Abstract
Lipoplexes containing a mixture of cationic phospholipids dioleoylethylphosphatidylcholine (EDOPC) and dilauroylethylphosphatidylcholine (EDLPC) are known to be far more efficient agents in transfection of cultured primary endothelial cells than are lipoplexes containing either lipid alone. The large magnitude of the synergy permits comparison of the physical and physico-chemical properties of lipoplexes that have very different transfection efficiencies, but minor chemical differences. Here we report that the superior transfection efficiency of the EDLPC/EDOPC lipoplexes correlates with higher surface activity, higher affinity to interact and mix with negatively charged membrane-mimicking liposomes, and with considerably more efficient DNA release relative to the EDOPC lipoplexes. Observations on cultured cells agree with the results obtained with model systems; confocal microscopy of transfected human umbilical artery endothelial cells (HUAEC) demonstrated more extensive DNA release into the cytoplasm and nucleoplasm for the EDLPC/EDOPC lipoplexes than for EDOPC lipoplexes; electron microscopy of cells fixed and embedded directly on the culture dish revealed contact of EDLPC/EDOPC lipoplexes with various cellular membranes, including those of the endoplasmic reticulum, mitochondria and nucleus. The sequence of events outlining efficient lipofection is discussed based on the presented data.
Keywords: cationic lipid, lipofection, membrane fusion, surface tension, DNA unbinding, gene therapy
1. Introduction
Synthetic cationic lipids have been the subject of intense examination in the areas of cell transfection and gene therapy during the past two decades. They are now considered the most promising non-viral gene carriers. A critical obstacle for clinical application of lipid-mediated DNA delivery (lipofection) is its unsatisfactorily low efficiency. It is believed that about 106 copies of plasmid must be delivered to the cell nucleus to achieve expression sufficient for a clinical effect, even though in a typical transfection experiment with cationic liposomes only 102-104 copies enter the nucleus and are expressed [1]. Progress in enhancing the lipofection efficacy is impeded because its mechanism is still largely unknown. One of the major obstacles to efficient transfection is the low level of DNA release from the complexes with cationic lipids (lipoplexes). Cationic lipid-DNA interactions are strong [2] and the only possibility for release of DNA under cellular conditions appears to be by neutralization of the cationic lipid charge with cellular anionic lipids, as first demonstrated in model experiments [3-7]. A similar process of lipid exchange between lipoplexes and cytoplasmic membranes resulting in DNA release into the cytoplasm was later observed in cultured cells too [8-10].
Dioleoylethylphosphatidylcholine (EDOPC), the cationic triester of DOPC, prepared by phosphate ethylation, was one of the first cationic phospholipids applied to transfection. It was recently found that mixing EDOPC with its shorter-chain analog, dilauroylethylphosphatidylcholine (EDLPC), dramatically increased transfection efficacy [11]. The largest improvement in transfection efficiency (up to 30-fold) was found with a 6:4 (w/w) mixture of EDLPC/EDOPC, when human umbilical artery endothelial cells (HUAEC) were transfected. This finding was particularly significant given that primary cells of endothelial tissues have been especially difficult to transfect [12]. In an effort to shed light on the mechanism of lipid-mediated DNA delivery, we examined the physical and physico-chemical properties of the EDLPC/EDOPC lipoplexes. As described here, we found that their superior transfection efficiency correlates with higher surface activity, higher affinity to interact and mix with negatively charged liposomes, and with considerably faster DNA release relative to the EDOPC lipoplexes1. All of these physical characteristics seem important in the general lipofection pathway. Experiments with model systems were complemented with observations on transfected HUAEC cells. Confocal microscopy of transfected HUAEC cells demonstrated more extensive DNA release into the cytoplasm and nucleoplasm for the EDLPC/EDOPC lipoplexes than for EDOPC lipoplexes; close contacts of EDLPC/EDOPC lipoplexes with various cellular membranes, including those of the endoplasmic reticulum, mitochondria and nucleus, were revealed by electron microscopy.
2. Materials and methods
2.1. Lipids
The triflate salts of 1,2-dioleoyl-sn-glycero-3-ethylphosphocholine (EDOPC) and 1,2-dilauroyl-sn-glycero-3-ethylphosphocholine (EDLPC) were synthesized as previously described [14], or these lipids were purchased as the chloride salt from Avanti Polar Lipids (Alabaster, AL). Dioleoylphosphatidylcholine (DOPC), dioleoylphosphatidylethanolamine (DOPE), dioleoylphosphatidylglycerol (DOPG), dioleoylphosphatidylserine (DOPS), cholesterol (Chol), cardiolipin (CL) (heart, Na-salt), phosphatidylinositol (PI) (bovine liver, Na-salt), and dioleoylphosphatidylethanolamine with covalently attached rhodamine (Rh-DOPE), all from Avanti Polar Lipids, were used without further purification. The phospholipids migrated as a single spot by thin-layer chromatography. BODIPY FL C12-HPC, a fluorescent derivative of PC that has spectral characteristics similar to fluorescein, was purchased from Molecular Probes (Eugene, OR). Lipids were stored at -20°C in chloroform.
2.2. Liposome and lipoplex preparation
Aliquots of lipids EDOPC or an EDLPC/EDOPC mixture were placed in borosilicate glass tubes, freed of chloroform with an argon stream, and kept under high vacuum for at least 1 h per mg lipid to remove any solvent residues. Subsequently, samples were hydrated with phosphate-buffered saline (PBS), pH 7.4 and vortexed for 5 min. For preparation of lipoplexes, the cationic liposomes were added to plasmid DNA pCMVSport-β-Gal DNA (from Clontech, Palo Alto, CA, propagated and purified by Bayou Biolabs, Harahan, LA) at the desired ratio, as indicated in the text. (Assuming an average nucleotide mol wt 330, the lipid/DNA weight ratios corresponding to isoelectric lipoplexes are 2.9:1 for EDOPC, 2.6:1 for EDLPC/EDOPC 6:4 w/w, and 2.4:1 for EDLPC; all lipoplex compositions used in this study were with some excess of cationic lipid.) For most of the experiments, the most efficient lipoplex composition, EDLPC/EDOPC 6:4 (w/w) [11] was compared to the EDOPC lipoplexes; in some experiments, other EDLPC/EDOPC ratios were also used, as indicated. In some experiments we used DNA covalently labeled with fluorescein-5-isothiocyanate (FITC) using the Label IT labeling kit (Mirus, Madison, WI) according to the protocol supplied by the manufacturer. DNA so labeled was purified on Microspin columns included in the kit, or by ethanol precipitation according to the suggested protocol. The number of labels per DNA molecule was measured as suggested by Mirus (www.genetransfer.com, FAQ Q26).
2.3. X-ray diffraction
Samples were prepared by adding pre-formed cationic liposomes (5 wt% dispersions) to the DNA aqueous solution, as previously described [15]. Samples were filled into glass capillaries (d = 1.5 mm) (Charles Super Co., Natick, MA), flame-sealed, and equilibrated for 2-3 days at room temperature before measurements. Small-angle X-ray diffraction (SAXD) measurements were performed at 37°C at Argonne National Laboratory, Advanced Photon Source, BioCAT (beamline 18-ID) or DND-CAT (beamline 5-IDD), using 12 keV X-rays. Exposure times were typically ∼0.5-1 sec. Some samples with longer exposure time were checked by thin layer chromatography after the experiments. Products of lipid degradation were not detected in these samples, and radiation damage of the lipids was not evident from their X-ray patterns.
2.4. Measurement of surface activity at the surface of lipid dispersions
Lipid dispersions (1 mg/mL lipid concentration) were prepared in 0.15 M sodium chloride solution. Subsequently, appropriate dilutions were made for measurements. Surface tension was measured via the detachment variation of the Wilhelmy method [16,17]. As a Wilhelmy surface, we used a roughened, 0.5 mm platinum wire. Teflon wells, d=0.8 cm, contained 75 μL and were mounted on a platform that underwent 2 mm vertical oscillations 4 times/min. The Wilhelmy wire was attached to the underside sensor connection of a Cahn RTL electrobalance. Maximum excursions of the recorder pen, which corresponded to the surface tension at zero contact angle and zero buoyancy of the wire, were recorded. The instrument was calibrated with clean and aspirated water, and initial values were set at 71.5-72 mN/m. The time course of change in surface tension of the air-water interface of the cationic lipid dispersions was followed using two different techniques: (i) by filling the well with 50 μg/ml lipid dispersion and cleaning the surface by aspiration immediately before the measurement was started [18]; (ii) by spreading of dispersion containing 5 μg cationic lipid over aqueous subphase in the well, immediately prior to taking measurements [19]. The two techniques gave virtually identical results with respect to the apparent equilibrium surface tension reached after ∼30-60 min, but because of its greater consistency, the second technique was found more appropriate for following the initial kinetics of the surface tension change.
2.5. Measurement of the kinetics of DNA release from lipoplexes by using flow fluorometry
This technology allows determination of the relative lipid content per particle and the lipid/DNA composition of individual particles in the lipoplex dispersion (except for very small vesicles and bare plasmid DNA, which are not detectable on our instrument) [20,21]. A conventional flow cytometer, FACSCalibur (Becton & Dickinson) was used. A laser beam is focused on a very small portion of a dilute, flowing stream and the fluorescence (at up to 3 emission wavelengths), as well as light scattering, are measured for each particle passing through the beam. It is distinctively useful because it allows access to meaningful information on the composition of a heterogeneous population. Although not previously used for such purposes, it was found extremely well suited to analyzing highly heterogeneous lipoplex ensembles. Cationic lipid was labeled with 2.5 wt% of the fluorescent label BODIPY FL C12-HPC. DNA samples were labeled with the high-affinity fluorescent dye, ethidium homodimer-2 (Ethd-2) at 60 bp/dye [20]; both labels were purchased from Molecular Probes (Eugene, OR). Negatively charged liposomes mimicking a typical membrane lipid composition (MM=DOPC:DOPE:DOPS:Chol 45:20:20:15, w/w) were prepared from unlabeled lipids. Particles were detected at the FL1 channel (515-545 nm spectral window) by their BODIPY-PC emission (λem=513 nm). The FL3 channel (spectral window >650 nm) provides information about the amount of DNA in the particle, since Ethd-2 strongly emits into this channel (λem=624nm). The FL3/FL1 ratio was calibrated with lipoplexes that had known DNA:lipid ratios of less than one, as previously described [20]. To test for DNA release, negatively charged liposomes (unlabeled) were added at 1:1 lipid weight ratio to the lipoplexes, and data were collected as a function of time. Typically, 10,000 particles were classified as to lipid content and composition (time for data collection was usually <1 min), and the data presented as a 3-D plot with relative lipid content per particle on the X axis, the charge ratio of DNA to lipid in the particle (DNA/lipid stoichiometry) on the Y axis, and the relative number of particles on the Z axis (see Results section, Fig. 5B). The short time needed for data collection makes it possible to study the formation and disintegration of lipoplexes over time.
Fig. 5.
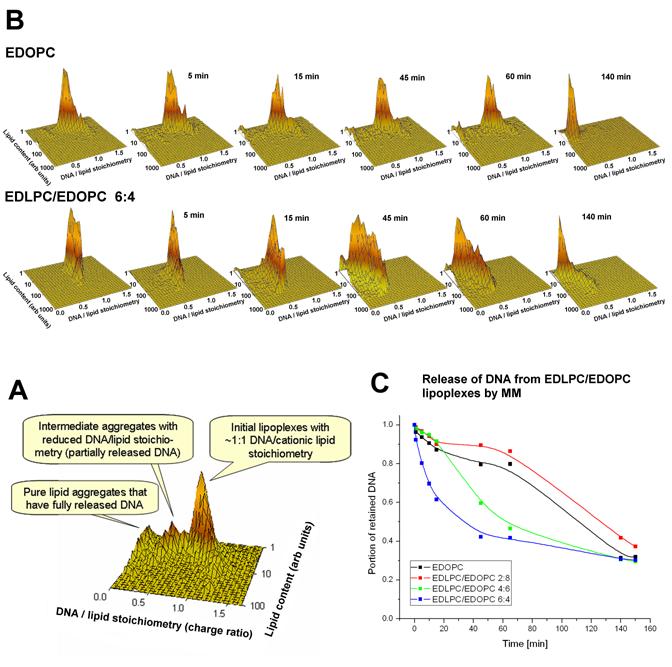
(A) Plots of DNA/lipid stoichiometry (charge ratio) vs. cationic lipid content showing the time-course of DNA unbinding from EDOPC (upper panel) or EDLPC/EDOPC 6:4 (lower panel) lipoplexes after addition of a negatively charged model membrane (MM) mixture (MM=DOPC/DOPE/DOPS/Chol 45:20:20:15 w/w). The stoichiometry (Y) axis represents the ratio of DNA to lipid charges in the particle. Lipoplexes contained a nearly isoelectric ratio of cationic lipid and DNA, labeled with 2.5% BODIPY-FL, and DNA, labeled with the high affinity label, ethidium homodimer-2 (EthD-2) at 60bp/dye. The panels show the stoichiometry vs. lipid content distributions after different times of incubation at room temperature, as indicated. Lipoplexes were initially prepared at near the isoelectric lipid/DNA ratio; negatively charged liposomes were added at a 1:1 weight ratio to the cationic lipid. Data on each panel were collected on 10,000 particles within 1 min. (B) Explanation of the plots; (C) Kinetics of DNA release (plotted as the portion of the initial DNA retained at different time points) after addition of negatively charged membrane-mimicking liposomes for different EDLPC/EDOPC compositions.
2.6. Lipid mixing assay
Lipid mixing was monitored with an assay based on fluorescence resonance energy transfer (FRET) between two lipid probes, as described earlier [22]. Lipoplexes containing 1 wt% 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI) and 1 wt% 3,3′-dioctadecyloxacarbocyanine perchlorate (DiO) were prepared according to the procedure described above. Negatively charged liposomes contained 80 mol% DOPC and 20 mol% of one of the negatively charged membrane lipids, namely PS, PI, or CL. Liposomes were prepared as described above. For some experiments, the liposome dispersions were extruded using an Avanti Mini-Extruder (Avanti Polar Lipids Alabaster, AL) equipped with a 0.1 μm polycarbonate membrane to prepare large unilamellar vesicles. Lipoplexes ware prepared according to standard protocol (see previous paragraph) and analyzed with an Alphascan fluorometer (Photon Technology International, Princeton, NJ); the final lipid concentration was 20 μg/ml. The lipoplexes were treated with unlabeled negatively charged liposomes, with constant stirring. Wavelengths were 489 nm for excitation and 506 nm for emission. The last step of an experiment was to measure the fluorescence in the presence of 0.2 wt % Triton X100. For calibration of the fluorescence scale, the initial residual fluorescence intensity before the addition of anionic lipid was set to zero and the intensity at infinite probe dilution obtained by lysis of the lipoplexes with Triton X-100 was set to 100%. To estimate the influence of charged lipids on the fluorescence of the probes (from possible effects of surface charge on extinction coefficients and quantum yield), we tested a sequence of cationic/anionic lipid mixtures representing the expected compositions of liposomes that would be obtained at different extents of lipid mixing. The fluorescence intensity as a function of the percentage of lipid mixing was essentially linear (scatter was approx. ±10%), thus providing a simple and unambiguous relationship between the parameters measured and the amount of lipid mixing [14,23,24].
2.7. Confocal microscopy
The procedure of lipoplex preparation was similar to that presented above, except that in some samples DNA was covalently labeled with fluorescein using the Label-IT kit, as described above. The protocol used was similar to that used for cell transfection [11], and included manipulation of cells under sterile conditions at 37°C. Cells were treated with lipoplexes containing FITC-labeled DNA and unlabeled lipids and— in separate experiments— with unlabeled DNA and Rh-DOPE-labeled lipids. 50 μl of lipoplex suspension was added to cells cultured in 200 μl EBM-2-MV (with 5% serum). Cells were treated for 2 h, after which they were washed with HBSS and incubated for 24 h in EBM-2-MV (with 5% serum) at 37°C, followed by 1 wt % glutaraldehyde fixation and microscopy. For fluorescein-labeled DNA, we used 488 nm laser excitation and collected emission at 500-540 nm. For rhodamine labeled lipid, we used 546 nm laser excitation and collected emission at 560-600 nm. In all experiments, the pinhole was 1.0 and 15-30 Z-sections were taken. Images were analyzed with Leica and Volocity (Improvision, Coventry, England) software.
2.8. Electron microscopy
HUAEC were grown to 80% confluence on Permanox plastic slides (Lab-Tek, distributed by Electron Microscopy Sciences, Hatfield, PA). After standard treatment of cells with lipoplex solution as described in the previous paragraph, the slides with attached cells were treated with 1 vol% glutaraldehyde for 60 min at intervals of 1 h, 3 h, 8 h and 24 h following transfection. Then the slides were covered with 1% agar for protection and the cells were postfixed with 1 wt% osmium tetroxide overnight at 4°C, and then with 1 wt% tannin for 3 h at 4°C. All fixative solutions were prepared in 0.1 M cacodylate buffer (pH 7.4). After dehydration in an acetone series, the slides were covered with PELCO, Eponate 12 (distributed by Ted Pella, Redding, CA). After resin polymerization, the embedded cells were removed from the surface of Permanox slides and re-embedded in Eponate blocks. Sections of 50-70 nm thickness were cut in the direction perpendicular to the cell layer on a MT6000-XL microtome (RMC, Tucson, AZ). Sections were stained with uranyl acetate and lead citrate and observed with a JEM-100CX (JEOL, Peabody, MA) electron microscope. A similar procedure was used for lipoplexes.
2.9. Cell viability assay
Cell viability was assayed 24 h after transfection with the MTT method [25] adapted for a microplate reader as follows: Briefly, 5 mg/ml MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) solution was added to cells in 96-well plates at 15 μl per well, and the plate was incubated at 37°C for 4h. Then, 100 μl acid-isopropanol (0.04M HCl in isopropanol) was added to each well and mixed thoroughly to dissolve the dark blue crystals. The plates were read on Spectra MAX PLUS microplate spectrophotometer (Molecular Devices, Sunnyvale, CA) with a test wavelength of 570 nm and a reference wavelength of 630 nm. The viability of untreated cells was set as 100%.
3. Results
3.1. Structure of lipoplexes
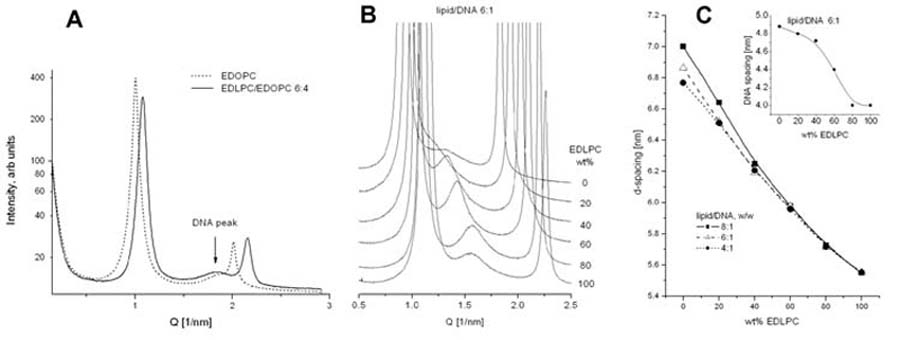
SAXD revealed that the EDOPC and EDLPC/EDOPC lipoplexes at different compositions are arranged in lamellar arrays, as shown by the sets of sharp reflections in the diffraction patterns (Fig. 1A,B). The same is true for the pure lipid samples - they also arrange into lamellar arrays, with ∼1-1.5 nm smaller repeat period than that of the lipoplexes - ∼5 nm for EDOPC and ∼4.2 nm for EDLPC (not illustrated, but see, e.g., [22,26]). The difference in the lamellar spacing induced by the presence of DNA is consistent with reports on lipoplexes of other EPCs [15,27-29]; the presence of the DNA strands between the lipid bilayers has been verified by the electron density profiles of the lipoplexes [22]. In addition to the sharp lamellar reflections, a low-intensity diffuse peak was also present in the lipoplex diffraction patterns. Its spacing was 3.3-3.4 nm for the EDOPC and EDLPC/EDOPC (6:4) lipoplexes at 4:1 lipid/DNA (w/w) ratio (Fig. 1A). Such peaks have been interpreted as reflecting the in-plane packing of the DNA strands intercalated between the lipid lamellae [27,30,31]. The arrangement of the DNA strands between the lipid bilayers has been found sensitive to the lipid/DNA stoichiometry of the lipoplex preparations [26,32] and is consistent with an expanding one-dimensional lattice of DNA chains; thus, the DNA chains confined between bilayers form a 2D smectic phase. [27,31]. The DNA interstrand repeat distance within the EDLPC/EDOPC 6:4 (w/w) lipoplexes increased from 3.4 nm to 4.4 nm when the lipid/DNA weight ratio was increased from 4:1 to 6:1 (Fig. 1A,B). The lamellar repeat period of the lipoplexes monotonously decreased with increasing EDLPC fraction (Fig. 1C). In spite of the pronounced difference in the chain lengths of the two cationic lipids, the SAXD patterns revealed no indication of phase separation. The DNA interstrand distance also decreased with increasing the EDLPC fraction (Fig. 1C, inset).
Fig. 1.
Small-angle X-ray diffraction profiles of: (A) EDOPC and EDLPC/EDOPC (6:4) lipoplexes, at 4:1 lipid/DNA weight ratio (arrow points to the peaks originating from the DNA-DNA in-plane correlation); (B) EDLPC/EDOPC lipoplexes at different lipid composition and 6:1 lipid/DNA weight ratio; in order to magnify the DNA diffraction peaks, the sharp lipid lamellar reflections are truncated. Diffraction data were collected for 1 sec at 37°C. (C) Lipid lamellar repeat distance and DNA spacing (inset) of EDLPC/EDOPC lipoplexes at different compositions.
Thin-section electron microscopy (Fig. 2) also revealed a multilamellar structure of both EDOPC lipoplexes and EDLPC/EDOPC 6:4 (w/w) lipoplexes. In transverse sections, the lipoplexes were typically seen to be composed of periodically arranged bilayers in concentric circles or in uniform spirals (Fig. 2). As demonstrated previously, these patterns represent bilayers alternating with single layers of parallel DNA molecules [22,31]. Our dynamic light scattering experiments showed that the mean diameter of the cationic liposomes was about 550 nm for EDOPC and about 400 nm for EDLPC/EDOPC 6:4 (w/w). In addition to the demonstration of similar structure and morphology with methods just described, fluorescence measurements on lipoplexes treated with increasing concentrations of NaCl [2] also revealed virtually equivalent DNA-cationic lipid binding energies for the EDOPC and EDLPC/EDOPC 6:4 lipoplexes (Koynova, unpublished data).
Fig. 2.
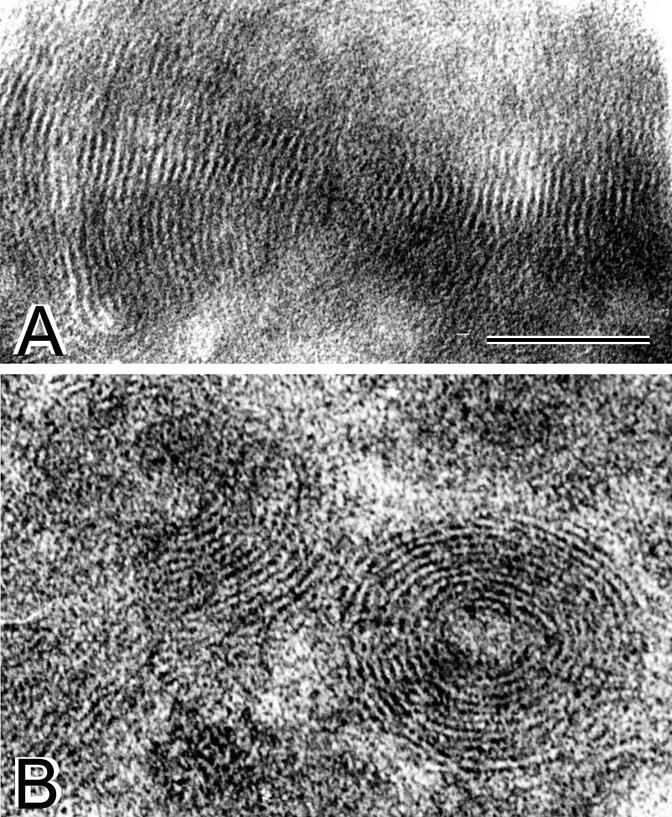
Thin-section electron microscopy of (A) EDOPC lipoplexes and (B) EDLPC/EDOPC lipoplexes. For both images, bar is 0.1 μm.
3.2. Surface activity of the cationic lipid dispersions
The rate of transfer of lipid molecules from the bulk to the air-water interface (dynamic surface activity [17]) was assessed by monitoring the changes in the surface tension of the liposome suspension with time [16]. The surface tension of EDLPC/EDOPC suspensions of different compositions dispersed in 0.15 M NaCl is presented as a function of time in Fig. 3. As judged from the initial rate of the surface tension decrease, the rate of transfer of lipid molecules to the air-water interface was greater for dispersions in which EDLPC was the dominant lipid. The apparent equilibrium surface tension (∼40 dyn/cm for EDOPC vs. <≈30 dyn/cm for EDLPC/EDOPC 6:4), as well as the equilibration time (∼45 min for EDOPC vs. ∼25 min for EDLPC/EDOPC 6:4), were also considerably lower for the mixtures that contained more EDLPC than EDOPC (Fig. 3, inset).
Fig. 3.
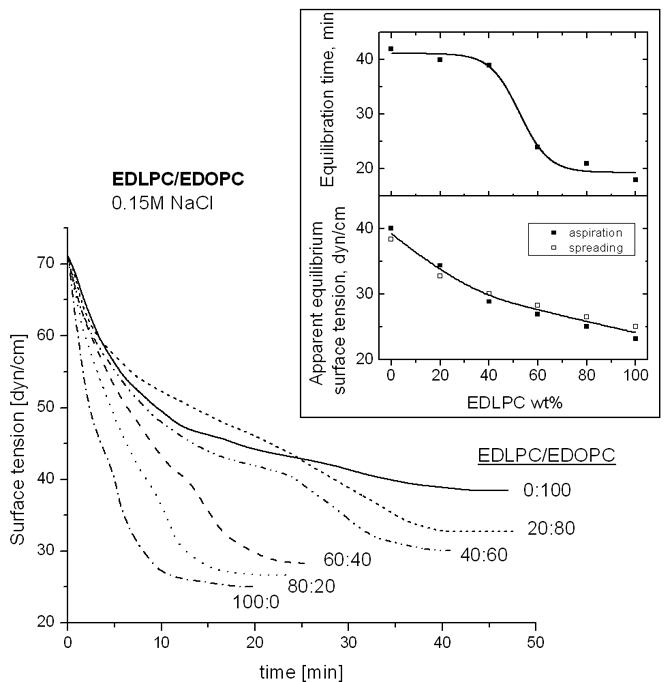
Time course of the change in surface tension of surfaces of dispersions of EDLPC/EDOPC at different lipid ratios recorded immediately after spreading a dispersion containing a total of 5 μg cationic lipid over the aqueous subphase. Inset: Equilibration time (upper panel) and apparent equilibrium surface tension (lower panel) at different EDLPC/EDOPC ratios. In the lower panel, data obtained both by suspension spreading and by surface aspiration (see Materials and Methods) are included.
3.3. Lipid mixing between lipoplexes and negatively charged liposomes
In the lipid mixing experiments, we used as a target liposomes containing 80 mol% DOPC and 20 mol% negatively charged membrane lipid (PS, PI, or CL), corresponding approximately to the amount of anionic lipids in cellular membranes [33,34]. The initial rates of lipid mixing with all three anionic membrane lipids tested were considerably higher for the EDLPC/EDOPC 6:4 lipoplexes than for the EDOPC lipoplexes and, in terms of the type of negatively charged lipid, initial rates were in the sequence CL>PS>PI for the extruded unilamellar anionic liposomes (Fig. 4). This trend was similar to that observed with multilamellar negatively charged liposomes (not illustrated), as well as with the trend previously observed for EDOPC lipoplexes [35].
Fig. 4.
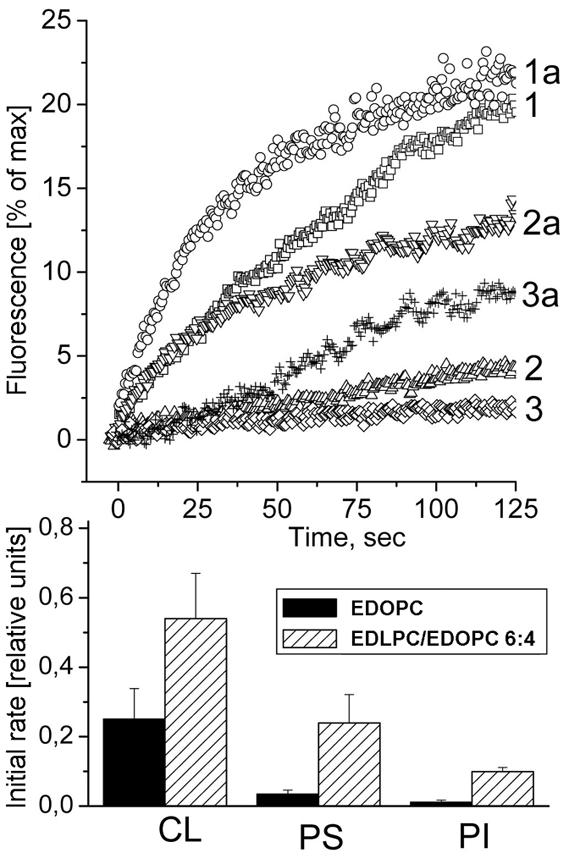
FRET assay for mixing between EDOPC lipoplexes (1,2 and 3) or EDLPC/EDOPC 6:4 lipoplexes (1a, 2a, and 3a) and DOPC unilamellar liposomes containing 20 mol % CL (1 and 1a), PS (2 and 2a), or PI (3 and 3a).
3.4. Kinetics of DNA release from lipoplexes induced by negatively charged membrane lipids
We applied flow-fluorometry [20] to examine, as a function of time, the release of DNA from EDOPC and EDLPC/EDOPC lipoplexes, induced by addition of negatively charged liposomes that had a composition mimicking natural membranes, MM = DOPC:DOPE:DOPS:Chol, 45:20:20:15 (w/w) [36]. The results are presented as plots of the DNA/cationic lipid stoichiometry (as a charge ratio) vs. the relative cationic lipid content of the individual particles (see Fig. 5B) at different times after the addition of the negatively charged liposomes (Fig. 5A). The lipoplexes were prepared at close to isoelectric conditions (DNA/lipid ∼1:1 charge ratio) and equilibrated for 30 min before the addition of the negatively charged membrane lipids; measurements were initiated immediately upon addition of the negatively charged liposomes. As seen from the leftmost plots on Fig. 5A, the EDLPC/EDOPC 6:4 lipoplexes are more homogeneous with respect to lipid/DNA composition and have higher lipid content than the EDOPC lipoplexes. The latter observation correlates with our dynamic light scattering experiments; the latter indicated that although EDLPC/EDOPC 6:4 liposomes are somewhat smaller than EDOPC liposomes (400 nm vs. 550 nm), the mixed lipid lipoplexes grew about 4x larger than the EDOPC lipoplexes (not illustrated). Shortly after addition of the negatively charged liposomes to the EDOPC lipoplexes, particles of intermediate DNA/cationic lipid stoichiometries could be detected, but even after 60 min the original 1:1 lipoplexes strongly dominated the distribution; only after >2h was a considerable decrease of the DNA/cationic lipid stoichiometry (i.e., DNA release) observed. In the case of EDLPC/EDOPC 6:4 lipoplexes, changes in composition occurred: within 5 min after addition of the negatively charged liposomes, the DNA/cationic lipid stoichiometry was shifted to lower values, and after 45-60 min, a considerable portion of the DNA appears to have been released from the lipoplexes. Similar experiments were also carried out on EDLPC/EDOPC 2:8 and 4:6 (w/w) lipoplexes (not illustrated). The flow-fluorometry data were used to assess the kinetics of the DNA release from the lipoplexes of different compositions. The data, calculated as the portion of the initially contained DNA that was retained in the lipoplexes after the addition of the negatively charged liposomes, are plotted as a function of time in Fig. 5C. The EDLPC/EDOPC 6:4 lipoplex formulation, which is the most active transfection agent [11], exhibited much faster DNA release than did lipoplexes of other compositions.
3.5. Confocal microscopy
The distribution of fluorescent DNA and lipid (Fig. 6) was examined in HUAEC cells fixed with glutaraldehyde 24 h after treatment with lipoplexes. Lipid was rendered fluorescent by inclusion of (Rh-DOPE, red) and DNA was covalently labeled (FITC-DNA, green). In cells treated with EDOPC lipoplexes, both DNA and lipid were localized in the perinuclear endosomes (Fig.6A,C). In contrast, the distribution of fluorescent compounds in cells treated with EDLPC/EDOPC lipoplexes was quite different (Fig.6B,D), clearly showing that DNA and lipid had both spread into the cytoplasm. In addition, DNA fluorescence was also detectable in the nucleoplasm. Incidentally, the latter observation is noteworthy, since it is not often that plasmid fluorescence in the nuclei has been observed. Somewhat surprisingly, some Rh-DOPE labeled lipid was also present in the nucleoplasm. 3-D reconstitution of fluorescence profiles revealed presence of Rh-DOPE fluorescent spots on the bottom of the nucleus; some fibrillar structures crossing the nucleus were also visible (Tarahovsky et al., unpublished data). We were unable to attribute the localization of fluorescent lipid to any of the nuclear regions.
Fig. 6.
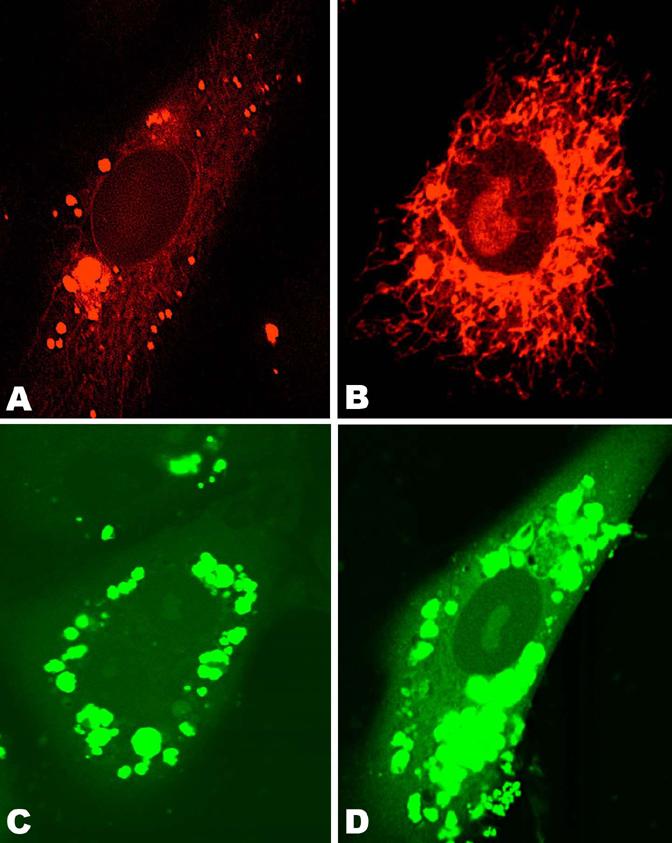
Confocal microscopy of HUAEC cells fixed 24h after treatment with lipoplexes that contained rhodamine-labeled lipid (A and B), and fluorescein-labeled DNA (C and D). Cells were treated with EDOPC lipoplexes (A and C) and EDLPC/EDOPC lipoplexes (B and D). EDOPC lipoplexes (A, C) remained in compact perinuclear endosomes, while EDLPC/EDOPC 6:4 lipoplexes (B, D) interacted with cellular membranes and released DNA into both cytoplasm and nucleus.
3.6. Electron microscopy of cells
For this study, a technique for transverse thin-sectioning of cell monolayers was developed in which cells were fixed and embedded directly on the plastic growth surface. The procedure excluded enzymatic treatment or centrifugation of cells, which could lead to changes of the initial shape and possible redistribution of cytoplasmic components. All cells were observed to have maintained their native spindle-like shape in transverse sections. In the micrograph shown in Fig. 7A, the bottom of the picture corresponds to the attachment site of the cultured cell to the plastic surface, although the plastic itself is not present, having been removed before sectioning. Electron microscopy of cells treated with lipoplexes revealed easily-recognizable endosomal compartments in the cytoplasm containing multilamellar lipoplexes, similar to that described previously by others [37-39].
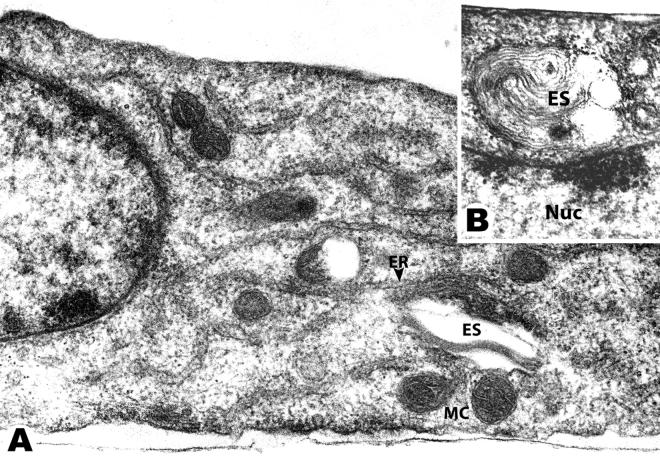
Fig. 7.
Electron micrographs of HUAEC cells treated with EDLPC/EDOPC 6:4 lipoplexes and fixed 8 hours later: Panel A shows a typical transverse section of a treated cell. Lipoplexes, perhaps with attached remnants of endosomal membrane (ES), were seen in contact with mitochondria (MC) and endoplasmic reticulum (ER). In panel B, multilamellar lipoplexes are seen in contact with the nucleus (Nuc). For all images, the bar corresponds to 0.5 μm.
EDOPC lipoplexes retained their multilamellar structure for longer than did lipoplexes prepared from the EDLPC/EDOPC 6:4 mixtures. In some cases, the multilamellar structure of the EDOPC lipoplexes could be seen as long as 24 h after transfection (not shown). The lipoplexes containing the EDLPC/EDOPC 6:4 mixture lost their multilamellar structure and after a few hours of residence within cells took on the appearance of vesicles with only a few layers of membranes. Endosomes containing EDLPC/EDOPC 6:4 lipoplexes were commonly seen interacting intimately with various cytoplasmic membranes (it should be recognized that we cannot distinguish between an endosome that completely encapsulates a lipoplex and one that has fused with the outer layers of a lipoplex such that its membrane has acquired cationic lipids). In particular, there were numerous examples of endosome/lipoplex contacts with mitochondria, endoplasmic reticulum (Fig. 7A), and nuclei (Fig. 7B).
4. Discussion
Understanding the mechanism of gene delivery by cationic liposomes is of utmost importance for effective gene therapy. A number of physical and physico-chemical factors have been suggested as lipofection modulators, but the specific route of DNA delivery by cationic lipid vectors is still mostly unknown and their efficiency of delivery is unsatisfactorily low for many applications. To date, the primary approach to improving transfection properties of cationic lipids has involved synthesizing new kinds of cationic amphiphiles or including non-cationic helper lipids in lipoplex formulations. Another strategy, more recent and particularly effective, is to combine two cationic lipid derivatives having the same head group but different hydrocarbon chains. Such combinations often synergistically enhance transfection and allow optimizing activity by merely varying the ratio of the two components. For example, some compositions of the cationic lipid binary mixture EDLPC/EDOPC transfected DNA into cells over 30-times more efficiently than either compound separately [11]. Because of the magnitude of this synergy and the fact that it involved homologs of the same molecules, this system appeared appropriate for analyzing the origin of the activity difference between them by comparing the physical and physico-chemical properties of lipoplexes with very different transfection efficiencies, but minor chemical differences.
In an earlier publication, we described a correlation between the delivery efficiency of these DNA carriers and the mesomorphic phases they form after interaction with anionic membrane lipids. Specifically, formulations that are particularly effective DNA carriers form phases of highest negative interfacial curvature when mixed with negatively charged membrane lipids, whereas less effective formulations form phases of lower curvature under the same conditions [40]. In the present study, we examined further physical characteristics that might account for the transfection efficiency of superior lipoplexes, namely, their structure, surface activity, propensity to admix/fuse with negatively charged membrane lipids, and ability to eventually release DNA. Experiments with model systems were complemented with observations on transfected HUAEC cells.
EDOPC and EDLPC/EDOPC lipoplexes are structurally similar to each other, and to the majority of lipoplexes [27,30,31,41], consisting of lamellar lipid arrays with intercalated DNA threads (Fig. 1). The density of DNA packing is higher (lower interstrand distance) in lipoplexes in which EDLPC predominates (Fig. 1C, inset), as expected from the presumptive lower area per lipid molecule of the shorter-chain compound [42].
The lipid concentration of the dispersions used in the X-ray diffraction experiments (∼50mM) are certainly considerably higher than bulk concentrations in the cell, however, they may be similar to the local concentrations in the cell (i.e., at membrane-membrane contacts). In any case, their relevance to physiological concentrations with respect to phase and structural data obtained has been repeatedly checked by control experiments at low concentrations (Koynova, unpublished data, but also see, e.g., the inset of Fig.2A in ref.[31]).
In order for DNA to be released from lipoplexes and enter the cell nucleus where it is transcribed, the cationic lipid electrostatic charge must be neutralized. The unbinding of DNA from lipoplexes has been identified as one of the critical steps along the transfection route. Although there may be other possibilities, according to current understanding, it involves neutralization of cationic lipid by cellular anionic lipids. Indeed, addition of negatively charged liposomes to lipoplexes results in dissociation of DNA from the lipid [3,4,22,35,43]. Neutralization of cationic lipid carriers by anionic membrane lipids, required for DNA release, presupposes lipid transfer between cationic lipoplexes and negatively charged membranes, most likely by fusion of cell membranes with lipoplexes. Mixing of lipoplex lipids with cellular lipids was observed a number of years ago and interpreted as fusion [8]. Precisely how the lipoplex bilayers fuse with cell membranes is unclear [3,5], but there is no question that cationic and anionic membranes are capable of both fusion and hemifusion, and do so extremely rapidly, even at relatively low anionic charge densities [44]. Because escape of lipoplexes from endosomes prior to their entry into lysosomes is essential for efficient transgene expression, fusion of lipoplexes with endosomal membranes should facilitate DNA release from endosomes into the cytoplasm, and thus promote DNA expression. To quantify this fusion process (strictly, lipid mixing is what is measured and, indeed, lipid mixing is what is required for neutralization of the lipoplex lipid), we used a FRET assay involving a pair of fluorescent lipid dyes. It revealed that EDLPC/EDOPC lipoplexes fuse faster and more extensively with negatively charged liposomes (Fig. 4) than do EDOPC lipoplexes. Previously, it was reported that EDLPC/EDOPC lipoplexes exhibit higher fusion capacity with liposomes containing the anionic DOPG [11]. Here we report that the superior fusogenicity of the EDLPC/EDOPC mixed lipoplexes is also manifested with other anionic membrane lipid classes - CL, PS, and PI. Even though fusion of lipoplexes with membranes does not necessarily result in release of DNA from the lipoplexes, it is nevertheless likely to be beneficial for lipoplex escape from endosomes. That is, membrane fusion is probably an essential step in transfection, but DNA release from the lipoplex may be the rate-limiting step. Indeed, a lack of close correlation between transfection activity and lipoplex-membrane lipid mixing has been reported before [45-49]. Our recent experiments also showed that facile fusion with negatively charged liposomes characterizes a variety of cationic lipid mixtures (relative to single-component cationic liposomes), regardless of their efficacy as transfection agents [13]. This phenomenon is perhaps related to nonideal mixing, domain formation, and/or higher probability for packing defects in the cationic lipid mixtures relative to single component bilayers (packing defects should facilitate the fusion of bilayers, because bilayer fusion, at some stage in the process, must involve merging of the hydrophobic cores of the two participating membranes [50]). There is no doubt that two cationic lipids have more degrees of freedom in lipoplex-membrane interactions than does a single lipid Increased degrees of freedom, in turn allow for larger variation of membrane curvature, which is likely to be important in membrane fusion. The bilayer bending constant is known to be lowered in lipid mixtures as a result of the fact that a mixed bilayer may assume different compositions in the two opposing monolayers; the magnitude of the bending constant reduction increases with increasing difference between the two amphiphiles with respect to charge, head group size, and chain length [51]. Thus, mixed bilayers, especially those involving species of considerable difference in the chain length, would exhibit higher tendency to curve and eventually fuse. In any case, high fusogenicity is likely to be an attribute of, even though not a guarantee for, superior transfection activity. It is clear that extrapolating results from fusion experiments on lipid model systems to natural membranes that contain proteins and much more complex mixture of lipids requires care; hence we emphasize that our lipid mixing (FRET) experiments involved oppositely charged lipid aggregates (as do the lipoplex-membrane interactions), in which fusion is activated by electrostatic interactions. Moreover, observations through the microscope of giant vesicles and fluorescent lipoplexes verify that true fusion of the lipoplexes with negatively charged bilayer membranes does occur [44,52-54].
As mentioned above, lipid mixing, in fact, is what was measured by the FRET experiments. In principle, monomer lipid exchange between aggregates could produce generally similar FRET results. The presumptive higher CMC of the shorter-chain EDLPC may significantly accelerate the lipid molecular transfer (the term “CMC” is used here to indicate the minimal lipid concentration required for monomeric lipids to form a supramolecular assembly, regardless of its geometry). Indeed, a difference of even two methylene groups in each chain results in a ∼30-fold change of the CMC of phosphatidylcholines [55]., In the case of EDLPC and EDOPC, the difference is six CH2 groups, so a much higher monomer concentration is expected in the dispersions with dominant EDLPC content. The higher concentration of monomers was indeed found to significantly facilitate molecular transfer between aggregates, even when the aggregates are all positively charged [56].
The dynamic surface activity of the cationic lipid mixtures with dominating EDLPC content is also considerably higher than that of the EDOPC dispersion (Fig. 3). The different rates of monolayer formation at the air/water interface of dispersions of lipid vesicles may stem from a variety of different causes. Both individual molecules and aggregates may contribute to monolayer formation. In the former case, of course, the higher CMC of EDLPC might be responsible for accelerated monolayer formation. Diffusion of whole aggregates to the monolayer, with subsequent disruption and rearrangement to form an extended planar monolayer, may certainly also contribute to the monolayer formation, but since the liposome size is not significantly different for different lipid compositions, this mechanism could hardly account for the pronounced difference in the surface activity of the EDOPC and EDLPC/EDOPC aggregates. In fact, with respect to its surface activity, the pure EDLPC dispersions are even superior to the EDLPC/EDOPC 6:4 mixture of maximum transfection activity (Fig. 3). The reason for the lower efficiency of the EDLPC lipoplexes is probably their toxicity; the viability of EDLPC-treated cells was only 45%, and such cells are likely too compromised to express high levels of beta-galactosidase [11], so it may be that the higher solubility and higher CMC of EDLPC accounts for, or at least contributes to, its toxicity when used in the absence of EDOPC.
With respect to the relationship of the processes of molecular lipid exchange and lipoplex-membrane fusion to the lipid-mediated DNA delivery, we emphasize that they should not be considered independent processes in a system involving positive and negative lipid aggregates. In fact, initial molecular exchange between the cationic lipid aggregates and the negatively charged membranes would create cationic / anionic lipid mixtures and such mixtures are known for their strong disposition to form nonlamellar phases [35,57,58]. Thus, since the propensity for formation of curved nonlamellar phases is thought to play a critical role in fusion of lipid aggregates [59,60], molecular lipid exchange could be expected to trigger fusion between cationic and anionic lipid aggregates.
DNA release data revealed a correlation between the extent of transfection by different lipoplex formulations and the extent of DNA release that was induced by treating those lipoplex formulations with membrane-mimicking, negatively charged liposomes; the highly active transfection agent EDLPC/EDOPC 6:4 exhibited considerably faster DNA release relative to the less effective EDOPC (Fig. 5A,C) after interaction with the negatively charged membrane lipids. The more complete DNA release from the EDLPC/EDOPC 6:4 lipoplexes is a likely result of the propensity of that cationic lipid mixture to form phases of the highest negative interfacial curvature when mixed with negatively charged membrane lipids [40]. Indeed, DNA release has been shown to unambiguously correlate with the interfacial curvature of the phases that develop when cationic lipids of the lipoplexes interact with the negatively charged cellular lipids [61].
We have made some effort to relate our experiments on model systems to the behavior of the same lipoplex formulations in transfected cultured cells. It is commonly thought that during cell transfection with cationic liposomes, lipoplexes enter the cytoplasm by endocytosis and that the DNA is subsequently released into the cytoplasm and migrates to the nucleus [8,10,37,38,62,63]. Indeed, thin section electron microscopy (Fig. 7) revealed the presence of lipoplexes inside endosomes, as has been previously demonstrated [37-39]. Furthermore, the present data revealed instances of close contacts of lipoplexes with various cellular membranes including those of the mitochondria, endoplasmic reticulum and nucleus. Membrane contacts were more frequently observed in cells treated with lipoplexes containing the medium-chain cationic lipid (EDLPC/EDOPC 6:4 lipoplexes) than in cells treated with lipoplexes consisting only of the longer chain component (EDOPC). Confocal microscopy also confirmed a considerable preference of EDLPC/EDOPC 6:4 lipoplexes to exchange lipid with cytoplasmic membranes and to release DNA into the cytoplasm and the nucleus (Fig. 6).
Lipoplexes with mean diameters of about 400-500 nm and lamellar repeat distances of ∼5-6 nm, as observed in the present study, would contain some ∼20 membrane bilayers. It is thus obvious that lipid exchange between a multilamellar lipoplex and surrounding single bilayer of endosomal membrane is not sufficient to neutralize the cationic charge necessary for DNA unbinding and release from lipoplexes, and much more extensive intermembrane interactions are required for more complete DNA release. The contacts visualized by electron microscopy between the superior EDLPC/EDOPC 6:4 lipoplexes and various cellular membranes may therefore support the concept of gradual lipoplex peeling and DNA release.
The lipid composition of different organelles may influence lipoplex lipid exchange and fusion with different cellular membranes, and hence also the distribution of released DNA inside of cell. The facile fusion of lipoplexes with CL-containing liposomes (Fig.4) suggests the possibility of interaction and fusion of lipoplexes with mitochondrial membranes and, indeed, contacts of EDLPC/EDOPC 6:4 lipoplexes with mitochondrial membranes were observed by electron microscopy (Fig. 7 A). Although the extent to which CL is found on the external mitochondrial membrane remains to be settled [64], the high content of this anionic lipid in mitochondria as a whole has been clearly established [65].
In summary,
the EDLPC/EDOPC 6:4 mixture, which had superior transfection efficiency, was found to exhibit considerably higher surface activity than EDOPC, a characteristic likely to favor cationic lipid transfer to (negatively charged) membranes. Mixtures of anionic and cationic lipids are thus likely to be generated in cells treated with these lipoplexes; such mixtures are known for their strong disposition to form nonlamellar phases [57,58,61], which propensity is thought to play a critical role in the fusion ability of lipid aggregates [59,60]; indeed, actual fusion of lipoplexes with anionic liposomes has been demonstrated [54]. Furthermore, the EDLPC/EDOPC 6:4 mixture exhibits considerably higher ability to mix with negatively charged liposomes, as demonstrated by FRET. Lipoplex-membrane mixing contributes to the charge neutralization of cationic lipids by cellular anionic lipids, which is required for DNA unbinding [3,4,14].
DNA discharge measured subsequent to treating lipoplexes with negatively charged membrane-mimicking liposomes was considerably faster for the EDLPC/EDOPC 6:4 mixture than for EDOPC lipoplexes. An unambiguous correlation between DNA release and the phases formed by the mixtures of the cellular lipids with the cationic lipids of the lipoplexes has been reported earlier - specifically, the magnitude of the negative interfacial curvature of the phase assumed by the cationic-cellular lipid mixture correlated with release of DNA [61]. The EDLPC/EDOPC 6:4 described here conforms to that correlation in that it also developed highly curved nonbilayer phases when mixed with negatively charged membrane lipids [40].
thin-section electron microscopy of treated cells revealed contacts between EDLPC/EDOPC 6:4 lipoplexes and various cellular membranes, including those of the endoplasmic reticulum, mitochondria and nucleus. Because of the multilayered structure of the lipoplexes, multiple encounters between lipoplexes and various cellular membranes are expected to be needed for efficient release of lipoplex DNA.
Acknowledgments
NIH grant GM52329 provided primary funding for this research, and NIH grant GM57305 provided some important additional support. We thank Robert A. Lamb (Northwestern University) for access to the flow cytometer instrument, William Russin (Northwestern Univ.) for technical support in confocal microscopy, and Harsh Parikh and Yaeko Hiyama (Northwestern Univ.) for synthesis of cationic lipids. BioCAT is a NIH-supported Research Center, through Grant RR08630. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Energy Research under Contract No. W-31-102-Eng-38. DND-CAT is supported by E.I. DuPont de Nemours & Co., The Dow Chemical Company, NSF Grant DMR-9304725, and the State of Illinois through the Department of Commerce and the Board of Higher Education Grant IBHE HECA NWU 96. Use of APS was supported by the U.S. DOE, Basic Energy Sciences, Office of Energy Research, Contract No. W-31-102-Eng-38.
Footnotes
Lipoplexes of pure EDLPC exhibited enhanced toxicity: the viability of EDLPC-treated cells was only 45% [13]
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Tachibana R, Harashima H, Ide N, Ukitsu S, Ohta Y, Suzuki N, Kikuchi H, Shinohara Y, Kiwada H. Quantitative Analysis of Correlation Between Number of Nuclear Plasmids and Gene Expression Activity After Transfection With Cationic Liposomes. Pharm. Res. 2002;19:377–381. doi: 10.1023/a:1015162722295. [DOI] [PubMed] [Google Scholar]
- [2].Pozharski E, MacDonald RC. Lipoplex Thermodynamics: Determination of DNA-Cationic Lipoid Interaction Energies. Biophys. J. 2003;85:3969–3978. doi: 10.1016/S0006-3495(03)74811-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Xu YH, Szoka FC. Mechanism of DNA Release From Cationic Liposome/DNA Complexes Used in Cell Transfection. Biochemistry. 1996;35:5616–5623. doi: 10.1021/bi9602019. [DOI] [PubMed] [Google Scholar]
- [4].Zelphati O, Szoka FC. Mechanism of Oligonucleotide Release From Cationic Liposomes. Proc. Natl. Acad. Sci. U. S. A. 1996;93:11493–11498. doi: 10.1073/pnas.93.21.11493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Szoka FC, Xu YH, Zelphati O. How Are Nucleic Acids Released in Cells From Cationic Lipid-Nucleic Acid Complexes? Adv. Drug Delivery Rev. 1997;24:291. [Google Scholar]
- [6].Bhattacharya S, Mandal SS. Evidence of Interlipidic Ion-Pairing in Anion-Induced DNA Release From Cationic Amphiphile-DNA Complexes. Mechanistic Implications in Transfection. Biochemistry. 1998;37:7764–7777. doi: 10.1021/bi971772j. [DOI] [PubMed] [Google Scholar]
- [7].Harvie P, Wong FMP, Bally MB. Characterization of Lipid DNA Interactions. I. Destabilization of Bound Lipids and DNA Dissociation. Biophys. J. 1998;75:1040–1051. doi: 10.1016/S0006-3495(98)77593-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wrobel I, Collins D. Fusion of Cationic Liposomes With Mammalian-Cells Occurs After Endocytosis. Biochim. Biophys. Acta-Biomembranes. 1995;1235:296–304. doi: 10.1016/0005-2736(95)80017-a. [DOI] [PubMed] [Google Scholar]
- [9].Noguchi A, Furuno T, Kawaura C, Nakanishi M. Membrane Fusion Plays an Important Role in Gene Transfection Mediated by Cationic Liposomes. FEBS Lett. 1998;433:169–173. doi: 10.1016/s0014-5793(98)00837-0. [DOI] [PubMed] [Google Scholar]
- [10].Nakanishi M, Noguchi A. Confocal and Probe Microscopy to Study Gene Transfection Mediated by Cationic Liposomes With a Cationic Cholesterol Derivative. Adv. Drug Delivery Rev. 2001;52:197–207. doi: 10.1016/s0169-409x(01)00207-1. [DOI] [PubMed] [Google Scholar]
- [11].Wang L, MacDonald RC. New Strategy for Transfection: Mixtures of Medium-Chain and Long-Chain Cationic Lipids Synergistically Enhance Transfection. Gene Ther. 2004;11:1358–1362. doi: 10.1038/sj.gt.3302297. [DOI] [PubMed] [Google Scholar]
- [12].Teifel M, Heine LT, Milbredt S, Friedl P. Optimization of Transfection of Human Endothelial Cells. Endothelium-New York. 1997;5:21–35. doi: 10.3109/10623329709044156. [DOI] [PubMed] [Google Scholar]
- [13].Wang L, Koynova R, Parikh H, MacDonald RC. Transfection Activity of Binary Mixtures of Cationic O-Substituted Phosphatidylcholine Derivatives: The Hydrophobic Core Strongly Modulates Their Physical Properties and DNA Delivery Efficacy. Biophys. J. 2006;91 doi: 10.1529/biophysj.106.092700. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].MacDonald RC, Rakhmanova VA, Choi KL, Rosenzweig HS, Lahiri MK. O-Ethylphosphatidylcholine: A Metabolizable Cationic Phospholipid Which Is a Serum-Compatible DNA Transfection Agent. J. Pharm. Sci. 1999;88:896–904. doi: 10.1021/js990006q. [DOI] [PubMed] [Google Scholar]
- [15].Koynova R, MacDonald RC. Mixtures of Cationic Lipid O-Ethylphosphatidylcholine With Membrane Lipids and DNA: Phase Diagrams. Biophys. J. 2003;85:2449–2465. doi: 10.1016/s0006-3495(03)74668-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].MacDonald RC, Simon SA. Lipid Monolayer States and Their Relationships to Bilayers. Proc. Natl. Acad. Sci. U. S. A. 1987;84:4089–4093. doi: 10.1073/pnas.84.12.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].MacDonald RC, Gorbonos A, Mornsen MM, Brockman HL. Surface Properties of Dioleoyl-Sn-Glycerol-3-Ethylphosphocholine, a Cationic Phosphatidylcholine Transfection Agent, Alone and in Combination With Lipids or DNA. Langmuir. 2006;22:2770–2779. doi: 10.1021/la0524566. [DOI] [PubMed] [Google Scholar]
- [18].Qiu RZ, MacDonald RC. A Metastable State of High Surface-Activity Produced by Sonication of Phospholipids. Biochim. Biophys. Acta-Biomembranes. 1994;1191:343–353. doi: 10.1016/0005-2736(94)90185-6. [DOI] [PubMed] [Google Scholar]
- [19].Jordanova A, Lalchev Z, Tenchov B. Formation of Monolayers and Bilayer Foam Films From Lamellar, Inverted Hexagonal and Cubic Lipid Phases. Eur. Biophys. J. 2003;31:626–632. doi: 10.1007/s00249-002-0263-x. [DOI] [PubMed] [Google Scholar]
- [20].Pozharski E, MacDonald RC. Analysis of the Structure and Composition of Individual Lipoplex Particles by Flow Fluorometry. Anal. Biochem. 2005;341:230–240. doi: 10.1016/j.ab.2005.03.027. [DOI] [PubMed] [Google Scholar]
- [21].Pozharski E, MacDonald RC. Single Lipoplex Study of Cationic Lipid-DNA Self-Assembled Complex: Flow Cytometric Approach. Biophys. J. 2001;80:427A. [Google Scholar]
- [22].MacDonald RC, Ashley GW, Shida MM, Rakhmanova VA, Tarahovsky YS, Pantazatos DP, Kennedy MT, Pozharski EV, Baker KA, Jones RD, Rosenzweig HS, Choi KL, Qiu RZ, McIntosh TJ. Physical and Biological Properties of Cationic Triesters of Phosphatidylcholine. Biophys. J. 1999;77:2612–2629. doi: 10.1016/S0006-3495(99)77095-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ouedraogo G, Morliere P, Maziere C, Maziere JC, Santus R. Alteration of the Endocytotic Pathway by Photosensitization With Fluoroquinolones. Photochem. Photobiol. 2000;72:458–463. doi: 10.1562/0031-8655(2000)072<0458:aotepb>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- [24].Traore M, Sun RJ, Fawzi-Grancher S, Dumas D, Qing X, Santus R, Stoltz JF, Muller S. Kinetics of the Endocytotic Pathway of Low Density Lipoprotein (LDL) in Human Endothelial Cells Line Under Shear Stress: An in Vitro Confocal Microscopy Study. Clin. Hemorheol. Microcirc. 2005;33:243–251. [PubMed] [Google Scholar]
- [25].Mosmann T. Rapid Colorimetric Assay for Cellular Growth and Survival - Application to Proliferation and Cyto-Toxicity Assays. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- [26].Koynova R, MacDonald RC. Cationic O-Ethylphosphatidylcholines and Their Lipoplexes: Phase Behavior Aspects, Structural Organization and Morphology. Biochim. Biophys. Acta-Biomembranes. 2003;1613:39–48. doi: 10.1016/s0005-2736(03)00135-4. [DOI] [PubMed] [Google Scholar]
- [27].Koynova R, MacDonald RC. Columnar DNA Superlattices in Lamellar O-Ethylphosphatidylcholine Lipoplexes: Mechanism of the Gel-Liquid Crystalline Lipid Phase Transition. Nano Lett. 2004;4:1475–1479. [Google Scholar]
- [28].Koynova R, Wang L, MacDonald RC. An Intracellular Lamellar - Nonlamellar Phase Transition Rationalizes the Superior Performance of Some Cationic Lipid Transfection Agents. Proc. Natl. Acad. Sci. U. S. A. 2006;103:14373–14378. doi: 10.1073/pnas.0603085103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Rakhmanova VA, McIntosh TJ, MacDonald RC. Effects of Dioleoylphosphatidylethanolamine on the Activity and Structure of O-Alkyl Phosphatidylcholine-DNA Transfection Complexes. Cell. Mol. Biol. Lett. 2000;5:51–65. [Google Scholar]
- [30].Lasic DD, Strey H, Stuart MCA, Podgornik R, Frederik PM. The Structure of DNA-Liposome Complexes. J. Am. Chem. Soc. 1997;119:832–833. [Google Scholar]
- [31].Radler JO, Koltover I, Salditt T, Safinya CR. Structure of DNA-Cationic Liposome Complexes: DNA Intercalation in Multilamellar Membranes in Distinct Interhelical Packing Regimes. Science. 1997;275:810–814. doi: 10.1126/science.275.5301.810. [DOI] [PubMed] [Google Scholar]
- [32].Koltover I, Salditt T, Safinya CR. Phase Diagram, Stability, and Overcharging of Lamellar Cationic Lipid-DNA Self-Assembled Complexes. Biophys. J. 1999;77:915–924. doi: 10.1016/S0006-3495(99)76942-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Daum G. Lipids of Mitochondria. Biochim. Biophys. Acta. 1985;822:1–42. doi: 10.1016/0304-4157(85)90002-4. [DOI] [PubMed] [Google Scholar]
- [34].Siakotos AN, Rouser G, Fleische S. Isolation of Highly Purified Human and Bovine Brain Endothelial Cells and Nuclei and Their Phospholipid Composition. Lipids. 1969;4:234–239. doi: 10.1007/BF02532638. [DOI] [PubMed] [Google Scholar]
- [35].Tarahovsky YS, Koynova R, MacDonald RC. DNA Release From Lipoplexes by Anionic Lipids: Correlation With Lipid Mesomorphism, Interfacial Curvature, and Membrane Fusion. Biophys. J. 2004;87:1054–1064. doi: 10.1529/biophysj.104.042895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gennis RB. Biomembranes. Molecular Structure and Function. Springer-Verlag; New York: 1989. [Google Scholar]
- [37].Friend DS, Papahadjopoulos D, Debs RJ. Endocytosis and Intracellular Processing Accompanying Transfection Mediated by Cationic Liposomes. Biochim. Biophys. Acta-Biomembranes. 1996;1278:41–50. doi: 10.1016/0005-2736(95)00219-7. [DOI] [PubMed] [Google Scholar]
- [38].Zabner J, Fasbender AJ, Moninger T, Poellinger KA, Welsh MJ. Cellular and Molecular Barriers to Gene-Transfer by A Cationic Lipid. J. Biol. Chem. 1995;270:18997–19007. doi: 10.1074/jbc.270.32.18997. [DOI] [PubMed] [Google Scholar]
- [39].Zhou XH, Huang L. DNA Transfection Mediated by Cationic Liposomes Containing Lipopolylysine - Characterization and Mechanism of Action. Biochim. Biophys. Acta-Biomembranes. 1994;1189:195–203. doi: 10.1016/0005-2736(94)90066-3. [DOI] [PubMed] [Google Scholar]
- [40].Koynova R, Wang L, Tarahovsky Y, MacDonald RC. Lipid Phase Control of DNA Delivery. Bioconjug. Chem. 2005;16:1335–1339. doi: 10.1021/bc050226x. [DOI] [PubMed] [Google Scholar]
- [41].Boukhnikachvili T, AguerreChariol O, Airiau M, Lesieur S, Ollivon M, Vacus J. Structure of in-Serum Transfecting DNA-Cationic Lipid Complexes. FEBS Lett. 1997;409:188–194. doi: 10.1016/s0014-5793(97)00505-x. [DOI] [PubMed] [Google Scholar]
- [42].Marsh D. Handbook of Lipid Bilayers. CRC Press; 1990. [Google Scholar]
- [43].Ashley GW, Shida MM, Qiu R, Lahiri MK, Levisay PC, Jones RD, Baker KA, MacDonald RC. Phosphatidylcholinium Compounds: A New Class of Cationic Phospholipids With Transfection Activin and Unusual Physical Properties (Abstract) Biophys. J. 1996;70:88A. [Google Scholar]
- [44].Pantazatos DP, MacDonald RC. Directly Observed Membrane Fusion Between Oppositely Charged Phospholipid Bilayers. J. Membr. Biol. 1999;170:27–38. doi: 10.1007/s002329900535. [DOI] [PubMed] [Google Scholar]
- [45].Stegmann T, Legendre JY. Gene Transfer Mediated by Cationic Lipids: Lack of a Correlation Between Lipid Mixing and Transfection. Biochim. Biophys. Acta-Biomembranes. 1997;1325:71–79. doi: 10.1016/s0005-2736(96)00241-6. [DOI] [PubMed] [Google Scholar]
- [46].Mui B, Ahkong QF, Chow L, Hope MJ. Membrane Perturbation and the Mechanism of Lipid-Mediated Transfer of DNA into Cells. Biochim. Biophys. Acta-Biomembranes. 2000;1467:281–292. doi: 10.1016/s0005-2736(00)00226-1. [DOI] [PubMed] [Google Scholar]
- [47].Pires P, Simoes S, Nir S, Gaspar R, Duzgunes N, de Lima MCP. Interaction of Cationic Liposomes and Their DNA Complexes With Monocytic Leukemia Cells. Biochim. Biophys. Acta-Biomembranes. 1999;1418:71–84. doi: 10.1016/s0005-2736(99)00023-1. [DOI] [PubMed] [Google Scholar]
- [48].Leventis R, Silvius JR. Interactions of Mammalian-Cells With Lipid Dispersions Containing Novel Metabolizable Cationic Amphiphiles. Biochim. Biophys. Acta. 1990;1023:124–132. doi: 10.1016/0005-2736(90)90017-i. [DOI] [PubMed] [Google Scholar]
- [49].Harvie P, Wong FMP, Bally MB. Characterization of Lipid DNA Interactions. I. Destabilization of Bound Lipids and DNA Dissociation. Biophys. J. 1998;75:1040–1051. doi: 10.1016/S0006-3495(98)77593-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Papahadjopoulos D, Nir S, Duzgunes N. Molecular Mechanisms of Calcium-Induced Membrane-Fusion. J. Bioenerg. Biomembr. 1990;22:157–179. doi: 10.1007/BF00762944. [DOI] [PubMed] [Google Scholar]
- [51].Bergstrom M. Thermodynamics of Unilamellar Vesicles: Influence of Mixing on the Curvature Free Energy of a Vesicle Bilayer. J. Colloid Interface Sci. 2001;240:294–306. doi: 10.1006/jcis.2001.7624. [DOI] [PubMed] [Google Scholar]
- [52].Lei GH, MacDonald RC. Lipid Bilayer Vesicle Fusion: Intermediates Captured by High-Speed Microfluorescence Spectroscopy. Biophys. J. 2003;85:1585–1599. doi: 10.1016/S0006-3495(03)74590-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Pantazatos DP, Pantazatos SP, MacDonald RC. Bilayer Mixing, Fusion, and Lysis Following the Interaction of Populations of Cationic and Anionic Phospholipid Bilayer Vesicles. J. Membr. Biol. 2003;194:129–139. doi: 10.1007/s00232-003-2031-y. [DOI] [PubMed] [Google Scholar]
- [54].Pantazatos SP, MacDonald RC. Real-Time Observation of Lipoplex Formation and Interaction With Anionic Bilayer Vesicles. J. Membr. Biol. 2003;191:99–112. doi: 10.1007/s00232-002-1050-4. [DOI] [PubMed] [Google Scholar]
- [55].Smith R, Tanford C. Critical Micelle Concentration of L-Alpha-Dipalmitoylphosphatidylcholine in Water and Water/Methanol Solutions. J. Mol. Biol. 1972;67:75–83. doi: 10.1016/0022-2836(72)90387-7. [DOI] [PubMed] [Google Scholar]
- [56].Koynova R, MacDonald RC. Lipid Transfer Between Cationic Vesicles and Lipid-DNA Lipoplexes. Effect of Serum. Biochim. Biophys. Acta-Biomembranes. 2005;1714:63–70. doi: 10.1016/j.bbamem.2005.05.009. [DOI] [PubMed] [Google Scholar]
- [57].Tarahovsky YS, Arsenault AL, MacDonald RC, McIntosh TJ, Epand RM. Electrostatic Control of Phospholipid Polymorphism. Biophys. J. 2000;79:3193–3200. doi: 10.1016/S0006-3495(00)76552-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Lewis RNAH, McElhaney RN. Surface Charge Markedly Attenuates the Nonlamellar Phase-Forming Propensities of Lipid Bilayer Membranes: Calorimetric and P-31-Nuclear Magnetic Resonance Studies of Mixtures of Cationic. Anionic, and Zwitterionic Lipids, Biophys. J. 2000;79:1455–1464. doi: 10.1016/S0006-3495(00)76397-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Ellens H, Siegel DP, Alford D, Yeagle PL, Boni L, Lis LJ, Quinn PJ, Bentz J. Membrane-Fusion and Inverted Phases. Biochemistry. 1989;28:3692–3703. doi: 10.1021/bi00435a011. [DOI] [PubMed] [Google Scholar]
- [60].Siegel DP, Epand RM. The Mechanism of Lamellar-to-Inverted Hexagonal Phase Transitions in Phosphatidylethanolamine: Implications for Membrane Fusion Mechanisms. Biophys. J. 1997;73:3089–3111. doi: 10.1016/S0006-3495(97)78336-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Tarahovsky Y, Koynova R, MacDonald RC. Efficiency of Anionic Lipids to Release DNA From Lipoplexes Correlates With the Interfacial Curvature of the Mesomorphic Phases of Cationic/Anionic Lipid Mixtures. Biophys. J. 2004;86:37A. doi: 10.1529/biophysj.104.042895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Bally MB, Harvie P, Wong FMP, Kong S, Wasan EK, Reimer DL. Biological Barriers to Cellular Delivery of Lipid-Based DNA Carriers. Adv. Drug Delivery Rev. 1999;38:291–315. doi: 10.1016/s0169-409x(99)00034-4. [DOI] [PubMed] [Google Scholar]
- [63].Lechardeur D, Sohn KJ, Haardt M, Joshi PB, Monck M, Graham RW, Beatty B, Squire J, O’Brodovich H, Lukacs GL. Metabolic Instability of Plasmid DNA in the Cytosol: a Potential Barrier to Gene Transfer. Gene Ther. 1999;6:482–497. doi: 10.1038/sj.gt.3300867. [DOI] [PubMed] [Google Scholar]
- [64].Hovius R, Thijssen J, Vanderlinden P, Nicolay K, Dekruijff B. Phospholipid Asymmetry of the Outer-Membrane of Rat-Liver Mitochondria - Evidence for the Presence of Cardiolipin on the Outside of the Outer-Membrane. FEBS Lett. 1993;330:71–76. doi: 10.1016/0014-5793(93)80922-h. [DOI] [PubMed] [Google Scholar]
- [65].Mcmurray WC, Dawson RMC. Phospholipid Exchange Reactions Within Liver Cell. Biochem. J. 1969;112:91–108. doi: 10.1042/bj1120091. [DOI] [PMC free article] [PubMed] [Google Scholar]