Abstract
Stat1 plays an essential role in signal transduction and gene expression of various cytokines including interferons (IFNs). Although the mechanism of cytokine-induced activation of Stat1 and transcriptional regulation of Stat1 gene expression have been established, post-transcriptional regulation of Stat1 protein expression is not fully understood. Here we report identification of a mutant of Stat1 that has decreased expression levels by using inducible translocation trap (ITT), a reporter gene-based detection system of nuclear translocation. The substitution of serine for phenylalanine 172 (F172S) in the coiled-coil domain causes marked decrease in Stat1 protein expression in various cell lines without decreasing its mRNA levels. Our results suggest that the decrease is caused by translational/post-translational mechanisms independent of proteasome machinery. These results suggest a novel potential mechanism of determination of specificity of Stat proteins and showed that the ITT system is a powerful technique to identify mutants of nuclear translocating signal transducers.
Keywords: Stat1, protein expression, mutation analysis, inducible translocation trap
Introduction
Stat1 plays an essential role in IFNγ-mediated biological responses [1, 2]. Binding of IFNγto its receptor induces activation of the receptor-associated Jak1 and Jak2 by transphosphorylation. The activated Jak kinases phosphorylate the intracellular domain of the receptor, which serves as a docking site for Stat1. Stat1 is phosphorylated on tyrosine 701, undergoes dimerization through Src-homology-2 (SH2) domain, translocates to the nucleus and regulates gene expression by binding to γ-activated sequence elements. In addition to well-characterized DNA-binding, SH2, and transactivation domains, Stat proteins have amino (N)-terminal, coiled-coil, and linker domains [1]. The N-terminal domain is necessary for tetramer formation of Stat proteins and efficient target gene expression. The coiled-coil domain mediates protein-protein interaction. The linker domain is involved in determination of dissociation time of Stat1 from DNA [3].
Although mechanism of activation and transcriptional regulation of Stat1 have been established, post-transcriptional regulation of Stat1 expression is not fully understood. Using inducible translocation trap (ITT), a reporter gene-based detection system of nuclear translocation [4], we identified a functional Stat1 mutant that shows markedly decreased protein expression. The causative mutation, phenylalanine 172 to serine (F172S) in the coiled-coil domain, decreased the protein expression without decreasing mRNA expression. Different nucleotide substitutions for serine residue exerted identical effects. Proteasome inhibitor failed to increase F172S expression. These results suggest that the F172S mutation affects Stat1 expression by translational/post-translational mechanism independent of proteasome machinery. Since the phenylalanine is not conserved in Stat4 and Stat6 that have tight specificity, our results suggest a novel potential mechanism of determination of specificity of Stat proteins. In addition, our results showed that the ITT system is a powerful technique to identify mutants of nuclear translocating signal transducers.
Materials and Methods
Cell lines, plasmids, and gene transfer
BLG is a murine hematopoietic Ba/F3-derived cell line containing LexA-d1EGFP reporter gene [4]. Stat1-deficient mouse embryonic fibroblasts (MEFs) and Stat1-deficient U3A derived from human fibroblast cell line, 2fTGH, were described elsewhere [5, 6].
Stat1 cDNA amplified by RT-PCR from Ba/F3 mRNA was cloned into pLG vector as described previously [4]. For construction of pLG-Stat1 F172S, PCR product amplified with 5′-ACAGGATCCATGTCACAGTGGTTCGAGCTTCAG-3′ and 5′-TGCAAGTACTGTCATATTCATCTTG-3′ as primers and pLG-Stat1 wild-type (WT) [4] as template was digested with BamH I and Sca I and inserted into pLG-Stat1 WT cleaved with BamH I and Dra I. For construction of HA-Stat1 WT/pMX-puro or HA-Stat1 F172S/pMX-puro, Stat1 WT or F172S mutant cDNA from pLG-Stat1 WT or pLG-Stat1 F172S was inserted into pMX-puro [7] together with cDNA fragment encoding hemagglutinin (HA)-epitope.
Plasmids were transfected into COS cells with Lipofectamine 2000 (Invitrogen). Retroviral gene transfer was performed as described previously [4].
ITT assay
BLG cells transduced with pLG-Stat1 WT or pLG-Stat1 mutants were analyzed for GFP expression with FACS Calibur (BD Biosciences). GFP expression was quantified by FlowJo software (Tree Star).
Immunoblot Analysis
Preparation of whole cell lysates (WCL) and immunoblot analyses were described previously [4, 8, 9]. The Antibodies (Abs) used in this study are anti-HA (clone 12CA5, Roche Applied Science), anti-Stat1 (Upstate), and anti-IRF-1 (C-20, Santa Cruz Biotechnology). For proteasome inhibitor experiments, cells were incubated with MG132 (10 μM, Peptide Institute) for 5 hrs.
Northern blot analysis
Total RNA was purified with RNeasy Protect Mini-Kit (Qiagen). Northern blot analysis was performed as previously described [10] except that DNA probes were labeled with digoxigenin (DIG)-dUTP and hybrids were detected by enzyme immunoassay (Roche Applied Science). HA-Stat1 WT/pMX-puro digested with Nco I was used as a probe.
Results and Discussion
Identification of Stat1 F172S mutant by ITT
Recently, we reported the establishment of ITT, a reporter gene-based system to detect nuclear translocation of signaling proteins [4]. In ITT, a fusion protein consisting of a LexA DNA-binding domain (LexA DB), the transactivation domain (TA) of a transcriptional activator, and a test protein is expressed in cell lines containing a reporter gene with LexA-binding sites in its promoter. When the fusion protein translocates into the nucleus, the fusion protein binds LexA-binding sites of the reporter gene with LexA DB and induces reporter expression by the action of the TA. Thus, reporter gene expression reflects nuclear translocation of the fusion protein.
In the establishment of ITT, Stat1 cDNA amplified by RT-PCR from Ba/F3 mRNA was sucloned into pLG to express Stat1 fused with LexA DB and Gal4 TA. We expressed Stat1 fusion proteins in BLG, which is a Ba/F3-derived cell line containing LexA-d1EGFP reporter. Flowcytometric analysis revealed two distinct GFP expression patterns in the absence of IFNγ, depending on transduced cDNAs: high GFP expression, or low GFP expression (data not shown, see below). GFP expression in the absence of IFNγ reflects ligand-independent nucleo-cytoplasmic shuttling of Stat1 [11]. Sequence analysis revealed that cDNA species that conferred high GFP expression were WT Stat1, whereas cDNA species that conferred low GFP expression had three point mutations: serine 144 to asparagine (AGT to AAT: S144N), methionine 154 to isoleucine (ATG to ATA: M154I), and phenylalanine 172 to serine (TTT to TCT: F172S). It is likely that these mutations were introduced in RT-PCR.
We constructed three Stat1 mutants that harbor each of the above amino acid substitutions and inserted into pLG. For construction of the F172S mutant, the serine residue was encoded by AGT instead of TCT in the original triple mutant. Background GFP expression in BLG cells transduced with pLG-Stat1 WT were high [mean fluorescence intensity (MFI) of GFP (+) cells: 1110] (Fig. 1, left panel), whereas BLG cells transduced with pLG-Stat1 F172S showed markedly lower GFP expression [MFI of GFP (+) cells: 113] (Fig. 1, right panel) identical to the original triple mutant. GFP expression in BLG cells transduced with pLG Stat1 S144N and M154I were comparable to that of WT (data not shown). These data showed that the F172S mutation caused lower background GFP expression in ITT assay.
Figure 1.
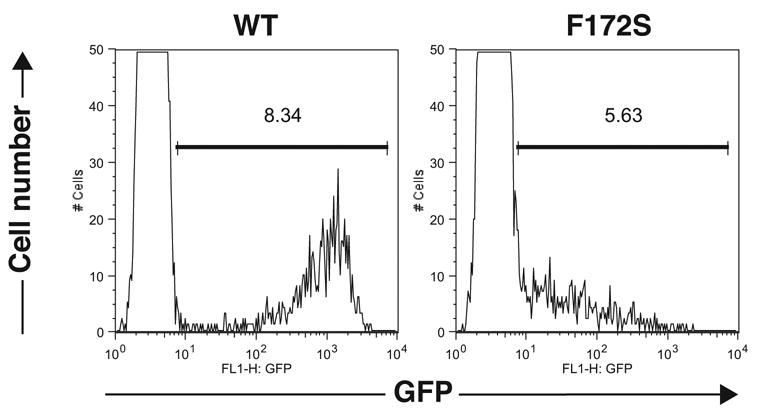
Lower GFP expression in BLG cells expressing LG-Stat1 F172S in the absence of IFNγ. GFP expression in BLG cells transduced with pLG-Stat1 WT [MFI of GFP (+) cells: 1110] (left panel) or pLG-Stat1 F172S [MFI of GFP (+) cells: 113] (right panel) are shown.
Reduced protein expression of the F172S mutant by translational/post-translational mechanisms
Lower background reporter expression by the F172S mutant in ITT assay reflects lower amounts of the LexA DB/Gal4 TA-fusion protein in the nucleus. To distinguish whether this is caused by (i) attenuated constitutive nuclear translocation or (ii) decreased protein expression, we expressed HA-tagged WT or the F172S mutant of Stat1 into COS cells. Protein expression was analyzed by immunoblot analysis with anti-HA Ab. Expression of WT Stat1 was clearly detected, whereas expression of the F172S mutant was barely detectable (Fig. 2A). mRNA levels of the F172S mutant were comparable to or even higher than those of WT (Fig. 2B), excluding the possibilities of lower transfection efficiency and lower mRNA expression.
Figure 2.
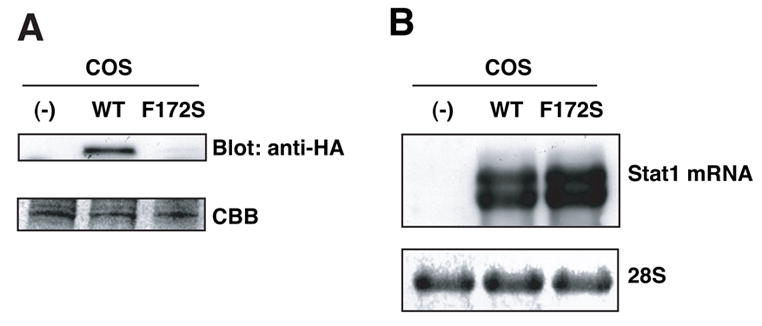
Lower protein expression of Stat1 F172S mutant in COS cells. (A) WCL from COS cells expressing HA-tagged WT Stat1 or the F172S mutant were subjected to immunoblot analysis with anti-HA Ab. Equal loading was confirmed by Coumassie Brilliant Blue (CBB) staining. (B) mRNA expression in transfected COS cells. Total RNA was subjected to Northern blot analysis with mouse Stat1 probe.
Consistent with the results of the COS cell experiment, protein expression of the F172S mutant was considerably lower than that of WT in Stat1-deficient MEFs [5] (Fig. 3A). In contrast, mRNA levels of the F172S mutant were higher than those of WT (Fig. 3B). Similar results were obtained in Stat1-deficient U3A cells (Fig. 4A). These results confirmed that the protein expression of the F172S mutant was markedly lower than that of WT and that the reduced protein expression is caused by translational or post-translational mechanisms.
Figure 3.
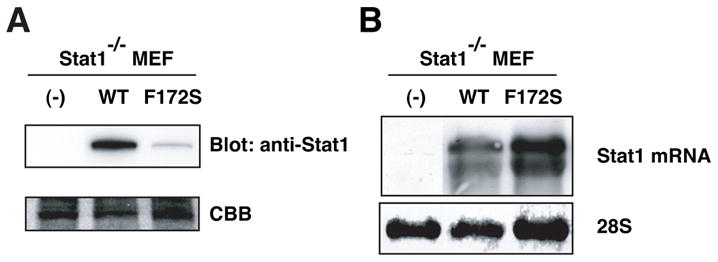
Lower protein expression of Stat1 F172S mutant in Stat1-deficient MEFs. (A) WCL from Stat1-deficient MEFs expressing WT Stat1 or the F172S mutant were subjected to immunoblot analysis with anti-Stat1 Ab. (B) Expression of mRNA in Stat1-deficient MEF-derived cells. Total RNA was subjected to Northern blot analysis with mouse Stat1 probe.
Figure 4.
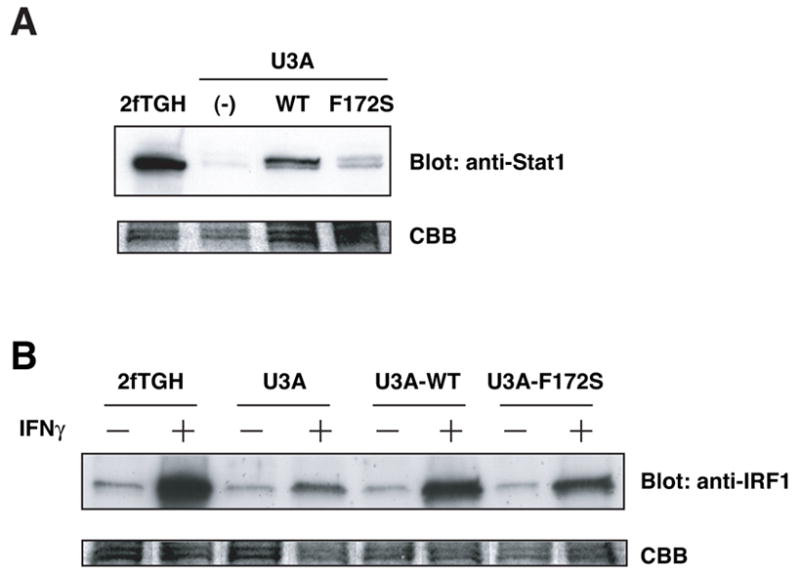
The F172S mutant can mediate IFNγ-induced gene expression. (A). Expression of WT Stat1 and the F172S mutant in U3A cells. WCL were subjected to immunoblot analysis with anti-Stat1 Ab. (B) IFNγ-induced IRF-1 expression in Stat1-reconstituted U3A cells. WCL were subjected to immunoblot blot analysis with anti-IRF-1 Ab.
Considering that two different codons (TCT and AGT) for the serine substitution for F172 exerted identical effects on GFP expression in ITT assay, we prefer the latter possibility because it is difficult to speculate potential mechanism of decreased translation by different nucleotide substitutions. Coiled-coil domain is one of the principal subunit oligomerization motifs in proteins [12]. In fact, interaction with several proteins with the coiled-coil domain of Stat1 has been reported [1]. It is possible that a molecule that induces protein degradation interacts with the F172S mutant with higher affinity than with WT. Recently, a novel ubiquitin E3 ligase, Stat-interacting LIM protein (SLIM), was shown to reduces Stat1 protein expression [13]. An interesting possibility is that interaction between SLIM and the Stat1 F172S mutant would be stronger than that between SLIM and WT Stat1.
The F172S mutant is functional in IFNγ-induced gene expression
Next, we examined whether the F172S mutant is functional. We stimulated 2fTGH, U3A, U3A expressing HA-Stat1 WT or HA-Stat1 F172S mutant with IFNγ. WCL were subjected to immunoblot analysis with anti-IRF-1 Ab. IRF-1 is one of the direct target genes of IFNγ-activated Stat1 [14, 15]. IFNγ induced expression of IRF-1 in 2fTGH cell line (Fig. 4B). In contrast, induction of IRF-1 by IFNγ in U3A cells was marginal. IFNγ stimulation induced IRF-1 expression in U3A cells expressing either WT Stat1 or the F172S mutant although IRF-1 expression in cells expressing the F172S mutant was slightly lower than that in cells expressing WT. These results showed that the F172S mutant is functional.
Proteasome-independent regulation of Stat1 protein expression in the absence of IFNγ
Proteasome is a ubiquitous protein degradation machinery involved in regulation of various important proteins [16]. To investigate potential involvement of proteasome in regulation of protein expression of WT Stat1 and the F172S mutant, we incubated Stat1-deficient MEFs expressing HA-Stat1 WT or HA-Stat1 F172S with a proteasome inhibitor, MG132. We did not detect effects of MG132 on protein expression levels of WT and the F172S mutant (Fig. 5). Similar results were obtained in COS cells (data not shown). These results suggest that proteasome machinery is involved in neither decreased expression of the F172S mutant nor steady-state expression of WT Stat1 in the absence of IFNγ. It was shown that the ubiquitin-proteasome pathway regulates IFNγ-activated Stat1 [17]. Therefore, it is likely that involvement of proteasome machinery is limited to degradation of Stat1 transported into the nucleus by IFNγ.
Figure 5.
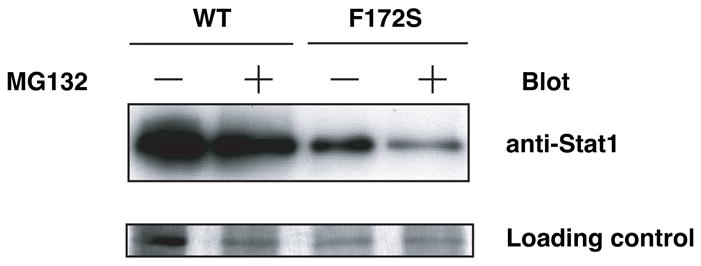
Proteasome-independent regulation of Stat1 protein expression in the absence of IFNγ. Stat1-deficient MEFs expressing HA-Stat1 WT or HA-Stat1 F172S were incubated with MG132 for 5 hrs. WCL were subjected to immunoblot analysis with anti-HA Ab. Silver staining is shown as a loading control.
Potential involvement of the coiled-coil domain of Stats in determination of specificity
In cytokine signaling, it is essentially important to ensure that each cytokine elicits specific activation of Stat proteins. For example, specific activation of Stat4 by IL-12 and Stat6 by IL-4 is essential for differentiation of naive T cells into Th1 and Th2 effector T cells, respectively [18–20]. Specificity of Stat activation is primarily determined by tyrosine-based motif in the cytoplasmic domains of cytokine receptors [1]. However, it is known that overexpression of Stat proteins abrogates their specific activation, suggesting importance of regulation of Stat expression levels to ensure their specificity.
Alignment of human and mouse Stats revealed that the F172 in Stat1 is conserved in Stat1, Stat2, Stat3, and Stat5, but not in Stat4 and Stat6 (Fig. 6). Stat4 and Stat6 are specifically activated by IL-12/IL-23 or IL-4/IL-13, respectively, whereas Stat1, Stat3, and Stat5 are activated by a variety of cytokines [1]. Since F172S mutation caused decreased Stat1 protein expression, an interesting possibility is that protein expression of Stat4 and Stat6 per mRNA might be lower than that of Stat1, Stat3, and Stat5. The potential lower expression of Stat4 and Stat6 might contribute to their tight specificity. In this regard, the phenylalanine residue is also conserved in Stat2, which is specifically activated by IFNα/β. In contrast to other Stats that can form homo- or heterodimers, Stat2 is activated only as a component of IFN-stimulated gene factor 3, heterotrimer of Stat1, Stat2, and IRF-9 [2]. Therefore, it is possible that specific activation of Stat2 does not require regulation of protein expression by this phenylalanine.
Figure 6.

Sequence alignment of the first α-helix in the coiled-coil domains of Stats. Blue: residues identical to human or mouse Stat; Yellow: residues related to human or mouse Stat1; Green: residues not conserved between mouse and human Stat1. The position of F172 in Stat1 is indicated with an asterisk.
In conclusion, we identified a functional Stat1 mutant that shows markedly decreased protein expression caused by translational/post-translational mechanisms independent of proteasome machinery. Since the phenylalanine is not conserved in Stat4 and Stat6 that have tight specificity, our results suggest a novel potential mechanism of specific activation of Stat proteins. The F172S mutant might be a useful tool in various research and clinical settings such as gene therapy of Stat1-deficient patients [21]. Lastly, this mutant was identified by ITT method, suggesting that ITT is a useful system to identify mutants of signaling molecules or for high throughput screening of modifiers of nuclear translocating signal transducers.
Acknowledgments
We thank Drs. T. Kitamura, D. E. Levy, and G. R. Stark for pMX-puro, Stat1-deficient MEF, and 2fTGH and U3A, respectively. This work is supported by NIH grant R01 AI059315 (H. F.).
References
- 1.Levy DE, Darnell JE., Jr Stats: Transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 2.Stark GR, Kerr IM, Williams BRG, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 3.Yang E, Henriksen MA, Schaefer O, Zakharova N, Darnell JE., Jr Dissociation time from DNA determines transcriptional function in a STAT1 linker mutant. J Biol Chem. 2002;277:13455–13462. doi: 10.1074/jbc.M112038200. [DOI] [PubMed] [Google Scholar]
- 4.Hoshino A, Matsumura S, Kondo K, Hirst JA, Fujii H. Inducible translocation trap: a system for detecting inducible nuclear translocation. Mol Cell. 2004;15:153–159. doi: 10.1016/j.molcel.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 5.Durbin JE, Hackenmiller R, Simon MC, Levy DE. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral diseases. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 6.Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 7.Kitamura T. New experimental approaches in retrovirus-mediated expression screening. Int J Hematol. 1998;67:351–359. doi: 10.1016/s0925-5710(98)00025-5. [DOI] [PubMed] [Google Scholar]
- 8.Fujii H, Ogasawara K, Otsuka H, Suzuki M, Yamamura K-i, Yokochi T, Miyazaki T, Suzuki H, Mak TW, Taki S, Taniguchi T. Functional dissection of the cytoplasmic subregions of the IL-2 receptor βc chain in primary lymphocyte populations. EMBO J. 1998;17:6551–6557. doi: 10.1093/emboj/17.22.6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsujino S, Di Santo JP, Takaoka A, McKernan TL, Noguchi S, Taya C, Yonekawa H, Saito T, Taniguchi T, Fujii H. Differential requirement of the cytoplasmic subregions of γc chain in T cell development and function. Proc Natl Acad Sci USA. 2000;97:10514–10519. doi: 10.1073/pnas.180063297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shibuya H, Yoneyama M, Ninomiya-Tsuji J, Matsumoto K, Taniguchi T. IL-2 and EGF receptors stimulate the hematopoietic cell cycle via different signaling pathways: demonstration of a novel role for c-myc. Cell. 1992;70:57–67. doi: 10.1016/0092-8674(92)90533-i. [DOI] [PubMed] [Google Scholar]
- 11.Kumar A, Commane M, Flickinger TW, Horvath CM, Stark GR. Defective TNF-α-induced apoptosis in Stat1-null cells due to low constitutive levels of caspases. Science. 1997;278:1630–1632. doi: 10.1126/science.278.5343.1630. [DOI] [PubMed] [Google Scholar]
- 12.Burkhard P, Stetefeld J, Strelkov SV. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 2001;11:82–88. doi: 10.1016/s0962-8924(00)01898-5. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka T, Soriano MA, Grusby MJ. SLIM is a nuclear ubiquitin E3 ligase that negatively regulates STAT signaling. Immunity. 2005;22:729–736. doi: 10.1016/j.immuni.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 14.Pine R, Canova A, Schindler C. Tyrosine phosphorylated p91 binds to a single element in the ISGF2/IRF1 promoter to mediate induction by IFNa and IFN©, and is likely to autoregulate the p91 gene. EMBO J. 1994;13:158–167. doi: 10.1002/j.1460-2075.1994.tb06245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sims SH, Cha Y, Romine MF, Gao PQ, Gottlieb K, Deisseroth AB. A novel interferon-inducible domain: structural and functional analysis of the human interferon regulatory factor 1 gene promoter. Mol Cell Biol. 1993;13:690–702. doi: 10.1128/mcb.13.1.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imai J, Yashiroda H, Maruya M, Yahara I, Tanaka K. Proteasomes and molecular chaperones: cellular machinery responsible for folding and destruction of unfolded proteins. Cell Cycle. 2003;2:585–590. [PubMed] [Google Scholar]
- 17.Kim TK, Maniatis T. Regulation of interferon-©-ativated STAT1 by the ubiquitin-proteasome pathway. Science. 1996;273:1717–1719. doi: 10.1126/science.273.5282.1717. [DOI] [PubMed] [Google Scholar]
- 18.Mosmann TR, Sad S. The expanding universe of T-cell subsets: Th1, Th2 and more. Immunol Today. 1996;17:138–146. doi: 10.1016/0167-5699(96)80606-2. [DOI] [PubMed] [Google Scholar]
- 19.Locksley RM, Fearson DT. The instructive role of innate immunity in the acquired immune response. Science. 1996;272:50–53. doi: 10.1126/science.272.5258.50. [DOI] [PubMed] [Google Scholar]
- 20.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol. 2003;3:133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- 21.Dupuis S, Dargemont C, Fieschi C, Thomassin N, Rosenzweig S, Harris J, Holland SM, Schreiber RD, Casanova JL. Impairment of mycobacterial but not viral immunity by a germline human STAT1 mutation. Science. 2001;293:300–303. doi: 10.1126/science.1061154. [DOI] [PubMed] [Google Scholar]


