R. Seidel, J. R. Sydor, B. Lüttenberg, and M. Engelhard. 1998. The photophobic receptor from Natronobacterium pharaonis: temperature and pH dependencies of the photocycle of sensory rhodopsin II. Biophys. J. 75:999–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lutz, A., A. A. Wegener, M. Engelhard, I. Boche, M. Otsuka, D. Oesterhelt, J. Wachtveitl, and W. Zinth. 2001. Primary reactions of sensory rhodopsins. Proc. Natl. Acad. Sci. USA. 98:962–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engelhard, M., B. Scharf, and F. Siebert. 1996. Protonation changes during the photocycle of sensory rhodopsin II from Natronobacterium pharaonis. FEBS Lett. 395:195–198. [DOI] [PubMed] [Google Scholar]
- 23.Furutani, Y., M. Iwamoto, K. Shimono, N. Kamo, and H. Kandori. 2002. FTIR spectroscopy of the M photointermediate in pharaonis phoborhodopsin. Biophys. J. 83:3482–3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furutani, Y., Y. Sudo, N. Kamo, and H. Kandori. 2003. FTIR spectroscopy of the complex between pharaonis phoborhodopsin and its transducer protein. Biochemistry. 42:4837–4842. [DOI] [PubMed] [Google Scholar]
- 25.Furutani, Y., M. Iwamoto, K. Shimono, A. Wada, M. Ito, N. Kamo, and H. Kandori. 2004. FTIR spectroscopy of the O photointermediate in pharaonis phoborhodopsin. Biochemistry. 43:5204–5212. [DOI] [PubMed] [Google Scholar]
- 26.Bergo, V., E. N. Spudich, J. L. Spudich, and K. J. Rothschild. 2003. Conformational changes detected in a sensory rhodopsin II-transducer complex. J. Biol. Chem. 278:36556–36562. [DOI] [PubMed] [Google Scholar]
- 27.Bergo, V., E. N. Spudich, K. J. Rothschild, and J. L. Spudich. 2005. Photoactivation perturbs the membrane-embedded contacts between sensory rhodopsin II and its transducer. J. Biol. Chem. 280:28365–28369. [DOI] [PubMed] [Google Scholar]
- 28.Sudo, Y., Y. Furutani, K. Shimono, N. Kamo, and H. Kandori. 2003. Hydrogen bonding alteration of Thr-204 in the complex between pharaonis phoborhodopsin and its transducer protein. Biochemistry. 42:14166–14172. [DOI] [PubMed] [Google Scholar]
- 29.Wegener, A.-A., I. Chizhov, M. Engelhard, and H.-J. Steinhoff. 2000. Time-resolved detection of transient movement of helix F I spin-labeled pharaonis sensory rhodopsin II. J. Mol. Biol. 301:881–891. [DOI] [PubMed] [Google Scholar]
- 30.Wegener, A.-A., J. P. Klare, M. Engelhard, and H.-J. Steinhoff. 2001. Structural insights into the early steps of receptor-transducer signal transfer in archaeal phototaxis. EMBO J. 20:5312–5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Losi, A., A. A. Wegener, M. Engelhard, W. Gärtner, and S. E. Braslavsky. 1999. Time-resolved absorption and photothermal measurements with recombinant sensory rhodopsin II from Natronobacterium pharaonis. Biophys. J. 77:3277–3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Losi, A., A. A. Wegener, M. Engelhard, W. Gärtner, and S. E. Braslavsky. 2000. Aspartate 75 mutation in sensory rhodopsin II from Natronobacterium pharaonis does not influence the production of the K-like intermediate but strongly affects its relaxation pathway. Biophys. J. 78:2581–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Losi, A., A. A. Wegener, M. Engelhard, and S. E. Braslavsky. 2001. Enthalpy-entropy compensation in a photocycle: the K-to-L transition in sensory rhodopsin II from Natronobacterium pharaonis. J. Am. Chem. Soc. 123:1766–1767. [DOI] [PubMed] [Google Scholar]
- 34.Nishida, S., T. Nada, and M. Terazima. 2004. Kinetics of intermolecular interaction during protein folding of reduced cytochrome c. Biophys. J. 87:2663–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishida, S., T. Nada, and M. Terazima. 2005. Hydrogen bonding dynamics during protein folding of reduced cytochrome c: temperature and denaturant concentration dependence. Biophys. J. 89:2004–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eitoku, T., Y. Nakasone, D. Matsuoka, S. Tokutomi, and M. Terazima. 2005. Conformational dynamics of phototropin 2 LOV2 domain with the linker upon photoexcitation. J. Am. Chem. Soc. 127:13238–13243. [DOI] [PubMed] [Google Scholar]
- 37.Terazima, M. 2006. Diffusion coefficients as a monitor of reaction kinetics of biological molecules. Phys. Chem. Chem. Phys. 8:545–557. [DOI] [PubMed] [Google Scholar]
- 38.Terazima, M., and N. Hirota. 1993. Translational diffusion of a transient radical studied by the transient grating method, pyrazinyl radical in 2-propanol. J. Chem. Phys. 98:6257–6262. [Google Scholar]
- 39.Terazima, M., K. Okamoto, and N. Hirota. 1993. Diffusion process of methyl red in organic solvents studied by the transient grating method. J. Phys. Chem. 97:5188–5192. [Google Scholar]
- 40.Terazima, M., K. Okamoto, and N. Hirota. 1993. Transient radical diffusion in photoinduced hydrogen abstraction reactions of benzophenone probed by the transient grating method. J. Phys. Chem. 97:13387–13393. [Google Scholar]
- 41.Terazima, M., H. Tomioka, K. Hirai, Y. Tanimoto, Y. Fujiwara, and Y. Akimoto. 1996. Translational diffusion of a carbine and radicals derived from carbenes. J. Chem. Soc. Faraday Trans. 92:2361–2368. [Google Scholar]
- 42.Terazima, M. 2000. Is the translational diffusion of organic radicals different from that of closed-shell molecules? Acc. Chem. Res. 33:687–694. [DOI] [PubMed] [Google Scholar]
- 43.Sakakura, M., I. Morishima, and M. Terazima. 2001. The structural dynamics and ligand releasing process after the photodissociation of sperm whale carboxymyoglobin. J. Phys. Chem. 105:10424–10434. [Google Scholar]
- 44.McCain, D. A., L. A. Amici, and J. L. Spudich. 1987. Kinetically resolved states of the Halobacterium halobium flagellar motor switch and moculation of the switch by sensory rhodopsin I. J. Bacteriol. 169:4750–4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Terazima, M., K. Okamoto, and N. Hirota. 1995. Translational diffusion of transient radicals created by the photoinduced hydrogen abstraction reaction in solution—anomalous size dependence in the radical diffusion. J. Chem. Phys. 102:2506–2515. [Google Scholar]
- 46.Okamoto, K., M. Terazima, and N. Hirota. 1995. Temperature dependence of diffusion intermediates probed by the transient grating method. J. Chem. Phys. 103:10445–10451. [Google Scholar]
- 47.Okamoto, K., N. Hirota, and M. Terazima. 1997. Diffusion process of the benzyl radical created by photodissociation probed by the transient grating method. J. Phys. Chem. 101:5269–5277. [Google Scholar]
- 48.Terazima, M., T. Hara, and N. Hirota. 1995. Reaction volume and enthalpy changes in photochemical reaction detected by the transient grating method; photodissociation of diphenylcyclopropenone. Chem. Phys. Lett. 246:577–582. [Google Scholar]
- 49.Hara, T., N. Hirota, and M. Terazima. 1996. New application of the transient grating method to a photochemical reaction: the enthalpy, reaction volume change, and partial molar volume measurements. J. Phys. Chem. 100:10194–10200. [Google Scholar]
- 50.Terazima, M. 2002. Molecular volume and enthalpy changes associated with irreversible photo-reactions. J. Photochem. Photobiol. C. 3:81–108. [Google Scholar]
- 51.Takeshita, K., Y. Imamoto, M. Kataoka, K. Mihara, F. Tokunaga, and M. Terazima. 2002. Structural change of site-directed mutants of PYP: new dynamics during pR state. Biophys. J. 83:1567–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nishioku, Y., M. Nakagawa, M. Tsuda, and M. Terazima. 2002. Energetics and volume changes of intermediates in the photolysis of octopus rhodopsin at physiological temperature. Biophys. J. 83:1136–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jung, K. H., E. N. Spudich, V. D. Trivedi, and J. L. Spudich. 2001. An archaeal photosignal-transducing module mediates phototaxis in Escherichia coli. J. Bacteriol. 183:6365–6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hippler-Mreyen, S., J. P. Klare, A. A. Wegener, R. Seidel, C. Herrmann, G. Schmies, G. Nagel, E. Bamberg, and M. Engelhard. 2003. Probing the sensory rhodopsin II binding domain of its cognate transducer by calorimetry and electrophysiology. J. Mol. Biol. 330:1203–1213. [DOI] [PubMed] [Google Scholar]
- 55.Sudo, Y., M. Iwamoto, K. Shimono, and N. Kamo. 2002. Association between a photo-intermediate of a M-lacking mutant D75N of pharaonis phoborhodopsin and its cognate transducer. J. Photochem. Photobiol. B. 67:171–176. [DOI] [PubMed] [Google Scholar]
- 56.Spudich, J. L. 1998. Vibrations on a molecular switch: transport and sensory signaling by archaeal rhodopsins. Mol. Microbiol. 28:1051–1058. [DOI] [PubMed] [Google Scholar]
- 57.Subramaniam, S., M. Gerstein, D. Oesterhelt, and B. Henderson. 1993. Electron diffraction analysis of structural changes in the photocycle of bacteriorhodopsin. EMBO J. 12:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vonck, J. 1996. A Three-dimensional difference map of the N-intermediate in the bacteriorhodopsin photocycle: part of the F helix tilts in the M to N transition. Biochemistry. 35:5870–5878. [DOI] [PubMed] [Google Scholar]
- 59.Rink, T., M. Pfeiffer, D. Oesterhelt, K. Gerwert, and H. J. Steinhoff. 2000. Unraveling photoexcited conformational changes of bacteriorhodopsin by time-resolved electron paramagnetic resonance spectroscopy. Biophys. J. 78:1519–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bordignon, B., J. P. Klare, J. Holterhues, S. Martell, A. Krasnaberski, M. Engelhard, and H. J. Steinhoff. 2006. Analysis of light-induced conformational changes of Natronomonas pharaonis sensory rhodopsin II by time resolved electron paramagnetic resonance spectroscopy. Photochem. Photobiol. In press. [DOI] [PubMed]
- 61.Hein, M., A. A. Wegener, M. Engelhard, and F. Siebelt. 2003. Time-resolved FTIR studies of sensory rhodopsin II (NpSRII) from Natronobacterium pharaonis: implications for proton transport and receptor activation. Biophys. J. 84:1208–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kataoka, M., H. Kamikubo, F. Tokunaga, L. S. Brown, Y. Yamazaki, A. Maeda, M. Sheves, R. Needleman, and J. K. Lanyi. 1994. Energy coupling in an ion pump. The reprotonation switch of bacteriorhodopsin. J. Mol. Biol. 243:621–638. [DOI] [PubMed] [Google Scholar]
- 63.Dioumaev, A. K., L. S. Brown, R. Needleman, and J. K. Lanyi. 1998. Partitioning of free energy gain between the photoisomerized retinal and the protein in bacteriorhodopsin. Biochemistry. 37:9889–9893. [DOI] [PubMed] [Google Scholar]
- 64.Furutani, Y., Y. Kamada, K. Sudo, K. Shimono, N. Kamo, and H. Kandori. 2005. Structural changes of the complex between Pharaonis phoborhodopsin and its cognate transducer upon formation of the M photointermediate. Biochemistry. 44:2909–2915. [DOI] [PubMed] [Google Scholar]
- 65.Durchschlag, H., and P. Zipper. 1997. Prediction of hydrodynamics parameters of biopolymers from small-angle scattering data. J. Appl. Crystallogr. 30:1112–1124. [Google Scholar]
- 66.Klare, J. P., E. Bordignon, M. Doebber, J. Fitter, J. Kriegsmann, I. Chizhov, H. J. Steinhoff, and M. Engelhard. 2006. Effects of solubilization on the structure and function of the sensory rhodopsin II/transducer complex. J. Mol. Biol. 356:1207–1221. [DOI] [PubMed] [Google Scholar]
- 67.Bordignon, E., J. P. Klare, M. Doebber, A. A. Wegener, S. Martell, M. Engelhard, and H. J. Steinhoff. 2005. Structural analysis of a HAMP domain the linker region of the phototransducer in complex with sensory rhodopsin II. J. Biol. Chem. 280:38767–38775. [DOI] [PubMed] [Google Scholar]
- 68.Tyn, M. T., and T. W. Gusek. 1990. Prediction of diffusion coefficients of proteins. Biotechnol. Bioeng. 35:327–338. [DOI] [PubMed] [Google Scholar]
- 69.Kunji, E. R. S., E. N. Spudich, R. Grisshammer, R. Henderson, and J. L. Spudich. 2001. Electron crystallographic analysis of two-dimensional crystals of sensory rhodopsin II: a 6.9 Å projection structure. J. Mol. Biol. 308:279–293. [DOI] [PubMed] [Google Scholar]
- 70.Luecke, H., B. Schobert, J. K. Lanyi, E. N. Spudich, and J. L. Spudich. 2001. Crystal structure of sensory rhodopsin II at 2.4 Å: insights into color tuning and transducer interaction. Science. 293:1499–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Van Holde. K. E., W. C. Johnson, and P. Shing Ho. 1998. Physical Biochemistry. Prentice-Hall, Englewood Cliffs, NJ.
- 72.Inoue, K., N. Baden, and M. Terazima. 2005. Diffusion coefficient and the secondary structure of poly-L-glutamic acid in aqueous solution. J. Phys. Chem. B. 109:22623–22628. [DOI] [PubMed] [Google Scholar]
- 73.Zhang, X.-N., and J. L. Spudich. 1998. HtrI is a dimer whose interface is sensitive to receptor photoactivation and His-166 replacements in sensory rhodopsin I. J. Biol. Chem. 273:19722–19728. [DOI] [PubMed] [Google Scholar]