SUMMARY
Objective
The purpose of the study was to explore the physiological and antigenic diversity of a large number of S. mitis biovar 1 isolates in order to begin to determine whether these properties contribute to species persistence.
Design
S. mitis biovar 1 was collected from four infants from birth to one year of age. At each of 8–9 visits 60 isolates each were obtained from the cheeks, tongue and incisors (once erupted) yielding 4,440 in total. These were tested for production of neuraminidase, β1-N-acetylglucosaminidase, β1-N-acetylgalactosaminidase, IgA1 protease, and amylase-binding. Antigenic diversity was examined by ELISA and Western immunoblotting using antisera raised against S. mitis biovar 1 NCTC 12261T and SK145.
Results
3,330 (75%) of the isolates were identified as S. mitis biovar 1 and 3,144 (94.4%) could be divided into four large phenotypic groups based on glycosidase production. 54% of the isolates produced IgA1 protease, but production was disproportionate among the phenotypes. Between 1/3 to 1/2 of the strains of each phenotype bound salivary α-amylase. Antisera against strains NCTC 12261T and SK145 displayed different patterns of reactivity with randomly selected representatives of the four phenotypes.
Conclusions
S. mitis biovar 1 is physiologically and antigenically diverse, properties which could aid strains in avoiding host immunity and promote re-colonization of a habitat or transfer to a new habitat.
INTRODUCTION
Streptococcus mitis biovar 1 is a pioneer in the human oral cavity and remains a major fraction of the commensal microbiota of the oropharynx.1–3 The persistence of this species suggests that it is ideally adapted to survive ecological pressures that might lead to its elimination. Understanding the reasons why these bacteria survive is important because it relates to their abilities to avoid or adapt to immunological, physiological, and other environmental pressures.4
Although this species persists, it is known that very few strains of S. mitis biovar 1 are stable in the mouth.5–7 Rather, their population exhibits clonal replacement.7 It is possible that this extensive genetic diversity 5–7 and any associated phenotypic diversity contribute to the survival of S. mitis biovar 1. Such phenotypic diversity could provide a range of strains each ‘best suited’ to a given environment.
Our interest is in understanding how S. mitis biovar 1 and other commensal oral bacteria survive mucosal immunity and whether immune pressure contributes to clonal replacement. We have shown that the level of SIgA antibodies reactive with S. mitis and other viridans streptococci decline over time, suggesting that the induction of a limited immune response may contribute to their survival.8
Hohwy et al., 7 suggest that the clones of S. mitis in one habitat are replaced by clones from other habitats in the oropharynx. They have shown quite clearly that mutation and recombination within a habitat are unlikely to account for clonal diversity. While other habitats may be the source of the transient clones at a specific site little is known about the reason why one clone would replace another. On shedding mucosal surfaces it could be argued that a “new” clone from saliva would replace bacteria lost on desquamated epithelial cells, however, this is not likely to be the case with bacteria associated with non-shedding tooth surfaces. This leaves open the possibility that the selection of strains possessing a specific phenotype best suited to the environment occurs at a given time, and that these strains then become established and grow to be a significant, but transient, part of the streptococcal population. The variations in phenotype that could contribute to such outgrowth could be many. Selection of species of oral streptococci based on single phenotypic characters such as acidurance and glucose uptake has been shown using mixed chemostat culture.9 Moreover, antigenic variation and certain physiological properties such as IgA1 protease production and α-amylase binding might increase the competitiveness of a given strain of streptococcus within a habitat and/or host. Therefore, study of the survival of species of oral streptococci in infants requires accurate definition of the phenotypes and physiological characters of individual strains of species to appreciate how a given characteristic might increase their fitness in the population. In addition, analysis of their antigenic relatedness could provide insights into relationships between survival and antigenic differences among strains.
METHODS
Study population
The study population comprised three males and one female (#3, #6, #8 and #10) all of whom were breast fed for the first three months postpartum. Two subjects were white (not of Hispanic Origin), one was Hispanic and one was Asian. The study population has been described in detail elsewhere.10–12 The Institutional Review Board of Georgetown University Medical Center approved the clinical protocol.
Sample collection, processing and culture
From the infants, swab samples of the oral mucosa were obtained 1–3 days, 2 weeks, 4 weeks and 2, 4, 6, 8, 10 and 12 months postpartum for a maximum of nine samples. Three of the infants missed the 2-week visit, giving a maximum of 8 samples. Two areas of the oral mucosa were sampled at each visit using separate swabs. The left and right buccal mucosae were sampled with one swab and the dorsum of the tongue was sampled with a second swab. As soon as teeth erupted (usually the lower central incisors) their labial surfaces were swabbed using a third swab. Sample collection, processing and culture were performed exactly as described previously.12
Reference strains of viridans streptococci
The reference strains used in this study and their biochemical and serological profiles are listed in Table 1.
TABLE 1.
Biochemical and serological profile of reference strains used in the study
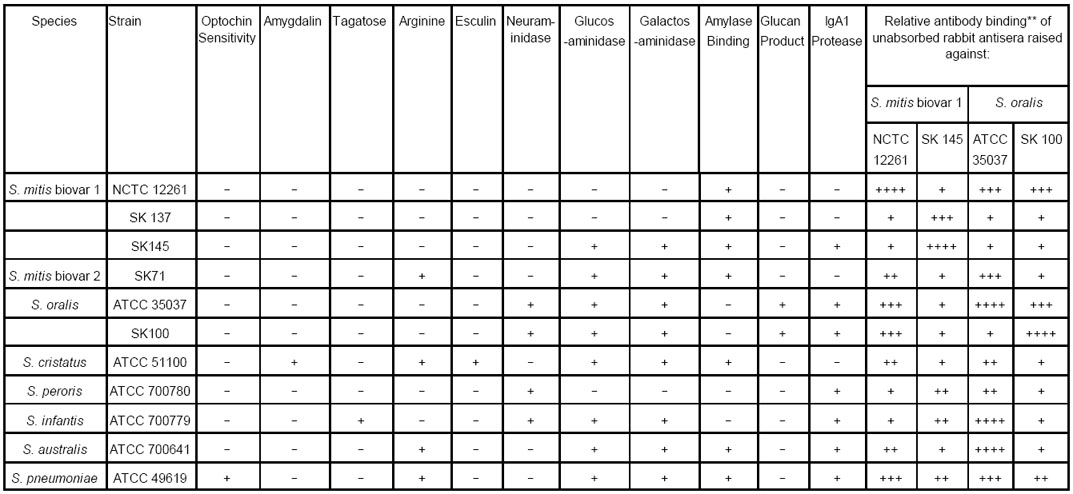
ATCC: American Type Culture Collection, Manassas, Virginia, U.S.A.
NCTC: National Collection of Type Cultures, Colindale, London, UK
SK: Dr. Mogens Kilian, Department of Medical Microbiology and Immunology, University of Aarhus, Aarhus, Denmark
Binding relative to positive control: 1+ = <25%; 2+ = 26–50%; 3+ = 51–75%; 4+ = >75%
Physiological tests
The characteristics examined were fermentation of amygdalin, tagatose and glucose; hydrolysis of arginine and esculin; production of neuraminidase, β1-N-acetylglucosaminidase, and β1-N-acetylgalactosaminidase; ability to bind α-amylase, production of IgA1 protease, production of extracellular polysaccharide from sucrose and sensitivity to optochin. All of the tests were performed as described previously 12–17
Creation of a random subset of the isolates
As it was not possible to perform DNA-DNA hybridization and serological analysis on every isolate a sampling frame was created by assigning a unique number to each of the 3,144 isolates and 48 isolates were chosen using a computer-generated random numbers list. It should be noted that, because preliminary analysis of the glycosidase production by the isolates showed them to fall into four main groups (see Results) we selected 12 random isolates from each phenotypic group.
DNA-DNA hybridization
In order to confirm the assignment of isolates in the four phenotypic groups to S. mitis biovar 1, DNA-DNA hybridization, using reference strains of S. mitis biovar 1 and S. oralis as controls, was performed as previously described.18,19
Rabbit antisera
Rabbit antisera were raised against S. mitis biovar 1 strains NCTC 11261T (Type strain) and SK145 and S. oralis strains ATCC 35037T (Type strain) and SK100 as described previously.20 The antisera were used without absorption.
Whole cell ELISA
The same set of randomly-selected isolates examined by DNA-DNA hybridization (see above) was examined by whole cell ELISA. Titration curves of the binding of rabbit IgG antibody raised against S. mitis biovar 1 strains NCTC 12261T and SK145 were performed; binding to S. oralis ATCC 35037T and SK100, a genetically closely-related species, were used as controls. Briefly, wells of 96 well microtiter plates were coated with 10 μg (dry weight) of washed whole cells of the selected isolates. After blocking with 0.1% bovine albumin in 0.1M PBS, pH 6.8, containing 0.1% Tween 20 (PBS Tween) duplicate wells were charged with serial two-fold dilutions (1:500 to 1:32,000) of the rabbit antisera diluted in PBS-Tween with 0.1% globulin-free albumin. Individual plates were used for each antiserum and included duplicate wells coated with the homologous strain. Pre-immune serum served as a control for natural antibodies reactive with the antigen. The plates were shaken at room temperature for 3 h, washed and then incubated for 1 h with swine anti-rabbit IgG conjugated with horseradish peroxidase (DAKO). The plates were washed in PBS Tween and developed with OPD. For each isolate tested the sum of the optical densities at each dilution was expressed as a percentage of the sum of the total optical density value of the homologous strain (set at 100%).22 For simplicity of presentation in Table 1 these values have been stratified as follows: + 0–25%; ++ 26–50%; +++ 51–75%; ++++ 76–100%.
Western immunoblotting
From the same set of randomly-selected isolates cell wall extracts were obtained by sonication as described previously.3 The extracts were separated by 12% sodium dodecyl sulphate-polyacrylamide gel electrophoresis using a BioRad Mini Protean II system (Bio-Rad, Hercules, CA, U.S.A.) and transferred to polyvinylidene difluoride membranes (Immobilon-P, Millipore, Bedford, MA, U.S.A.) using a Bio-Rad Trans-Blot SD system.8 Development of the blots was performed as previously described 8 except that unabsorbed rabbit antiserum to S. mitis biovar 1 NCTC 12261T and SK145 were used to probe the blots and bound rabbit antibodies were detected with swine anti-rabbit immunoglobulins conjugated with horseradish peroxidase (DAKO). Digital images of the blots were imported into BioNumerics (Applied Maths, Austin, TX, U.S.A.) and the profile of each isolate was compared by cluster analysis using UPGMA.
RESULTS
Physiological characteristics
The distribution of the isolates obtained from the four infants is shown in Table 2. Of the 4,440 isolates collected 3,330 (75%) that were negative for hydrolysis of arginine and esculin, fermentation of amygdalin and tagatose, resistant to optochin and unable to produce extracellular polysaccharide from sucrose 2 were assigned to S. mitis biovar 1. This assignment was confirmed by examination of a randomly-selected subset of isolates by DNA-DNA hybridization (data not shown). Based on the production of neuraminidase, β1-N-acetylglucosaminidase, and β1-N-acetylgalactosaminidase 3,144 of these 3,330 isolates (94.4%) could be divided into four large groups (Table 3). Group 1 comprised isolates that were negative for the three glycosidases (233 isolates); Group 2 those that were negative for neuraminidase but positive for β1-N-acetylglucosaminidase, and β1-N-acetylgalactosaminidase (685 isolates), Group 3 those that were positive for neuraminidase, only (1,172 isolates); and Group 4, those that were positive for all three glycosidases (1,054 isolates).
TABLE 2.
Breakdown of the study population and number of isolates collected from each infant
| Infant # | Sex | Race | *BreastFed/Formula Fed | Number Of Visits | Teeth Eruption (months) | Number of Isolates | |||
|---|---|---|---|---|---|---|---|---|---|
| Total | Cheeks | Tongue | Teeth | ||||||
| 3 | M | White | B | 8 | 6 | 1200 | 480 | 480 | 240 |
| 6 | F | White | B | 8 | 12 | 1020 | 480 | 480 | 60 |
| 8 | M | Hispanic | B | 9 | >12 | 1080 | 540 | 540 | 0 |
| 10 | M | Asian | B | 8 | 8 | 1140 | 480 | 480 | 180 |
Breast -fed for the first three months postpartum
TABLE 3.
Division of S. mitis biovar 1 into four groups based on production of Neuraminidase, β-N-acetylglucosaminidase and β-N-acetylgalactosaminidase and their distribution on cheeks, tongue and teeth
| Group | Numbers and percentages of isolates of each phenotype obtained from each surface | Neuraminidase | β-N-acetyl-Glucosaminidase | β-N-acetyl-Galactosaminidase | Amylase Binding (% positive) | IgA1 Protease (% positive) | |||
|---|---|---|---|---|---|---|---|---|---|
| Total | Cheek | Tongue | Tooth | ||||||
| 1 | 233 | 104(45) | 118(51) | 11(5) | − | − | − | 36 | 80 |
| 2 | 685 | 233(34) | 384(56) | 68(10) | − | + | + | 46 | 12 |
| 3 | 1,172 | 588(50) | 559(48) | 25(2) | + | − | − | 39 | 39 |
| 4 | 1,054 | 565(54) | 387(37) | 102(10) | + | + | + | 38 | 91 |
Binding of salivary α–amylase was similar among the four groups, ranging from 36% in Group 1 to 46% in Group 2. All four groups contained isolates that produced IgA1 protease and overall 54% produced this enzyme. However, the percentages of strains positive for this enzyme differed markedly between the groups. In Group 4, 91%, and in Group 1, 80% of the isolates produced IgA1 protease whereas in Groups 2 and 3 only 12% and 39% of the isolates, respectively, produced this enzyme.
Whole cell ELISA
Because of the close genetic relationship betweeen S. mitis biovar 1 and S. oralis we examined the reactivity of antisera raised against the type strains of both of these species together with antisera raised against a well characterized strain of S. mitis biovar 1 (SK145) and S. oralis (SK100). Unabsorbed rabbit antisera raised against S. mitis biovar 1 NCTC 12261T and SK145 and S. oralis ATCC 35037T and SK100 displayed different patterns of reactivity when incubated with the reference strains and representatives of the four phenotypic groups (Table 1). The patterns of reactivity for the serum raised against S. mitis biovar 1 NCTC 12261T was discordant with that of S. mitis biovar 1 SK145 as was the case for S. oralis strains ATCC 35037T and SK100.
Antiserum against NCTC 12261T gave weak reactions against the standard strains of S. mitis biovar 1, S. peroris and S. infantis but cross-reacted strongly with S. oralis ATCC 35037T. As expected antiserum against S.oralis ATCC 35037T gave only weak reactions against S. mitis biovar 1 strains SK137, and SK145. However, it gave a strong reaction with S. mitis NCTC 12261T cells and also S. infantis, and S. australis. Its weak reaction with S. oralis strain SK100 was also surprising.
In contrast to the antisera raised against the Type strains, the antisera raised against S. mitis biovar 1 SK145 and S. oralis SK100 were more discriminating. Significantly, compared to the antiserum against ATCC 35037T, the antiserum against SK100 reacted weakly with S.cristatus, S. peroris, S. infantis, and S. australis (Table 1). However, it was noteworthy that the antiserum against S. oralis SK100 reacted strongly with cells of S. mitis biovar 1 NCTC 12261T, mimicking the reaction of the antiserum against ATCC 35037T. In addition, antiserum against SK145 gave mid-range (20–60%) reactions with cells of S. peroris and S. infantis. Each of the antisera gave positive reactions with cells of S. pneumoniae.
As the antisera against SK145 and SK100 were more discriminatory than those raised against the type strains of S. mitis and S. oralis we used them in a whole cell ELISA to examine 12 randomly selected strains of each of the phenotypic groups (Fig. 1). Nine of the 12 phenotype 0-0-0 strains bound the antiserum against SK145 at 100% and only strain #40 showed equivalent binding between the antisera against SK145 and SK100. None of the strains exceeded 58% binding of the antiserum against SK100 (Fig. 1A). For the 0-1-1 phenotype, 11 of the 12 randomly selected isolates bound the antiserum against SK145 at 89% or better. In fact, 10 of these 11 isolates bound at 100%. None of the strains in this group exceeded 30% binding of the antiserum against SK100 (Fig. 1B). Although the majority of strains (10/12) of the1-0-0 phenotype exhibited 100% binding of the antiserum against SK145, strains #6 #28 and #48 also bound the antiserum against SK100 at a high level (82%, 86% and 100%, respectively) (Fig. 1C). Similarly, for phenotype 1-1-1, although 8 of the 12 strains bound the antiserum against SK145 at 100%, strains #33 and #34 also bound the antiserum against SK100 equivalently (Fig. 1D).
FIGURE 1.
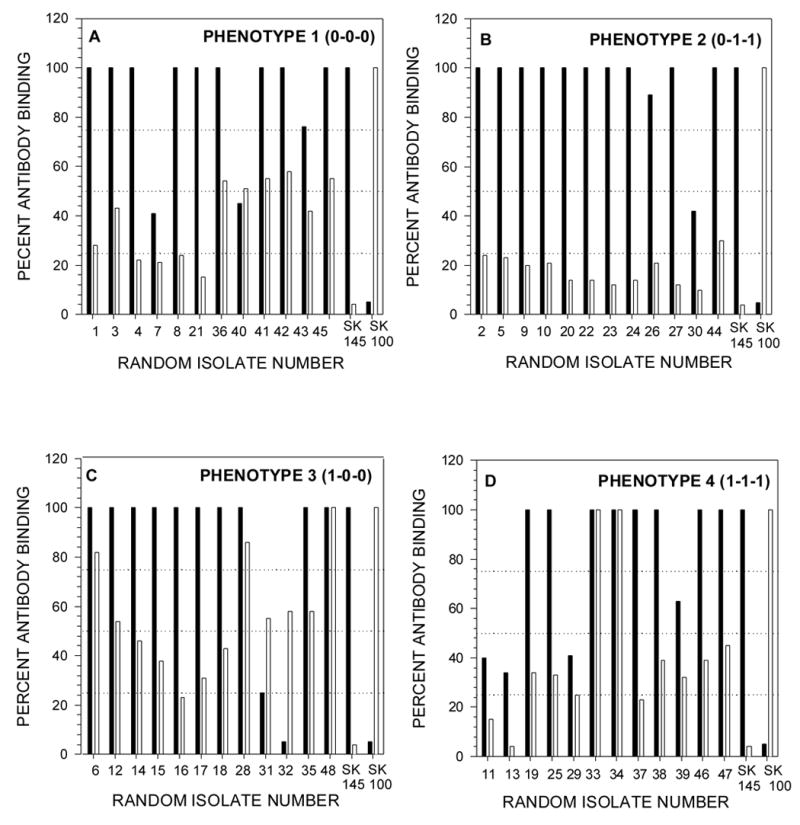
A sampling frame was created by assigning a unique number to each isolate and then, within each phenotypes, 12 isolates were randomly chosen using a computer generated random numbers list. Wells of microtiter plates were coated with washed whole cells of the selected isolates and an ELISA was performed as described in Materials and Methods using serial twofold dilutions (1:500 to 1:32,000) of rabbit antisera raised against S. oralis, SK100 and S. mitis biovar 1, SK145. Individual plates were used for each antiserum which always included the homologous strain. Pre-immune serum served to control for natural antibodies reactive with the antigen. For each isolate tested the sum of the absorbance value at each dilution was expressed as a percentage of the sum of the absorbance value at each dilution of the homologous strain. Filled bars represent SK145 antiserum and open bars represent SK100 antiserum. The interrupted horizontal lines are set at 25%, 50%, and 75% of the antibody binding of the homologous control. In the text isolates of the respective phenotypes are assigned the prefix A, B, C or D.
Overall, 37 of the 48 randomly selected isolates (77%) bound the antiserum against SK145 as well as the homologous strain. An additional strain bound the antiserum against SK145 at a level of 89% of the homologous control. In contrast, only 5 of the 48 strains (10%) bound the antiserum against S. oralis SK100 above 80% of the positive control. Of the 48 strains, three (C48, D33 and D34), were unusual in that they bound both the antiserum against SK145 and the antiserum against SK100 at 100%.
Western immunoblotting
Western immunoblots were run on cell wall extracts of the same 48 selected strains tested by ELISA. The blots were developed using antisera against S. mitis biovar 1 strains NCTC 12261T and SK145 and S. oralis strains ATCC 35037T and SK100. Blots developed with the two sera raised against S. mitis gave complex patterns of bands with the extracts. However, in complete contrast to the results of ELISA, none of the extracts showed any reaction when the blots were developed with the antisera against S. oralis under identical conditions.
The patterns of antigen reactivity of the anti-S. mitis sera are shown in Figures 2A (anti-NCTC 12261T serum) and Figure 2B (anti-SK145 serum). The dendrograms shown in Figures 2A and 2B indicate that while both antisera react well with the cell wall extracts they differ in the manner in which they group the isolates and in the magnitude of inter-isolate similarities. Antiserum raised against S. mitis biovar 1 NCTC 12261T (Fig. 2A) defines an overall similarity of 36% for the 48 isolates, whereas antiserum raised against S. mitis biovar 1 strain SK145 defines an overall similarity of 73%, essentially twice that of the antiserum raised against strain NCTC 12261T (Fig. 3). Thus, the antigenic patterns of the 48 isolates are closer to that of strain SK145 than strain NCTC 12261T. These data are generally consistent with those obtained from the whole-cell ELISA.
FIGURE 2.
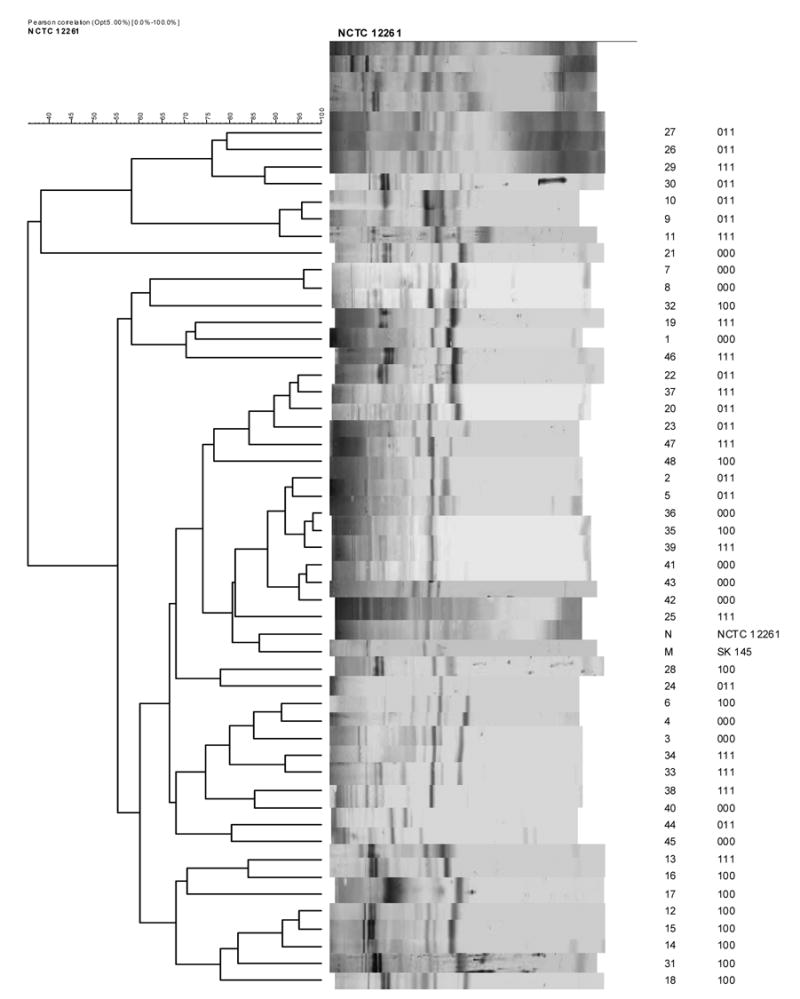

Western immunoblots were run on cell wall extracts of the same isolates tested by ELISA. The blots were developed using antisera against S. mitis biovar 1 strains NCTC 12261T (Figure 2A) and SK 145 (Figure 2B). Antisera to S. oralis strains ATCC 35037T and SK100 were non-reactive in Western blotting. Digital images of the blot strips were compared by cluster analysis using UPGMA. To the right of the strips is shown the identity of the isolate and its phenotype.
The cell wall profiles of the randomly-selected isolates did not cluster according to their biochemical phenotype with complete fidelity showing that considerable diversity may exist in antigenic profile among strains within the same biochemical phenotype. Having said that, groups of isolates (as many as 6 of 12) of the same biochemical phenotype were clustered together indicating that strains exhibiting the same biochemical profile can be closely similar in antigenic profile (Figures 2A and 2B).
The anomalous strains in Figure 1 that gave relatively low whole-cell binding values when reacted with antiserum to S. mitis biovar 1 SK145 (#43, #30, #31, #32, #11, #13, #29 and #39) or bound antiserum to S. oralis at above 80% (#6, #28, #48, D33 and #34) were, with the exception of #33 and #34 (both phenotype 1-1-1), not clustered together but distributed throughout the dendograms. In addition, these strains often formed pairs and clusters at high similarities with other isolates. This suggests that the antigens of these strains were closely similar to those of the isolates that reacted strongly in the whole cell ELISA assay.
DISCUSSION
Continued examination of the potential roles of salivary SIgA antibodies reactive with S. mitis biovar 1and physiological adaptation in clonal replacement of this bacterium required information about the phenotypic and antigenic diversity among strains colonizing a specific habitat in an infant. Consequently, we obtained over 1000 isolates of S. mitis biovar 1 from the mouth of each of four infants during the first year of life. In this way we could be reasonably certain that we had an accurate representation of strains harbored in the oral cavity of these subjects. As different oral streptococcal species and biovars can exhibit the same colonial morphology 22, colonies were picked at random from non-selective primary isolation plates. 3,330 isolates were assigned to S. mitis biovar 1 on the basis that they failed to ferment amygdalin and tagatose, failed to hyrolyzed arginine and esculin, did not produce extracellular polysaccharide from sucrose 2, and were resistant to optochin. The assignment of these isolates to S. mitis biovar 1 was confirmed by DNA-DNA hybridization performed on a computer-generated random set of isolates.
Based on glycosidase production 3,144 (94.4%) of the isolates fell into four main phenotypic groups (Table 3). However, it should be noted that, based on the glycosidase profile of the Type strain of S. peroris, it is possible that representatives of this species may be contained in phenotypic group 3.
The ability to bind α-amylase and produce IgA1 protease have been proposed as useful characteristics that discriminate species of viridans streptococci 13,14,22 as well as ecological determinants for these bacteria. 23–25 However, α-amylase binding was a variable feature of isolates in each of the four phenotypic groups. Furthermore, while the production of IgA1 protease is a defining characteristic of S. oralis, S. sanguinis and S. pneumoniae this enzyme was also produced by almost two-thirds of S. mitis biovar 1 isolated from the mouth.1,3,24,25 We examined the strains in each of the phenotypic groups for the production of this enzyme and found that almost all of Group 4 isolates (91%) produced IgA1 protease. However, a preponderance (80%) of Group 1 isolates as well as 12% of Group 2 isolates, and 39% of Group 3 isolates also produced IgA1 protease. In addition, we found that the Type strain of S. peroris and, also, the Type strains of S. infantis and S. australis produced IgA1 protease, a property not previously ascribed to these species.
Using antiserum against strains SK145 and SK100 (Fig 1), 37 of the 48 randomly-selected isolates exhibited 100% binding with the antiserum against strain SK145. Of these 37 only three also exhibited 100% binding of the antiserum against strain SK100. These results suggest that 34 of the 48 strains examined form a group antigenically-related to SK145. Previously, using Rantz and Randall extracts and a range of antisera it has been shown that the antigenic profiles of strains of S. mitis biovar 1 are different, suggesting that S. mitis biovar 1 includes a range of antigenic types. 6 Despite this diversity it appears that among our randomly-selected isolates from the four phenotypic groups we have identified a significant oral group of S. mitis isolates that carry antigens distinct from the Type strain NCTC 12261T.
All of the antisera reacted with whole cells of S. pneumoniae (Table 1). S. mitis biovar 1 SK137 has been shown to carry a specific teichoic acid-like antigen and also the group O antigen in common with S. pneumoniae. 26 This antigen may also be carried by the immunizing strains and contribute to the antibody binding. Given the high degree of binding of antiserum against SK145 by SK137 cells and its weak reaction with the other three sera (Table 1) this strain could also be included in the antigenic group represented by SK145.
The results from Western blotting of cell wall extracts confirm the close antigenic similarity of all of the isolates to SK145. Significantly, none of the cell wall extracts from these isolates reacted with the antiserum against S. oralis SK100 in Western blots. The positive reaction of these strains in whole cell ELISA with antiserum to S. oralis (Figure 1) could be based on carbohydrate or protein antigens that were not represented in the cell wall extract transferred to the polyvinylidene difluoride membranes. Similarly, those strains that gave low whole cell binding with antiserum against strain SK145 may lack significant protein or possibly wall carbohydrate antigens of the type described for S. mitis biovar1 strain SK137.
At this time little can be said about the identity of the antigens responsible for placing organisms into the SK145 antigenic group. Based on the information available for SK137 26,, it is likely that strains of S. mitis biovar 1 can carry both antigenic wall carbohydrate 6 and teichoic acid. Consequently, at least two carbohydrate-based antigens may be responsible for defining the SK145 Group of S. mitis biovar 1 along with protein antigens known to be common in streptococci.
The results of the serology in this study identify an antigenic group of strains of S. mitis biovar 1 based on strain SK145 that probably also includes strain SK137. This result is similar to that described by Hohwy and Kilian 6 where, by using antiserum against S. oralis SK2 (ATCC 10557) they could separate strains of S. oralis from those of S. mitis biovar 1. Such a result may indicate that a group of antigenically identical or closely similar strains, quite distinct from S. mitis biovar 1, exists within the S. oralis population. Relatively few of the 48 strains tested gave an antigenic profile similar to the Type strains of S. mitis biovar 1 and S. oralis, although such strains were identified (Fig. 1). Moreover, the Type strains of S. infantis and S. australis also reacted strongly with antiserum against S. oralis ATCC 35037, indicating common antigens.
If the group of strains based on binding to antibody against SK145 has an identical antigenic profile and is common and numerically significant in infants’ mouths it is difficult to envisage that antigenic drift or antigenic variation that reduces the effectiveness of host antibody is related to clonal replacement of S. mitis biovar 1 strains. However, the serology carried out in the present study is limited and provides no information about the nature of the antigens responsible for antibody binding. Thus, it remains possible that changes in surface antigens could aid strains in avoiding host immunity and promote recolonization of a habitat or transfer to a new habitat.
Our collection of well-defined isolates allows us to determine whether strains are shared between infants, persist in a given infant, or change their habitat within an infant. Furthermore, saliva samples collected in parallel with the isolates permit us to determine which antigens are recognized by salivary SIgA antibodies and whether their specificity changes over time. The results from such analyses can be used to select strains for comparison of their physiology, possibly including competition experiments in chemostats.
Acknowledgments
This work was supported by Public Health Service grant NIH DE08178 from the National Institute of Dental Research. G.H.B. is supported by grant MT 7611 from the Medical Research Council of Canada. The authors wish to thank Manju Chauhan and Shelley Tunwall for excellent technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Frandsen EV, Pedrazzoli V, Kilian M. Ecology of viridans streptococci in the oral cavity and pharynx. Oral Microbiol Immunol. 1991;6:129–133. doi: 10.1111/j.1399-302x.1991.tb00466.x. [DOI] [PubMed] [Google Scholar]
- 2.Kononen E, Jousimies-Somer H, Bryk A, Kilp T, Kilian M. Establishment of streptococci in the upper respiratory tract: longitudinal changes in the mouth and nasopharynx up to 2 years of age. J Med Microbiol. 2002;51:723–730. doi: 10.1099/0022-1317-51-9-723. [DOI] [PubMed] [Google Scholar]
- 3.Pearce C, Bowden GH, Evans M, Fitzsimmons SP, Johnson J, Sheridan MJ, Wientzen R, Cole MF. Identification of pioneer viridans streptococci in the oral cavity of human neonates. J Med Microbiol. 1995;42:67–72. doi: 10.1099/00222615-42-1-67. [DOI] [PubMed] [Google Scholar]
- 4.Bowden GH, Hamilton IR. Survival of oral bacteria. Crit Rev Oral Biol Med. 1998;9:54–85. doi: 10.1177/10454411980090010401. [DOI] [PubMed] [Google Scholar]
- 5.Fitzsimmons SP, Evans M, Pearce C, Sheridan MJ, Wientzen R, Bowden G, Cole MF. Clonal diversity of Streptococcus mitis biovar 1isolates form the oral cavity of human neonates. Clin Diagn Lab Immunol. 1996;3:517–522. doi: 10.1128/cdli.3.5.517-522.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hohwy J, Kilian M. Clonal diversity of the Streptococcus mitis biovar 1 population in the human oral cavity and pharynx. Oral Microbiol Immunol. 1995;10:19–25. doi: 10.1111/j.1399-302x.1995.tb00113.x. [DOI] [PubMed] [Google Scholar]
- 7.Hohwy J, Reinholdt J, Kilian M. Population dynamics of Streptococcus mitis in its natural habitat. Infect Immun. 2001;69:6055–6063. doi: 10.1128/IAI.69.10.6055-6063.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole MF, Bryan S, Evans MK, Pearce CL, Sheridan MJ, Sura PA, Wientzen R, Bowden GHW. Humoral immunity to commensal oral bacteria in human infants: Salivary secretory immunoglobulin A antibodies reactive with Streptococcus mitis biovar 1, Streptococus oralis, Streptococcus mutans, and Enterococcus faecalis during the first two years of life. Infect Immun. 1999;67:1878–1886. doi: 10.1128/iai.67.4.1878-1886.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDermid AS, McKee AS, Ellwood DC, Marsh PD. The effect of lowering the pH on the composition and metabolism of a community of nine oral bacteria in a chemostat. J Gen Microbiol. 1986;132:1205–1214. doi: 10.1099/00221287-132-5-1205. [DOI] [PubMed] [Google Scholar]
- 10.Fitzsimmons SP, Evans MK, Sheridan MJ, Wientzen R, Cole MF. Immunoglobulin A subclasses in infant's saliva and in saliva and milk from their mothers. J Pediatr. 1994;124:566–573. doi: 10.1016/s0022-3476(05)83135-x. [DOI] [PubMed] [Google Scholar]
- 11.Kirchherr JL, Bowden GH, Richmond DA, Sheridan MJ, Wirth KA. Clonal diversity and turnover of Streptococcus mitis bv1 on shedding and nonshedding oral surfaces of human infants during the first year of life. Clin Diagn Lab Immunol. 2005;12:1184–1190. doi: 10.1128/CDLI.12.10.1184-1190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirchherr JL, Bowden GH, Richmond DA, Sheridan MJ, Wirth KA. Distribution of Streptococcus mitis biovar 1 phenotypes on shedding and non-shedding oral surfaces of human infants during the first year of life. Microb Ecol Hlth Dis. 2005;17:138–145. [Google Scholar]
- 13.Douglas CWI, Pease AA, Whiley RA. Amylase-binding as a discriminator among oral streptococci. FEMS Microbiol Lett. 1990;66:193–197. doi: 10.1016/0378-1097(90)90281-t. [DOI] [PubMed] [Google Scholar]
- 14.Kilian M, Nyvad B. Ability to bind salivary α-amylase discriminates certain viridans streptococcal species. J Clin Microbiol. 1990;28:2576–2577. doi: 10.1128/jcm.28.11.2576-2577.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reinholdt J, Kilian M. A sensitive enzyme-linked immunosorbent assay for IgA protease activity. J Immunol Methods. 1983;63:367–376. doi: 10.1016/s0022-1759(83)80010-6. [DOI] [PubMed] [Google Scholar]
- 16.Bailey RW, Oxford AE. Pre-requisites for dextran production by Streptococcus bovis. Nature (Lond) 1958;182:185–186. doi: 10.1038/182185a0. [DOI] [PubMed] [Google Scholar]
- 17.Hehre EJ, Neill JM. Formation of serologically reactive dextrans by streptococci form subacute bacterial endocarditis. J Exp Med. 1946;83:147–162. [PMC free article] [PubMed] [Google Scholar]
- 18.Ezaki T, Hashimoto Y, Yabuuchi E. Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are usd to determine genetic relatednessamong bacterial strains. Int J Syst Bacteriol. 1989;39:224–229. [Google Scholar]
- 19.Kawamura Y, Whiley RA, Shu S-E, Ezaki T, Hardie JM. Genetic approaches to the identification of the mitis group within the genus Streptococcus. Microbiology. 1999;145:2605–2613. doi: 10.1099/00221287-145-9-2605. [DOI] [PubMed] [Google Scholar]
- 20.Hardie JM, Bowden GH. Some serological cross-reactions between Streptococcus mutans, S sanguis, and other dental plaque streptococci. J Dent Res. 1976;55:C50–58. doi: 10.1177/002203457605500325011. Spec No. [DOI] [PubMed] [Google Scholar]
- 21.Bowden GHW. Serological Identification. In: Goodfellow M, O'Donnell AG, editors. Handbook of New Bacterial Systematics. Vol. 1993. London: Academic Press; pp. 429–455. [Google Scholar]
- 22.Kilian M, Mikkelsen L, Henrichsen J. Taxonomic study of viridans streptococci: Description of Streptococcus gordonii sp. nov. and emended descriptions of Streptococcus sanguis (White and Niven 1946), Streptococcus oralis (Bridge and Sneath 1982), and Streptococcus mitis (Andrewes and Horder 1906) . Int J Syst Bacteriol. 1989;39:471–484. [Google Scholar]
- 23.Scannapieco FA, Torres G, Levine M. Salivary α-amylase: role in dental plaque and caries formation. Crit Rev Oral Biol Med. 1993;4:301–307. doi: 10.1177/10454411930040030701. [DOI] [PubMed] [Google Scholar]
- 24.Kilian M, Holmgren K. Ecology and nature of immunoglobulin A1 protease-producing streptococci in the human oral cavity and pharynx. Infect Immun. 1981;31:868–873. doi: 10.1128/iai.31.3.868-873.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kilian M, Reinholdt J, Nyvad B, Frandsen EV, Mikkelsen L. IgA1 proteases of oral streptococci: ecological aspects. Immunol Invest. 1989;18:161–170. doi: 10.3109/08820138909112235. [DOI] [PubMed] [Google Scholar]
- 26.Bergstrom N, Jansson PE, Kilian M, Skov, Sorensen UB. Structures of two cell wall-associated polysaccharides of a Streptococcus mitis biovar 1 strain. A unique teichoic acid-like polysaccharide and the group O antigen which is a C-polysaccharide in common with pneumococci. Eur J Biochem. 2000;267:7147–7157. doi: 10.1046/j.1432-1327.2000.01821.x-i2. [DOI] [PubMed] [Google Scholar]


