Abstract
A Caenorhabditis elegans ORF encoding the presumptive condensing enzyme activity of a fatty acid elongase has been characterized functionally by heterologous expression in yeast. This ORF (F56H11.4) shows low similarity to Saccharomyces cerevisiae genes involved in fatty acid elongation. The substrate specificity of the C. elegans enzyme indicated a preference for Δ6-desaturated C18 polyunsaturated fatty acids. Coexpression of this activity with fatty acid desaturases required for the synthesis of C20 polyunsaturated fatty acids resulted in the accumulation of arachidonic acid from linoleic acid and eicosapentaenoic acid from α-linolenic acid. These results demonstrate the reconstitution of the n-3 and n-6 polyunsaturated fatty acid biosynthetic pathways. The C. elegans ORF is likely to interact with endogenous components of a yeast elongation system, with the heterologous nematode condensing enzyme F56H11.4 causing a redirection of enzymatic activity toward polyunsaturated C18 fatty acid substrates.
Unsaturated fatty acids are essential components required for normal cellular function, being involved in roles ranging from membrane fluidity to acting as signal molecules (1, 2). In particular, the class of fatty acids known as the polyunsaturated fatty acids (PUFAs) has attracted considerable interest as pharmaceutical and nutraceutical compounds (2, 3). PUFAs can be defined as fatty acids of 18 carbons or more in length, containing two or more double bonds. These double bonds are inserted by specific fatty acid desaturase enzymes that have been the subject of intense research in recent years (4, 5). The PUFAs can be classified into two groups, n-6 or n-3, depending on the position (n) of the double bond nearest the methyl end of the fatty acid (1, 2, 5). Thus, γ-linolenic acid (18:3Δ6,9,12) is classified as an 18:3, n-6 PUFA, whereas α-linolenic acid (18:3Δ9,12,15) is an 18:3, n-3 PUFA. Many PUFAs are also essential fatty acids, being required in the diet for normal development in mammals that cannot synthesize the primary essential fatty acid-PUFA linoleic acid (18:2, n-6; ref. 2). More recently, interest has focused on the C20 fatty acid arachidonic acid (20:4, n-6), which has been shown to be involved in neonatal health, including retinal and brain development (6).
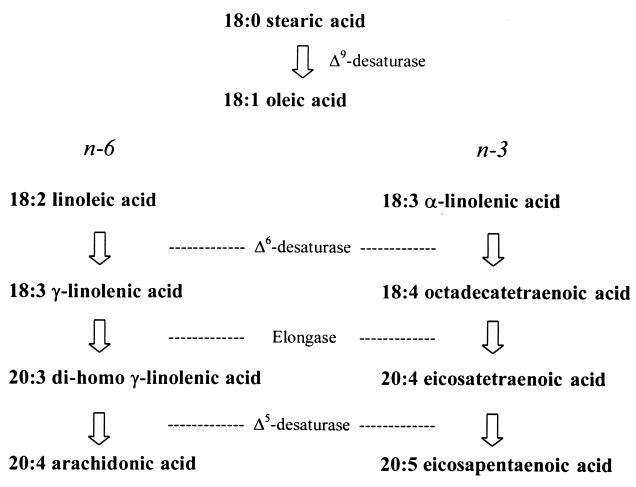
C20 fatty acids such as 20:4, n-6 are synthesized by sequential desaturation, elongation, and further desaturation of dietary 18:2, n-6 (2, 5), and a schematic representing a generalized pathway for PUFA synthesis is shown in Fig. 1. Although the two desaturases (Δ6- and Δ5-fatty acid desaturases) required for this pathway have been cloned recently from a number of different sources (7–14), no genes have been characterized for the C2 elongation of the C18 PUFA. Biochemical characterization of mammalian elongation systems (most notably from liver microsomes) has indicated that the “elongase” actually consists of four enzyme activities, being made up of a condensing enzyme, a β-ketoreductase, a dehydrase, and an enoyl reductase (reviewed in ref. 15). In higher plants, the FAE1 (fatty acid elongation 1) gene from Arabidopsis thaliana is required for the accumulation of the monounsaturated C22 fatty acid erucic acid (22:1, n-9; ref. 16). The FAE1 gene product is predicted to have a condensing enzyme activity, and the deduced amino acid sequence shows some limited similarity to other condensing enzymes such as chalcone synthase and stilbene synthase (16). Although FAE1 is normally expressed only in seed tissues, ectopic expression of FAE1 in nonseed tissue (or heterologous expression in yeast) resulted in the accumulation of erucic acid (17).
Figure 1.
Scheme representing the biosynthesis of PUFAs in animals. Primary n-6 and n-3 fatty acids (in the form of linoleic and α-linolenic acid) must be provided from dietary intake because of an inability to desaturate oleic acid further. The enzyme activities required for this pathway are shown.
To reconstitute the long-chain PUFA biosynthetic pathway in a heterologous host such as yeast, it was necessary to identify enzymes involved in PUFA elongation. Previously, we and others (18, 19) have observed a high degree of similarity between the Δ6-fatty acid desaturase (responsible for the first step in PUFA synthesis by the desaturation of 18:2, n-6) and a desaturase that introduces double bonds into the long-chain base of sphingolipids. We reasoned that the relationship between fatty acid and sphingolipid modification might also extend to the elongation reaction. Two yeast genes (ELO2 and ELO3) involved in the formation of the very long-chain saturated fatty acid moiety of sphingolipids have been characterized at the genetic and biochemical levels (20). We therefore used conserved motifs contained within these ELO genes to identify potential ORFs encoding enzymes involved in fatty acid elongation in the PUFA-accumulating organism Caenorhabditis elegans, which has also been the subject of a completed genome sequencing program (21). These ORFs were characterized functionally by heterologous expression in yeast, allowing the identification of activities involved in PUFA elongation.
Materials and Methods
Identification of C. elegans ORFs of Interest.
The C. elegans genome sequence was searched for sequences related to the yeast ELO2/3 genes via the blast suite of programs by using those yeast ORFs as search templates (http://www.sanger.ac.uk/Projects/C_elegans/blast_server.shtml). The conserved motifs H-X-X-H-H and M-Y-X-Y-Y were also used in conjunction with the ELO2/3 ORFs to search databases with phi-blast. PCR primers were designed to amplify the predicted ORFs of interest with cDNA purified from a C. elegans N2 mixed-stage library used as a template, according to previously described methods (8, 12). PCR amplification was carried out with standard protocols (7, 8, 10, 12).
Cloning of Desaturase and Elongase Genes in Yeast Expression Vectors.
C. elegans ORFs F56H11.4 and F41H10.8 were cloned by PCR into the pYES2 vector (Invitrogen). F56H11.4 was amplified with primers 56h114.for (5′-GCGGGTACCATGGCTCAGCATCCGCTC-3′) and 56h114.rev (5′-GCGGGATCCTTAGTTGTTCTTCTTCTT-3′), and F41H10.8 was amplified with primers 41h108.for (5′-GCGGGTACCATGCCACAGGGAGAAGTC-3′) and 41h108.rev (5′-GCGGGATCCTTATTCAATTTTTCTTTT-3′). Amplified sequences were then restricted with KpnI and BamHI (underlined in the forward and reverse primers, respectively), purified with the Qiagen PCR purification kit (Chatsworth, CA), and ligated into a KpnI/BamHI-cut pYes2. An ORF encoding the Mortierella alpina Δ5-fatty acid desaturase (10) was amplified with primers Mad5.for (5′-GCGAATTCACCATGGGTACGGACCAAGGA-3′) and Mad5.rev (5′GCGGAGCTCCTACTCTTCCTTGGGACG-3′), restricted with EcoRI and SacI, gel purified as described (10), and ligated into a EcoRI/SacI-cut pESC-TRP vector (Stratagene) to generate pESC/Δ5. An ORF encoding the borage Δ6-fatty acid desaturase (7) was restricted from pGEM3 with BamHI and XhoI and ligated into a BamHI/XhoI-cut pESC-TRP vector to generate pESC/Δ6. A double construct was also generated by ligating the BamHI/XhoI borage Δ6 insert into the pESC/Δ5 construct described above, generating pESC/(Δ5,Δ6). All PCR amplifications were carried out under standard conditions (7).
Functional Characterization in Yeast.
ORFs encoding putative elongation activities and desaturase constructs were introduced in Saccharomyces cerevisiae W303-1A by using a lithium acetate-based method, and expression of the transgenes was induced by addition of galactose to 2% (wt/vol) as described (8, 10, 12). Yeast transformants containing pYES2-derived constructs were grown on synthetic dextrose minimal medium minus uracil; pESC-derived constructs were grown on synthetic dextrose minimal medium minus tryptophan. Cotransformed yeast (containing both pYES2 and pESC derivatives) was grown on synthetic dextrose minimal medium minus uracil and tryptophan. Before induction, cultures were grown at 22°C with shaking in the presence of 2% (vol/vol) raffinose and supplemented with 0.5 mM of appropriate fatty acid substrate in the presence of 1% tergitol-Nonidet P-40 (Sigma). All cultures were then grown for a further 48 h unless indicated. All analysis was performed on triplicate samples and replicated three times.
Fatty Acid Analysis.
Total fatty acids extracted from yeast cultures were analyzed by GC of methyl ester derivatives. Lipids were extracted and transmethylated with methanolic HCl, and the fatty acid methyl esters were analyzed as described (7, 8, 10).
GC-MS Analysis.
Identification of induced peaks was carried out by coinjection with known standards (such as 20:3, n-6; 20:4, n-6; etc.), followed by characterization via GC-MS (Kratos Analytical Instruments MS80RFA) operating at an ionization voltage of 70 eV with a scan range of 40–500 Da, as described (8, 10, 12).
Results
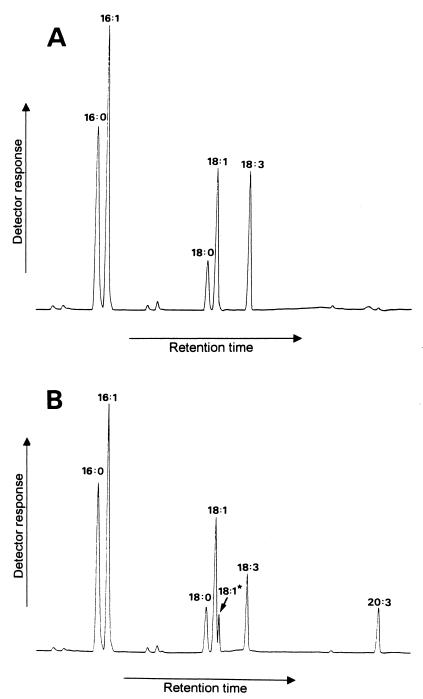
The entire C. elegans genome was searched for ORFs that showed low levels of similarity to the yeast ELO genes, so as to identify related sequences or paralogues rather than actual homologues. A number of C. elegans ORFs were identified by this search and then subjected to functional characterization by heterologous expression of the appropriate cDNA in yeast. Previously, we have used this method successfully to identify a number of fatty acid desaturases from organisms including C. elegans (8, 12). PCR was used to amplify the coding sequence of the ORFs of interest by using a C. elegans mixed-stage cDNA library as a template. To identify the elongation reaction responsible for the synthesis of di-homo-γ-linolenic acid (20:3, n-6) from 18:3, n-6, ORFs were expressed in the presence of this (exogenous) substrate. One cDNA ORF tested in this manner displayed a high level of elongation activity on the 18:3, n-6 substrate, converting 44% to 20:3, n-6 (Fig. 2). The identity of this elongation product was confirmed by GC comparison with a known standard, followed by GC-MS analysis. The DNA sequence and deduced amino acid sequence of this functional clone identified it as being encoded by the C. elegans gene F56H11.4. Fig. 3 indicates that, when compared with the deduced amino acid sequence of the yeast ELO genes, similarity was confined to a few short amino acid motifs. It was also clear that the deduced amino acid sequence of F56H11.4 (288 amino acid residues) showed no similarity to any previously described condensing enzymes (e.g., FAE1 or chalcone synthase). F56H11.4 does not contain the putative NADPH-binding site initially identified in the yeast ELO1 gene product (ref. 22; which is also absent in ELO2/3; ref. 20) and seems to contain multiple membrane-spanning domains. The protein is likely to be located (and retained) in the endoplasmic reticulum, as indicated by the presence of a C-terminal dilysine retention signal. We also observed some similarity with a mouse gene, Cig30 (23), which has been implicated in the recruitment of brown adipose tissue in liver tissue, as well as a potential human homologue encoded by a gene located on chromosome 4q25, bacterial artificial chromosome 207d4 (Fig. 3).
Figure 2.
Identification of fatty acid elongation activity in yeast expressing a C. elegans ORF. Chromatograms of fatty acid methyl esters from yeast transformed with the control (empty) plasmid pYES2 (A) or with ORF F56H11.4 in pYES2 (B). Exogenous substrate in the form of 18:3, n-6 (annotated as 18:3) was supplied to the cultures. Two additional peaks are observed in B; these peaks were identified (against known standards) as 20:3, n-6 and 18:1, n-7 and are annotated in B as 20:3 and 18:1* (in the latter case, this annotation is to distinguish this 18:1, n-7 fatty acid from 18:1, n-9). Detection was by flame ionization, with peak identification by subsequent GC-MS.
Figure 3.
Comparison of C. elegans ORF F56H11.4 with related sequences. The entire coding sequence of F56H11.4 (accession no. Z68749) was compared with the S. cerevisiae ELO ORFs (ELO1, Z49471; ELO2, CAA42301; ELO3, AAC28398) and a related mouse gene, Cig30 (U97107). The most closely related C. elegans ORFs, F41H10.8 (U61954) and F56H11.3 (Z68749) are also shown, as is part of a related human gene from chromosome IV (present on bacterial artificial chromosome clone B207d4; AC004050). The GenBank accession numbers are given for all sequences.
The range of fatty acids synthesized by C. elegans may require a number of different elongation reactions (24). We therefore determined the substrate specificity of the F56H11.4 activity by using a range of exogenously supplied fatty acids. This determination revealed (Table 1) that 18:3, n-6 is the major substrate, with a number of other fatty acids being elongated at a lower efficiency. Although most of these substrates are PUFAs, we were surprised to observe the (F56H11.4-directed) elongation of palmitoleic acid (16:1, n-7) to vaccenic acid (18:1, n-7). The biosynthetic pathway for 18:1, n-7 is unclear, but our data indicate that it may be synthesized by elongation of Δ9-monounsaturated C16 fatty acid. In some treatments, decreased levels of monounsaturated fatty acids resulted from the addition of exogenous C18 PUFAs, via their previously documented metabolic repression of the OLE1 Δ9-fatty acid desaturase (25). We also observed that, although expression of F56H11.4 conferred the ability to elongate C18 PUFAs, this ability was not observed for C20 PUFAs, even though C20 PUFAs are readily taken up yeast (10, 12).
Table 1.
Fatty acid composition of yeast expressing C. elegans ORF F56H11.4
| Fatty acid | mol percentage of fatty acids
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Control (pYES2)
|
F56H11.4
|
||||||||||
| −
|
−
|
18:3, n-6
|
18:2, n-6
|
18:3, n-3
|
20:5, n-3
|
||||||
| − | + | − | + | − | + | − | + | − | + | − | |
| 16:0 | 17.5 ± 3.3 | 19.9 ± 3.5 | 20.5 ± 4.1 | 27.7 ± 1.4 | 29.8 ± 0.2 | 22.9 ± 1.5 | 23.9 ± 1.0 | 19.1 ± 0.7 | 20.2 ± 1.1 | 23.4 ± 0.2 | 24.2 ± 1.0 |
| 16:1, n-7 | 53.2 ± 7.2 | 40.9 ± 3.1 | 49.4 ± 3.2 | 32.5 ± 4.4 | 34.4 ± 1.8 | 21.2 ± 2.2 | 24.8 ± 4.9 | 18.1 ± 1.5 | 19.6 ± 2.5 | 26.9 ± 0.7 | 34.5 ± 1.5 |
| 18:0 | 4.5 ± 0.7 | 4.7 ± 0.9 | 4.9 ± 0.5 | 5.6 ± 0.5 | 5.6 ± 0.3 | 5.1 ± 0.3 | 4.4 ± 0.1 | 5.0 ± 0.3 | 4.5 ± 0.1 | 5.3 ± 0.2 | 5.4 ± 0.2 |
| 18:1, n-9 | 24.8 ± 3.9 | 24.9 ± 1.4 | 25.2 ± 2.3 | 16.9 ± 0.9 | 16.1 ± 0.3 | 11.2 ± 2.4 | 10.7 ± 1.5 | 10.1 ± 1.1 | 9.9 ± 1.2 | 15.4 ± 0.4 | 17.8 ± 1.1 |
| 18:1, n-7 | — | 9.6 ± 0.6 | — | 3.9 ± 0.6 | — | 3.2 ± 0.6 | — | 3.1 ± 0.4 | — | 6.2 ± 0.3 | — |
| 18:2, n-6 | — | — | — | — | — | 34.4 ± 4.2 | 36.2 ± 5.6 | — | — | — | — |
| 18:3, n-3 | — | — | — | — | — | — | — | 43.1 ± 3.9 | 45.8 ± 4.8 | — | — |
| 18:3, n-6 | — | — | — | 7.5 ± 1.2 | 14.0 ± 0.3 | — | — | — | — | — | — |
| 20:2, n-6 | — | — | — | — | — | 2.0 ± 0.9 | — | — | — | — | — |
| 20:3, n-6 | — | — | — | 5.8 ± 0.9 | — | — | — | — | — | — | — |
| 20:3, n-3 | — | — | — | — | — | — | — | 1.5 ± 0.1 | — | — | — |
| 20:5, n-3 | — | — | — | — | — | — | — | — | — | 22.8 ± 0.7 | 20.1 ± 2.4 |
| Percentage elongated | |||||||||||
| 18:3, n-6 | — | — | — | 44 | — | — | — | — | — | — | — |
| 18:2, n-6 | — | — | — | — | — | 5.5 | — | — | — | — | — |
| 18:3, n-3 | — | — | — | — | — | — | — | 3.4 | — | — | — |
| 20:5, n-3 | — | — | — | — | — | — | — | — | — | 0 | — |
Substrate specificity of C. elegans ORF F56H11.4 expressed in yeast in the presence of different fatty acids; 18:3, n-6; 18:2, n-6; 18:3, n-3; or 20:5, n-3 fatty acids were added to the cultures before induction with galactose. Analysis by GC of fatty acid methyl esters was carried out on either induced (+) or noninduced (−) triplicated samples; induction time was 48 h. The fatty acid composition of control transformed yeast (empty pYES2 vector) is also indicated. All values are expressed as mol percentage of total fatty acids. Standard deviations are given (n = 4), as is the percentage elongation of different substrates.
The C. elegans ORF F56H11.4 is located on chromosome IV at 4.32 centimorgans, with a related sequence (F56H11.3; 51% similarity) 1,824 bp downstream. We also observed another C. elegans gene (F41H10.8), also present on chromosome IV, which shows a slightly higher level (53%) of similarity to F56H11.4 (Fig. 3). However, when a PCR product encoding ORF F41H10.8 was expressed in yeast in a manner identical to that used for F56H11.4, the former failed to direct the elongation of any fatty acids, despite the provision of a range of substrates (data not shown).
To reconstitute the PUFA biosynthetic pathway in a heterologous system, we expressed the PUFA elongating activity F56H11.4 in yeast in conjunction with either the Δ6- or Δ5-fatty acid desaturases previously isolated and characterized by our group (8, 12). Expression of the Δ6-fatty acid desaturase and F56H11.4 was carried out in the presence of two different substrates (18:2, n-6 or 18:3, n-3), but the Δ5-fatty acid desaturase and the elongase were expressed in the presence of 18:3, n-6 only. The results shown in Table 2 indicated that it was possible to combine a desaturase and an elongation activity in yeast to generate significant amounts of final “product.” In the case of F56H11.4 and the Δ6-fatty acid desaturase, the reactions proved highly efficient with the production of 4.5% of 20:3, n-6 from the 18:2, n-6 substrate; 25% of the 18:2, n-6 substrate was desaturated to 18:3, n-6, which was then elongated to 20:3, n-6 at a similar level of efficiency (18%). This elongation efficiency is lower than the percentage conversion observed for 18:3, n-6 when supplied exogenously (Table 1), indicating that in vivo production (as opposed to exogenous supplementation) of substrates for elongation may be rate-limiting. If 18:3, n-3 was used as a substrate, 27% was initially Δ6-desaturated to yield octadecatetraenoic acid (18:4, n-3), but only 8% was elongated subsequently to yield eicosatetraenoic acid (20:4, n-3). Thus, the conversion efficiency of 18:3, n-3 to the final C20 tetraenoic PUFA was only about 2.2%. These data indicate that the reconstituted elongase activity is capable of accepting both forms (n-6 and n-3) of essential fatty acid, although with slightly different efficiencies. We also verified that the C20 PUFAs synthesized in our yeast expression system were generated by the Δ6-desaturation of C18 substrates, which were elongated subsequently, because the Δ6-desaturase showed no activity on 20:2, n-6 or 20:3, n-3 substrates (Table 2). The combination of the Δ5-desaturase and F56H11.4 also demonstrated that these two enzymes could work in tandem, although the efficiency of this overall conversion was low (3.3% 20:4, n-6 from 18:3, n-6) because of the previously observed low activity of the Δ5-desaturase enzyme (10, 12). Thus, although nearly 45% of the 18:3, n-6 substrate was elongated to 20:3, n-6, only 7.5% was subsequently desaturated to 20:4, n-6 (Table 2).
Table 2.
Fatty acid composition of yeast coexpressing C. elegans ORF F56H11.4 and a fatty acid desaturase
| Fatty acid | mol percentage of fatty acids
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Δ6
|
F56H11.4 + Δ6
|
F56H11.4 + Δ5
|
||||||
| 20:2, n-6
|
20:3, n-3
|
18:2, n-6
|
18:3, n-3
|
18:3, n-6
|
||||
| + | − | + | − | + | − | + | − | |
| 16:0 | 24.7 ± 1.3 | 25.2 ± 1.5 | 18.7 ± 0.6 | 23.7 ± 0.5 | 17.4 ± 0.7 | 21.0 ± 1.3 | 27.9 ± 4.2 | 29.8 ± 3.8 |
| 16:1, n-7 | 46.0 ± 2.8 | 43.7 ± 3.7 | 18.9 ± 1.2 | 24.6 ± 0.7 | 5.3 ± 0.6 | 9.1 ± 0.9 | 24.6 ± 3.4 | 25.1 ± 3.2 |
| 16:2, n-7 | 5.2 ± 1.2 | 4.1 ± 1.4 | 0.6 ± 0.1 | — | 0.4 ± 0.1 | — | — | — |
| 18:0 | 4.8 ± 0.4 | 5.1 ± 0.4 | 4.0 ± 0.3 | 5.1 ± 0.1 | 6.2 ± 0.7 | 5.4 ± 0.2 | 5.6 ± 0.8 | 5.4 ± 0.7 |
| 18:1, n-9 | 15.3 ± 1.1 | 16.1 ± 1.2 | 12.2 ± 1.4 | 11.2 ± 0.4 | 5.7 ± 0.8 | 6.0 ± 0.4 | 12.7 ± 2.9 | 13.0 ± 2.5 |
| 18:1, n-7 | — | — | 7.7 ± 0.7 | — | 2.6 ± 0.3 | — | 2.9 ± 0.9 | — |
| 18:2, n-6 | — | — | 25.0 ± 3.2 | 35.4 ± 2.1 | — | — | — | — |
| 18:3, n-3 | — | — | — | — | 42.3 ± 3.3 | 58.5 ± 4.7 | — | — |
| 18:3, n-6 | — | — | 7.9 ± 2.2 | — | — | — | 13.2 ± 3.6 | 19.2 ± 3.5 |
| 18:4, n-3 | — | — | — | — | 15.3 ± 1.8 | — | — | — |
| 20:2, n-6 | 4.0 ± 0.3 | — | 3.3 ± 0.5 | — | — | — | — | — |
| 20:3, n-6 | — | — | 1.7 ± 0.2 | — | — | — | 9.8 ± 1.8 | — |
| 20:3, n-3 | — | 5.8 ± 0.5 | — | — | 3.4 ± 0.4 | — | — | — |
| 20:4, n-6 | — | — | — | — | — | — | 0.8 ± 0.2 | — |
| 20:4, n-3 | — | — | — | — | 1.4 ± 0.2 | — | — | — |
| Percentage elongated | ||||||||
| 18:3, n-6 | — | — | 17.7 | — | — | — | 44.5 | — |
| 18:4, n-3 | — | — | — | — | 8.4 | — | — | — |
| 18:2, n-6 | — | — | 8.7 | — | — | — | — | — |
| 18:3, n-3 | — | — | — | — | 5.4 | — | — | — |
Fatty acid composition of yeast transformed with F56H11.4 and either the borage Δ6-fatty acid desaturase or the M. alpina Δ5-fatty acid desaturase. Exogenous substrates were 18:3, n-6; 18:3, n-3 (Δ6); or 18:3, n-6 (Δ5). The Δ6-desaturase also was expressed alone in the presence of either 20:2, n-6 or 20:3, n-3 substrates. The fatty acid compositions of transformed yeast were analyzed as described for Table 1.
Finally, the production of either 20:4, n-6 or eicosapentaenoic acid (20:5, n-3) yeast from dienoic or trienoic C18 substrates was achieved via expression of all three enzymes (the two desaturases and the F56H11.4 PUFA elongating activity simultaneously. As shown in Table 3, small but significant amounts of 20:4, n-6 were produced when the yeast was supplied with the dienoic substrate 18:2, n-6. The identity of this C20 PUFA was verified by GC-MS, indicating that the conversion efficiency of 18:2, n-6 to 20:4, n-6 was 0.65%. When 18:3, n-3 was used as a substrate, 12.5% of the (Δ6-desaturated and elongated) 20:4, n-3 fatty acid was Δ5-desaturated, resulting in a total conversion of 0.3% of the substrate 18:3, n-3 to 20:5, n-3 (the identity of which was also confirmed by GC-MS).
Table 3.
Fatty acid composition of yeast expressing C. elegans ORF F56H11.4 and both fatty acid desaturases required for PUFA biosynthesis
| Fatty acid | mol percentage fatty acids
|
|||
|---|---|---|---|---|
| F56H11.4 + Δ5 + Δ6
| ||||
| 18:2, n-6
|
18:3, n-3
|
|||
| + | − | + | − | |
| 16:0 | 19.1 ± 0.9 | 22.8 ± 1.3 | 17.1 ± 0.9 | 21.8 ± 0.3 |
| 16:1, n-7 | 17.2 ± 0.6 | 23.4 ± 0.3 | 5.2 ± 0.8 | 10.6 ± 0.9 |
| 16:2, n-7 | 0.4 ± 0.2 | — | 0.2 ± 0.1 | — |
| 18:0 | 4.2 ± 0.1 | 5.1 ± 0.1 | 5.6 ± 0.5 | 5.4 ± 0.1 |
| 18:1, n-9 | 12.8 ± 1.5 | 10.5 ± 0.7 | 6.0 ± 0.8 | 6.3 ± 0.6 |
| 18:1, n-7 | 8.0 ± 1.7 | — | 2.5 ± 0.8 | — |
| 18:2, n-6 | 25.8 ± 2.1 | 38.2 ± 1.6 | — | — |
| 18:3, n-3 | — | — | 47.6 ± 3.4 | 55.9 ± 1.4 |
| 18:3, n-6 | 6.8 ± 1.4 | — | — | — |
| 18:4, n-3 | — | — | 10.8 ± 1.9 | — |
| 20:2, n-6 | 3.65 ± 1.2 | — | — | — |
| 20:3, n-6 | 1.4 ± 0.3 | — | — | — |
| 20:3, n-3 | — | — | 3.4 ± 0.2 | — |
| 20:4, n-6 | 0.25 ± 0.1 | — | — | — |
| 20:4, n-3 | — | — | 1.4 ± 0.3 | — |
| 20:5, n-3 | — | — | 0.2 ± 0.1 | — |
| Percentage elongated | ||||
| 18:3, n-6 | 19.5 | — | — | — |
| 18:4, n-3 | — | — | 12.9 | — |
| 18:2, n-6 | 9.6 | — | — | — |
| 18:3, n-3 | — | — | 5.4 | — |
Fatty acid composition of yeast cotransformed with F56H11.4 and both the Δ5- and Δ6-fatty acid desaturases; induction time was 96 h. Either 18:2, n-6 or 18:3, n-3 was supplied as exogenous substrates. Analysis was performed as described for Table 1.
Discussion
We have identified a C. elegans ORF F56H11.4 that is capable of directing the C2 elongation of γ-linolenic acid when heterologously expressed in yeast. The expression of a single ORF might not be expected to result in fatty acid elongation, because it is generally considered that the elongation activity consists of four distinct enzymes (15), and it is highly unlikely that all four activities are contained within a single polypeptide. Therefore, the “elongase” activity observed with the expression of F56H11.4 in yeast is likely to arise from an interaction with other endogenous components of a yeast elongase (which presumptively would interact normally with the ELO gene products). Interestingly, expression of the Arabidopsis FAE1 ORF in yeast results in the accumulation of very long-chain monounsaturated fatty acids (via the elongation of C18:1; ref. 17). The authors considered it likely that FAE1 (encoding the condensing enzyme subunit of the elongase) was able to interact with endogenous reductases and dehydrase to form a functional elongase complex (17). Whether FAE1 and F56H11.4 reconstitute their distinct elongase activities by interacting with common yeast components is an interesting (but open) question. The C. elegans ORF described in the present study does not show any similarity to FAE1, even over short domains, although FAE1 shows some limited similarity to other condensing enzymes. In fact, the only obvious motifs present in F56H11.4 are a “histidine box” normally found in enzymes such as desaturases (4), several conserved tyrosine residues, and a potential C-terminal endoplasmic reticulum-retention/recycling motif. In view of the presence of a number of predicted transmembrane domains in F56H11.4, it is probable that this ORF encodes an integral (endo)membrane protein, which is in good agreement with previous biochemical studies on mammalian elongases (15). Although, intuitively, it seems likely that C. elegans ORF F56H11.4 encodes a condensing enzyme, this likelihood awaits biochemical confirmation. The lack of similarity with previously described condensing enzymes (such as FAE1) may indicate that several distinct forms of this enzyme exist. There are at least seven ORFs related to F56H11.4 present in the C. elegans genome, which may represent other distinct (condensing) enzymes. Based on our data, these different condensing enzyme activities might determine the nature and substrate specificity of an elongase complex.
We also observed that F56H11.4 is in close proximity to a related gene (F56H11.3; 51% similarity). The presence of related (but distinct) enzymes involved in PUFA synthesis in close physical proximity has been observed previously by our group in the case of the C. elegans Δ5- and Δ6-fatty acid desaturases, which are less than 1 kilobase apart (5, 8, 12). That the Δ5- and Δ6-fatty acid desaturases and the PUFA elongase both map to very similar regions of chromosome IV (4.41, 4.42, and 4.32 centimorgans, respectively; less than 300 kilobases apart) may also be of significance. It is also clear that the PUFA elongating activity F56H11.4 and its proximal partner (F56H11.3) contain two conserved intron/exon junctions; similarly, the presence of conserved intron/exon junctions in the Δ5- and Δ6-fatty acid desaturases has also been reported (12, 26). One rationale for these presumptive gene-duplication events is that they drive the evolution of new enzyme activities (as in the case of the fatty acid desaturase).
In respect to the substrates used by the reconstituted PUFA elongation activity, the underlying mechanisms that allow this enzyme to elongate 18:3, n-6 (16:1, n-7), although they discriminate against 18:2, n-6 and 18:3, n-3, remain to be elucidated. Also, although the elongation of 16:1, n-7 was unexpected, this elongation may account for the significant levels of 18:1, n-7 observed in C. elegans (24). Nevertheless, it is clear that the fatty acid elongation activity described in this study does display substrate preference. Elongation of substrates such as 18:3, n-3 or 18:2, n-6 is less efficient than that observed for 18:3, n-6. The efficiency of elongation for tetraenoic fatty acid 18:4, n-3 is also twice that for 18:3, n-3, although only slightly greater (35%) than that for 18:2, n-6. Thus, the F56H11.4 enzyme recognizes a range of substrates commensurate with it having a major role in PUFA biosynthesis but displays a clear preference for Δ6-desaturated (n-6 or n-3) C18 fatty acids. This result is also in agreement with the reported profile of C. elegans PUFAs (24).
The coexpression of the elongase with the Δ5- and Δ6-fatty acid desaturases resulted in the production of C20 PUFAs, with both 20:4, n-6 and 20:5, n-3 being detected, although the latter was at a lower level. Previously, Browse and coworkers (27) functionally characterized a C. elegans ω3-fatty acid desaturase with specificity for both C18 and C20 substrates that was capable of producing 20:5, n-3 from 20:4, n-6. Thus, the biosynthetic pathway for 20:5, n-3 could be via either a final methyl-directed desaturation of 20:4 n-6 or the Δ5-desaturation of the 20:4, n-3 fatty acid.
Conclusions
We have isolated an enzymatic component of the C. elegans elongase responsible for the synthesis of C20 PUFAs. This ORF shows limited similarity to yeast genes required for medium-chain (ELO1) and sphingolipid very long-chain (ELO2/3) saturated fatty acid elongation but displays a high degree of substrate specificity for C18 trienoic and tetraenoic fatty acids. It is likely that F56H11.4 encodes a condensing enzyme activity that interacts with endogenous yeast enzyme components to reconstitute a functional elongase.
Expression of the C. elegans fatty acid elongation activity in yeast in conjunction with PUFA-specific fatty acid desaturases resulted in the accumulation of trienoic, tetraenoic, and pentaenoic C20 fatty acids from dienoic and trienoic C18 precursors. Thus, it is possible to reconstitute this metabolic pathway in a heterologous host, providing a biotechnological method for the synthesis of these valuable compounds.
Acknowledgments
We are grateful to Prof. Yuji Kohara (National Institute for Genetics, Mishima, Japan) for providing the C. elegans cDNA library. Institute of Arable Crops Research receives grant-aided support from the Biotechnology and Biological Sciences Research Council (United Kingdom).
Abbreviation
- PUFA
polyunsaturated fatty acid
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.110140197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.110140197
References
- 1.Gill I, Valivety R. Trends Biotechnol. 1997;15:401–409. doi: 10.1016/S0167-7799(97)01076-7. [DOI] [PubMed] [Google Scholar]
- 2.Broun P, Gettner S, Somerville C. Annu Rev Nutr. 1999;19:197–216. doi: 10.1146/annurev.nutr.19.1.197. [DOI] [PubMed] [Google Scholar]
- 3.Horrobin D F. Rev Contemp Pharmacother. 1990;1:1–45. [Google Scholar]
- 4.Shanklin J, Cahoon E B. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:611–641. doi: 10.1146/annurev.arplant.49.1.611. [DOI] [PubMed] [Google Scholar]
- 5.Napier J A, Michaelson L V, Stobart A K. Curr Opin Plant Biol. 1999;2:123–127. doi: 10.1016/s1369-5266(99)80025-9. [DOI] [PubMed] [Google Scholar]
- 6.Jimenez J, Boza J, Suarez M D, Gil A. J Nutr Biochem. 1997;8:217–223. [Google Scholar]
- 7.Sayanova O, Smith M A, Lapinskas P, Stobart A K, Dobson G, Christie W W, Shewry P R, Napier J A. Proc Natl Acad Sci USA. 1997;94:4211–4216. doi: 10.1073/pnas.94.8.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Napier J A, Hey S J, Lacey D J, Shewry P R. Biochem J. 1998;330:611–614. doi: 10.1042/bj3300611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho H P, Nakamura M T, Clarke S D. J Biol Chem. 1999;274:471–477. doi: 10.1074/jbc.274.1.471. [DOI] [PubMed] [Google Scholar]
- 10.Michaelson L V, Lazarus C M, Griffiths G, Napier J A, Stobart A K. J Biol Chem. 1998;273:19055–19059. doi: 10.1074/jbc.273.30.19055. [DOI] [PubMed] [Google Scholar]
- 11.Knutzon D S, Thurmond J M, Huang Y-S, Chaudhary S, Bobik E G, Chan G M, Kirchner S J, Mukerji P. J Biol Chem. 1998;273:29360–29366. doi: 10.1074/jbc.273.45.29360. [DOI] [PubMed] [Google Scholar]
- 12.Michaelson L V, Napier J A, Lazarus C M, Griffiths G, Stobart A K. FEBS Lett. 1998;439:215–218. doi: 10.1016/s0014-5793(98)01385-4. [DOI] [PubMed] [Google Scholar]
- 13.Cho H P, Nakamura M, Clarke S D. J Biol Chem. 1999;274:37335–37339. doi: 10.1074/jbc.274.52.37335. [DOI] [PubMed] [Google Scholar]
- 14.Girke T, Schmidt H, Zahringer U, Reski R, Heinz E. Plant J. 1998;15:39–48. doi: 10.1046/j.1365-313x.1998.00178.x. [DOI] [PubMed] [Google Scholar]
- 15.Cinti D L, Cook L, Nagi M H, Suneja S K. Prog Lipid Res. 1992;31:1–51. doi: 10.1016/0163-7827(92)90014-a. [DOI] [PubMed] [Google Scholar]
- 16.James D W, Lim E, Keller J, Plooy I, Ralston E, Dooner H K. Plant Cell. 1995;7:309–319. doi: 10.1105/tpc.7.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Millar A A, Kunst L. Plant J. 1997;12:121–131. doi: 10.1046/j.1365-313x.1997.12010121.x. [DOI] [PubMed] [Google Scholar]
- 18.Napier J A, Sayanova O, Sperling P, Heinz E. Trends Plant Sci. 1999;4:2–5. [Google Scholar]
- 19.Sperling P, Zahringer U, Heinz E. J Biol Chem. 1998;273:28590–28596. doi: 10.1074/jbc.273.44.28590. [DOI] [PubMed] [Google Scholar]
- 20.Oh C-S, Toke D A, Mandala S, Martin C E. J Biol Chem. 1997;272:17373–17384. doi: 10.1074/jbc.272.28.17376. [DOI] [PubMed] [Google Scholar]
- 21.The C. elegans Sequencing Consortium. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 22.Toke D A, Martin C E. J Biol Chem. 1996;271:18413–18422. doi: 10.1074/jbc.271.31.18413. [DOI] [PubMed] [Google Scholar]
- 23.Tvrdik P, Asadi A, Kozak L P, Nedergaard J, Cannon B, Jacobsson A. J Biol Chem. 1997;272:31738–31746. doi: 10.1074/jbc.272.50.31738. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka T, Ikita K, Ashida T, Motoyama Y, Yamaguchi Y, Satouchi K. Lipids. 1996;31:1173–1178. doi: 10.1007/BF02524292. [DOI] [PubMed] [Google Scholar]
- 25.Stukey J E, McDonough V M, Martin C E. J Biol Chem. 1989;264:16537–16544. [PubMed] [Google Scholar]
- 26.Watts J L, Browse J. Arch Biochem Biophys. 1999;362:175–182. doi: 10.1006/abbi.1998.1024. [DOI] [PubMed] [Google Scholar]
- 27.Spychalla J P, Kinney A J, Browse J. Proc Natl Acad Sci USA. 1997;94:1142–1147. doi: 10.1073/pnas.94.4.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]