Abstract
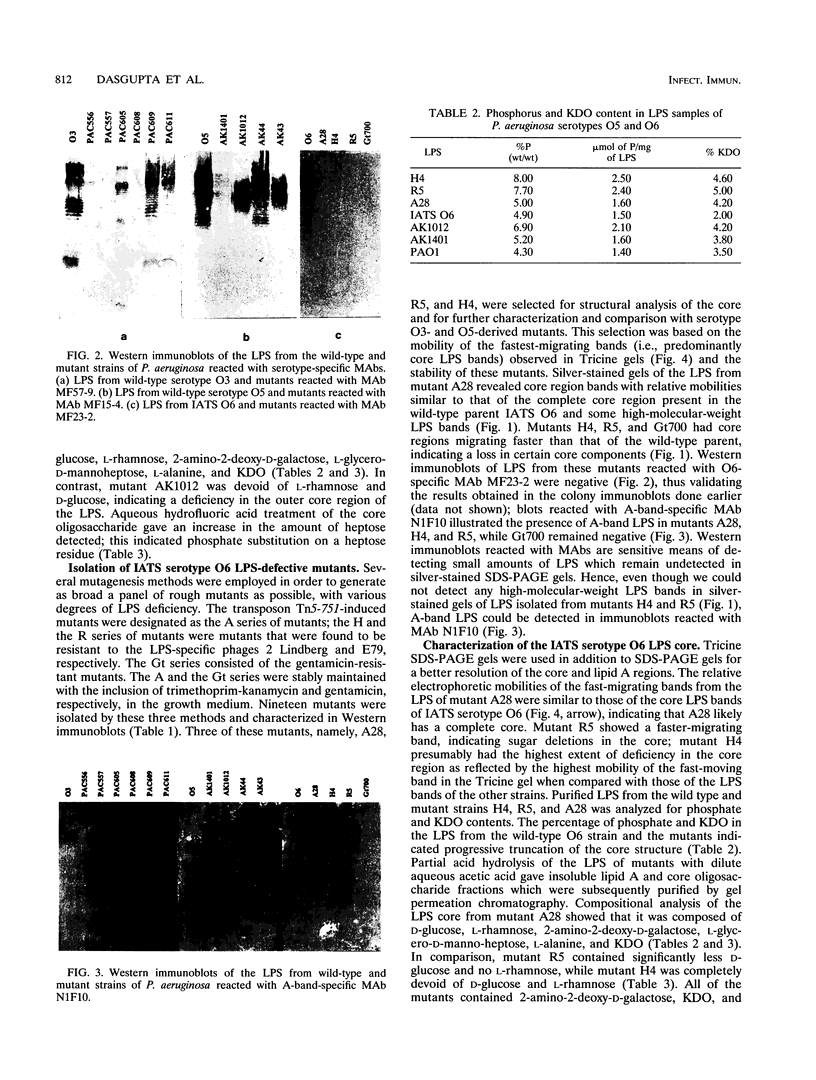
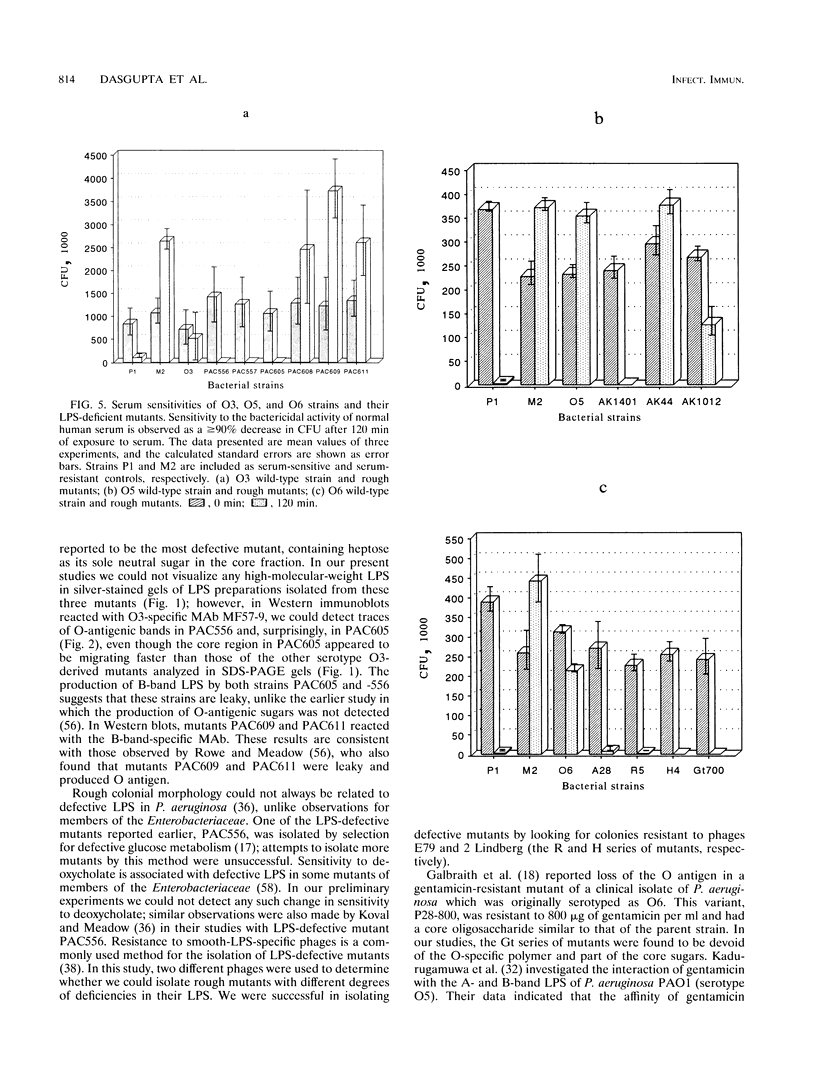
Well-characterized rough mutants are important for the understanding of structures, functions, and biosynthesis of lipopolysaccharide (LPS) in gram-negative organisms. In this study, three series of Pseudomonas aeruginosa LPS-deficient mutants, namely PAC strains derived from serotype O3, AK strains derived from strain PAO1 (serotype O5), and serotype O6-derived mutants were subjected to biochemical analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and silver staining as well as immunochemical characterization using LPS-specific monoclonal antibodies. The O-side-chain deficiency among the O6-derived mutants was also examined, and three mutants, A28, R5, and H4, were subsequently chosen for the elucidation of component sugars of the core structure of serotype O6 LPS. LPS of strain A28 has L-rhamnose and proportionally higher amounts of D-glucose, a feature shared by the O5-derived mutant, strain AK1401 (previously demonstrated as a mutant with a core-plus-one O repeat). In contrast strains R5 and H4 were shown to be devoid of L-rhamnose and have low and undetectable amounts of D-glucose, respectively, which indicated their core deficiency. The LPS-deficient or -sufficient characteristics of the P. aeruginosa strains examined correlated will with serum sensitivity data. This report represents a comprehensive analysis of rough mutants derived from O3 and O5 strains that have been used by others in many studies and a first look at the core oligosaccharide region of serotype O6 LPS obtained with the O6-derived mutants generated in this study.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ackermann H. W., Cartier C., Slopek S., Vieu J. F. Morphology of Pseudomonas aeruginosa typing phages of the Lindberg set. Ann Inst Pasteur Virol. 1988 Oct-Dec;139(4):389–404. doi: 10.1016/s0769-2617(88)80075-3. [DOI] [PubMed] [Google Scholar]
- Angus B. L., Fyfe J. A., Hancock R. E. Mapping and characterization of two mutations to antibiotic supersusceptibility in Pseudomonas aeruginosa. J Gen Microbiol. 1987 Oct;133(10):2905–2914. doi: 10.1099/00221287-133-10-2905. [DOI] [PubMed] [Google Scholar]
- Berry D., Kropinski A. M. Effect of lipopolysaccharide mutations and temperature on plasmid transformation efficiency in Pseudomonas aeruginosa. Can J Microbiol. 1986 May;32(5):436–438. doi: 10.1139/m86-082. [DOI] [PubMed] [Google Scholar]
- Bodey G. P., Bolivar R., Fainstein V., Jadeja L. Infections caused by Pseudomonas aeruginosa. Rev Infect Dis. 1983 Mar-Apr;5(2):279–313. doi: 10.1093/clinids/5.2.279. [DOI] [PubMed] [Google Scholar]
- Botzenhart K., Wolz C., Döring G. Cross-colonization and routes of infection assessed with a DNA probe. Antibiot Chemother (1971) 1991;44:8–12. doi: 10.1159/000420290. [DOI] [PubMed] [Google Scholar]
- Brade H., Galanos C. Isolation, purification, and chemical analysis of the lipopolysaccharide and lipid A of Acinetobacter calcoaceticus NCTC 10305. Eur J Biochem. 1982 Feb;122(2):233–237. doi: 10.1111/j.1432-1033.1982.tb05871.x. [DOI] [PubMed] [Google Scholar]
- Burnette W. N. "Western blotting": electrophoretic transfer of proteins from sodium dodecyl sulfate--polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981 Apr;112(2):195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Capobianco J. O., Darveau R. P., Goldman R. C., Lartey P. A., Pernet A. G. Inhibition of exogenous 3-deoxy-D-manno-octulosonate incorporation into lipid A precursor of toluene-treated Salmonella typhimurium cells. J Bacteriol. 1987 Sep;169(9):4030–4035. doi: 10.1128/jb.169.9.4030-4035.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester I. R., Meadow P. M., Pitt T. L. The relationship between the O-antigenic lipopolysaccharides and serological specificity in strains of Pseudomonas aeruginosa of different O-serotypes. J Gen Microbiol. 1973 Oct;78(2):305–318. doi: 10.1099/00221287-78-2-305. [DOI] [PubMed] [Google Scholar]
- Cryz S. J., Jr, Pitt T. L., Fürer E., Germanier R. Role of lipopolysaccharide in virulence of Pseudomonas aeruginosa. Infect Immun. 1984 May;44(2):508–513. doi: 10.1128/iai.44.2.508-513.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewry D. T., Symes K. C., Gray G. W., Wilkinson S. G. Studies of polysaccharide fractions from the lipopolysaccharide of Pseudomonas aeruginosa N.C.T.C. 1999. Biochem J. 1975 Jul;149(1):93–106. doi: 10.1042/bj1490093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubray G., Bezard G. A highly sensitive periodic acid-silver stain for 1,2-diol groups of glycoproteins and polysaccharides in polyacrylamide gels. Anal Biochem. 1982 Jan 15;119(2):325–329. doi: 10.1016/0003-2697(82)90593-0. [DOI] [PubMed] [Google Scholar]
- Fensom A. H., Meadow P. M. Evidence for two regions in the polysaccharide moiety of the lipopolysaccharide of Pseudomonas aeruginosa 8602. FEBS Lett. 1970 Jul 29;9(2):81–84. doi: 10.1016/0014-5793(70)80318-0. [DOI] [PubMed] [Google Scholar]
- Galbraith L., Wilkinson S. G., Legakis N. J., Genimata V., Katsorchis T. A., Rietschel E. T. Structural alterations in the envelope of a gentamicin-resistant rough mutant of Pseudomonas aeruginosa. Ann Microbiol (Paris) 1984 Sep-Oct;135B(2):121–136. doi: 10.1016/s0769-2609(84)80020-4. [DOI] [PubMed] [Google Scholar]
- Godfrey A. J., Shahrabadi M. S., Bryan L. E. Distribution of porin and lipopolysaccharide antigens on a Pseudomonas aeruginosa permeability mutant. Antimicrob Agents Chemother. 1986 Nov;30(5):802–805. doi: 10.1128/aac.30.5.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman N., Schmetz M. A., Foulds J., Klima E. N., Jimenez-Lucho V. E., Leive L. L., Joiner K. A., Jiminez V. Lipopolysaccharide size and distribution determine serum resistance in Salmonella montevideo. J Bacteriol. 1987 Feb;169(2):856–863. doi: 10.1128/jb.169.2.856-863.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. E., Carey A. M. Outer membrane of Pseudomonas aeruginosa: heat- 2-mercaptoethanol-modifiable proteins. J Bacteriol. 1979 Dec;140(3):902–910. doi: 10.1128/jb.140.3.902-910.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. E., Mutharia L. M., Chan L., Darveau R. P., Speert D. P., Pier G. B. Pseudomonas aeruginosa isolates from patients with cystic fibrosis: a class of serum-sensitive, nontypable strains deficient in lipopolysaccharide O side chains. Infect Immun. 1983 Oct;42(1):170–177. doi: 10.1128/iai.42.1.170-177.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock P. J., Brown T. M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983 Apr;154(1):269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell K. F., Kropinski A. M. Coliphage T7 receptors are present in Pseudomonas aeruginosa rough lipopolysaccharides. Biochem Biophys Res Commun. 1981 Apr 30;99(4):1185–1190. doi: 10.1016/0006-291x(81)90744-0. [DOI] [PubMed] [Google Scholar]
- Jarrell K. F., Kropinski A. M. Isolation and characterization of a bacteriophage specific for the lipopolysaccharide of rough derivatives of Pseudomonas aeruginosa strain PAO. J Virol. 1981 May;38(2):529–538. doi: 10.1128/jvi.38.2.529-538.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell K. F., Kropinski A. M. Pseudomonas aeruginosa bacteriophage phi PLS27-lipopolysaccharide interactions. J Virol. 1981 Nov;40(2):411–420. doi: 10.1128/jvi.40.2.411-420.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell K., Kropinski A. M. Identification of the cell wall receptor for bacteriophage E79 in Pseudomonas aeruginosa strain PAO. J Virol. 1977 Sep;23(3):461–466. doi: 10.1128/jvi.23.3.461-466.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell K., Kropinski A. M. The chemical composition of the lipopolysaccharide from Pseudomonas aeruginosa strain PAO and a spontaneously derived rough mutant. Microbios. 1977;19(76):103–116. [PubMed] [Google Scholar]
- Kadurugamuwa J. L., Lam J. S., Beveridge T. J. Interaction of gentamicin with the A band and B band lipopolysaccharides of Pseudomonas aeruginosa and its possible lethal effect. Antimicrob Agents Chemother. 1993 Apr;37(4):715–721. doi: 10.1128/aac.37.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkhanis Y. D., Zeltner J. Y., Jackson J. J., Carlo D. J. A new and improved microassay to determine 2-keto-3-deoxyoctonate in lipopolysaccharide of Gram-negative bacteria. Anal Biochem. 1978 Apr;85(2):595–601. doi: 10.1016/0003-2697(78)90260-9. [DOI] [PubMed] [Google Scholar]
- Knirel YuA, Vinogradov E. V., Kocharova N. A., Paramonov N. A., Kochetkov N. K., Dmitriev B. A., Stanislavsky E. S., Lányi B. The structure of O-specific polysaccharides and serological classification of Pseudomonas aeruginosa (a review). Acta Microbiol Hung. 1988;35(1):3–24. [PubMed] [Google Scholar]
- Knirel Y. A. Polysaccharide antigens of Pseudomonas aeruginosa. Crit Rev Microbiol. 1990;17(4):273–304. doi: 10.3109/10408419009105729. [DOI] [PubMed] [Google Scholar]
- Koval S. F., Meadow P. M. The isolation and characterization of lipopolysaccharide-defective mutants of Pseudomonas aeruginosa PAC1. J Gen Microbiol. 1977 Feb;98(2):387–398. doi: 10.1099/00221287-98-2-387. [DOI] [PubMed] [Google Scholar]
- Kropinski A. M., Chan L. C., Milazzo F. H. The extraction and analysis of lipopolysaccharides from Pseudomonas aeruginosa strain PAO, and three rough mutants. Can J Microbiol. 1979 Mar;25(3):390–398. doi: 10.1139/m79-060. [DOI] [PubMed] [Google Scholar]
- Kropinski A. M., Jewell B., Kuzio J., Milazzo F., Berry D. Structure and functions of Pseudomonas aeruginosa lipopolysaccharide. Antibiot Chemother (1971) 1985;36:58–73. doi: 10.1159/000410472. [DOI] [PubMed] [Google Scholar]
- Lam J. S., Graham L. L., Lightfoot J., Dasgupta T., Beveridge T. J. Ultrastructural examination of the lipopolysaccharides of Pseudomonas aeruginosa strains and their isogenic rough mutants by freeze-substitution. J Bacteriol. 1992 Nov;174(22):7159–7167. doi: 10.1128/jb.174.22.7159-7167.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam J. S., MacDonald L. A., Lam M. Y., Duchesne L. G., Southam G. G. Production and characterization of monoclonal antibodies against serotype strains of Pseudomonas aeruginosa. Infect Immun. 1987 May;55(5):1051–1057. doi: 10.1128/iai.55.5.1051-1057.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam M. Y., McGroarty E. J., Kropinski A. M., MacDonald L. A., Pedersen S. S., Høiby N., Lam J. S. Occurrence of a common lipopolysaccharide antigen in standard and clinical strains of Pseudomonas aeruginosa. J Clin Microbiol. 1989 May;27(5):962–967. doi: 10.1128/jcm.27.5.962-967.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesse A. J., Campagnari A. A., Bittner W. E., Apicella M. A. Increased resolution of lipopolysaccharides and lipooligosaccharides utilizing tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis. J Immunol Methods. 1990 Jan 24;126(1):109–117. doi: 10.1016/0022-1759(90)90018-q. [DOI] [PubMed] [Google Scholar]
- Lightfoot J., Lam J. S. Molecular cloning of genes involved with expression of A-band lipopolysaccharide, an antigenically conserved form, in Pseudomonas aeruginosa. J Bacteriol. 1991 Sep;173(18):5624–5630. doi: 10.1128/jb.173.18.5624-5630.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P. V., Wang S. Three new major somatic antigens of Pseudomonas aeruginosa. J Clin Microbiol. 1990 May;28(5):922–925. doi: 10.1128/jcm.28.5.922-925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J. W., Barclay G. R., Micklem L. R., Poxton I. R., Govan J. R. Production and characterisation of mouse monoclonal antibodies reactive with the lipopolysaccharide core of Pseudomonas aeruginosa. J Med Microbiol. 1992 May;36(5):358–365. doi: 10.1099/00222615-36-5-358. [DOI] [PubMed] [Google Scholar]
- Porat R., Mosseri R., Kaplan E., Johns M. A., Shibolet S. Distribution of polysaccharide side chains of lipopolysaccharide determine resistance of Escherichia coli to the bactericidal activity of serum. J Infect Dis. 1992 May;165(5):953–956. doi: 10.1093/infdis/165.5.953. [DOI] [PubMed] [Google Scholar]
- Rella M., Mercenier A., Haas D. Transposon insertion mutagenesis of Pseudomonas aeruginosa with a Tn5 derivative: application to physical mapping of the arc gene cluster. Gene. 1985;33(3):293–303. doi: 10.1016/0378-1119(85)90237-9. [DOI] [PubMed] [Google Scholar]
- Rivera M., McGroarty E. J. Analysis of a common-antigen lipopolysaccharide from Pseudomonas aeruginosa. J Bacteriol. 1989 Apr;171(4):2244–2248. doi: 10.1128/jb.171.4.2244-2248.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe P. S., Meadow P. M. Structure of the Core oligosaccharide from the lipopolysaccharide of Pseudomonas aeruginosa PAC1R and its defective mutants. Eur J Biochem. 1983 May 2;132(2):329–337. doi: 10.1111/j.1432-1033.1983.tb07366.x. [DOI] [PubMed] [Google Scholar]
- Sadoff J. C., Wright D. C., Futrovsky S., Sidberry H., Collins H., Kaufmann B. Characterization of mouse monoclonal antibodies directed against Pseudomonas aeruginosa lipopolysaccharides. Antibiot Chemother (1971) 1985;36:134–146. doi: 10.1159/000410478. [DOI] [PubMed] [Google Scholar]
- Sanderson K. E., Van Wyngaarden J., Lüderitz O., Stocker B. A. Rough mutants of Salmonella typhimurium with defects in the heptose region of the lipopolysaccharide core. Can J Microbiol. 1974 Aug;20(8):1127–1134. doi: 10.1139/m74-175. [DOI] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Shearer B. G., Legakis N. J. Pseudomonas aeruginosa: evidence for the involvement of lipopolysaccharide in determining outer membrane permeability to carbenicillin and gentamicin. J Infect Dis. 1985 Aug;152(2):351–355. doi: 10.1093/infdis/152.2.351. [DOI] [PubMed] [Google Scholar]
- Speert D. P., Campbell M. E. Hospital epidemiology of Pseudomonas aeruginosa from patients with cystic fibrosis. J Hosp Infect. 1987 Jan;9(1):11–21. doi: 10.1016/0195-6701(87)90089-2. [DOI] [PubMed] [Google Scholar]
- Speert D. P., Eftekhar F., Puterman M. L. Nonopsonic phagocytosis of strains of Pseudomonas aeruginosa from cystic fibrosis patients. Infect Immun. 1984 Mar;43(3):1006–1011. doi: 10.1128/iai.43.3.1006-1011.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanislavsky E. S., Makarenko T. A., Kozhenova T. E. Specific and non-specific mouse protection induced by different chemotypes of the Pseudomonas aeruginosa lipopolysaccharides. FEMS Microbiol Immunol. 1992 Oct;5(4):181–189. doi: 10.1111/j.1574-6968.1992.tb05900.x. [DOI] [PubMed] [Google Scholar]
- Stieritz D. D., Holder I. A. Experimental studies of the pathogenesis of infections due to Pseudomonas aeruginosa: description of a burned mouse model. J Infect Dis. 1975 Jun;131(6):688–691. doi: 10.1093/infdis/131.6.688. [DOI] [PubMed] [Google Scholar]
- Terashima M., Uezumi I., Tomio T., Kato M., Irie K., Okuda T., Yokota S., Noguchi H. A protective human monoclonal antibody directed to the outer core region of Pseudomonas aeruginosa lipopolysaccharide. Infect Immun. 1991 Jan;59(1):1–6. doi: 10.1128/iai.59.1.1-6.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson R. G., Gemski P., Jr, Stocker B. A. Non-smooth mutants of Salmonella typhimurium: differentiation by phage sensitivity and genetic mapping. J Gen Microbiol. 1972 May;70(3):527–554. doi: 10.1099/00221287-70-3-527. [DOI] [PubMed] [Google Scholar]
- Wilkinson S. G. Composition and structure of lipopolysaccharides from Pseudomonas aeruginosa. Rev Infect Dis. 1983 Nov-Dec;5 (Suppl 5):S941–S949. doi: 10.1093/clinids/5.supplement_5.s941. [DOI] [PubMed] [Google Scholar]
- Wilkinson S. G., Galbrath L. Studies of lipopolysaccharides from Pseudomonas aeruginosa. Eur J Biochem. 1975 Mar 17;52(2):331–343. doi: 10.1111/j.1432-1033.1975.tb04001.x. [DOI] [PubMed] [Google Scholar]
- Yokota S., Ochi H., Ohtsuka H., Kato M., Noguchi H. Heterogeneity of the L-rhamnose residue in the outer core of Pseudomonas aeruginosa lipopolysaccharide, characterized by using human monoclonal antibodies. Infect Immun. 1989 Jun;57(6):1691–1696. doi: 10.1128/iai.57.6.1691-1696.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]