Abstract
The kinase activity of Abl is known to be regulated by a putative trans-acting inhibitor molecule interacting with the Src homology (SH) 3 domain of Abl. Here we report that the kinase-deficient Src (SrcKD) directly inhibits the tyrosine phosphorylation of Cbl and other cellular proteins by Abl. We found that both the SH2 and SH3 domains of SrcKD are necessary for the suppressor activity toward the Abl kinase phosphorylating Cbl. To suppress the Cbl phosphorylation by Abl, the interaction between the SH3 domain of SrcKD and Cbl is required. This interaction between SrcKD and Cbl is regulated by a closed structure of Cbl. The binding of Abl to the extreme carboxyl-terminal region of Cbl unmasks the binding site of SrcKD to Cbl. This results in a ternary complex that inhibits the Abl-mediated phosphorylation of Cbl by steric hindrance. These results illustrate a mechanism by which the enzymatically inactive Src can exert a biological function in vivo.
The function of many protein tyrosine kinases is regulated by phosphorylation and dephosphorylation of the kinase molecule (1) and intramolecular binding of Src homology (SH) 2 and SH3 domains (2, 3).
On the other hand, the c-Abl (Abl) tyrosine kinase has been considered to be regulated by a kinase inhibitory molecule binding to the SH3 domain of Abl through a proline-rich motif PxxP of the inhibitor (4–6).
In transformed cells induced by the activated forms of the Abl kinase, c-Cbl (Cbl) is one of major substrates (7). Cbl is a 120-kDa adaptor protein that functions as a negative regulator of the tyrosine kinases in cells (8, 9). In osteoclasts, Cbl has been shown to bind to the c-Src (Src) tyrosine kinase and act downstream of Src (10).
The protooncogene c-src encodes a 60-kDa protein containing an SH3 domain, an SH2 domain, and a tyrosine kinase domain (11). c-src null mutant mice develop osteopetrosis as a result of osteoclast dysfunction (12). Schwartzberg et al. (13) have demonstrated that the expression of the kinase-deficient Src (SrcKD) in the c-src null mutant mice rescues the osteoclast function, indicating that a kinase-independent function for Src could have an important biological consequence in vivo.
In this study, we found that the phosphorylation of Cbl by the Abl kinase is inhibited by SrcKD. The results suggest that the complex formation between Cbl and SrcKD inhibits the function of Abl, illustrating another kinase-independent function of Src in vivo.
Materials and Methods
Plasmid Construction.
Wild-type (WT) Src and the mutants (SrcKD, SrcKD/SH3, and SrcKD/SH2) were cloned into pcDNA3 (Invitrogen). The SrcKD/SH3 mutant was produced by PCR with mutated primers by introducing in the SH3 domain of SrcKD the W118 to K inactivating mutation (14). The SrcKD/SH2 mutant was constructed from the Src SH2 mutant (W148R) (15). The SH3 domain of Src (between amino acids 86 and 138) was expressed as a glutathione S-transferase (GST)-fusion protein in the pEBG vector (gift from Bruce Mayer, Harvard, Boston).
WT murine type IV Abl and its mutants, AblKD (K290 M) and AblFNC (gifts from David Baltimore, Caltech, Pasadena, CA, and Bruce Mayer), were cloned into pcDNA3. The SH3-deficient mutant, AblSH3M, was produced by PCR with mutated primers by introducing in the SH3 domain of Abl the W118 to K-inactivating mutation. The mutant Abl461-End (between amino acids 461 and 1097), Abl461–756 (between amino acids 461 and 756), and Abl756-End (between amino acids 756 and 1097) were cloned in pFLAGCMV2 (Kodak).
Cbl hemagglutinin-tagged at the amino terminus was cloned into the BamHI site of pcDNA3. The Cbl857 stop mutant (16) was created by introducing the stop codon after amino acids 856 by PCR. WT Cbl and the fragments Cbl437–647 (between amino acids 437 and 647) and Cbl648–946 (between amino acids 648 and 946) produced by PCR were cloned into pEBG. A mutant of Cbl containing CblW202K and one containing the Cbl SH3 like sequence spanning between amino acids 160 and 207 were cloned in pEBG. pEBG CasSH3 was a gift from Kathrin Kirsch, The Rockefeller University. Every PCR product was confirmed by sequencing.
The following constructs expressing the SH3 domains as a GST fusion protein in bacteria were obtained: Src, Fyn, Yes (gifts from Marius Sudol, Mount Sinai Medical School, New York), Abl, Crk (gifts from Stephan Feller, Bavarian Julius-Maximilians University, Würzburg, Germany), Csk (gift from Hisataka Sabe, Osaka Bioscience Institute, Japan), Grb2 (gift from Albert Wong, Kimmel Cancer Institute, Philadelphia), and Nck (gift from Margaret Chou, University of Pennsylvania, Philadelphia).
Cells and Transfection.
293T cells were grown in DMEM supplemented with 10% FCS and antibiotics. Transient transfections of 293T cells with mammalian expression vectors (pcDNA3, pFLAGCMV2, and pEBG) bearing Abl, Cbl, and Src constructs were performed by a MBS calcium phosphate method (Stratagene) using 1 μg of each plasmid. In each transfection, total amounts of DNA were balanced by adding appropriate amounts of the pcDNA3 empty vector.
Antibodies.
Antibodies were obtained as follows: 8E9 anti-Abl (PharMingen), C15 anti-Cbl (Santa Cruz Biotechnology), 327 anti-Src (gift of Joan Brugge, Harvard, Boston), M2 anti-FLAG (Kodak), Y11 anti-hemagglutinin (Santa Cruz Biotechnology), B-14 anti-GST (Santa Cruz Biotechnology), and 4G10 anti-phosphotyrosine (Upstate Biotechnology).
Immunoprecipitation and Western Blotting.
The method of immunoprecipitation and immunoblotting has been described (17). Briefly, cells were lysed on the dish in lysis buffer (1% Triton X-100/10 mM Tris⋅HCl, pH 7.6/50 mM NaCl/30 mM sodium pyrophosphate/50 mM sodium fluoride/20 mM β-glycerophosphate/1 mM EDTA/1 mM EGTA/1 mM sodium vanadate/20 μg/ml aprotinin/1 mM PMSF) 24 h after transfection. Proteins were immunoprecipitated by incubating the lysates with specific antibodies for 2 h or overnight and then collected on protein G Sepharose beads (Amersham Pharmacia). For in vitro binding assay between the GST-SH3 domains and CblWT, 10 μg of the GST-SH3 fusion proteins was incubated overnight with 50 μg of 293T cell lysate containing CblWT. The immunoprecipitates were analyzed by 8–12% SDS/PAGE followed by a transfer onto Immobilon-P membrane filters (Millipore). The membranes were blocked and incubated with an antibody. After a secondary antibody, the complexes were visualized with enhanced chemiluminescence solutions (NEN).
Results
Cbl Tyrosine Phosphorylation by the Abl Kinase Is Blocked by the SrcKD.
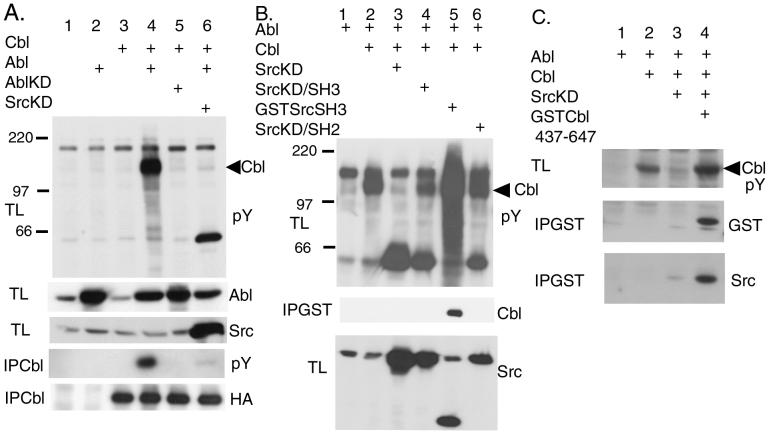
Because the tyrosine phosphorylation of Cbl was shown to be involved in the signaling of the activated Abl kinase (7), we investigated whether the nononcogenic form of the Abl kinase also is involved in the tyrosine phosphorylation of Cbl. By expressing Abl and Cbl in 293T cells, we found that Cbl and other cellular proteins were tyrosine-phosphorylated by Abl (Fig. 1A, lane 4). The coexpression of the kinase-deficient Abl with Cbl did not result in tyrosine phosphorylation of Cbl (Fig. 1A, lane 5), indicating the requirement of the kinase activity of Abl for the Cbl phosphorylation. To examine whether the Abl kinase directly phosphorylates Cbl, we coexpressed SrcKD with Cbl and Abl in 293T cells because SrcKD could serve as a dominant negative mutant for the endogenous Src kinase (Fig. 1A, lane 6). The coexpression of SrcKD blocked the tyrosine phosphorylation of Cbl as well as other cellular proteins (Fig. 1A, lane 6). This raised two possibilities. First, the Abl kinase activates the endogenous Src kinase, which in turn phosphorylates Cbl. Second, SrcKD acts more directly with the Abl kinase to suppress its activity. To distinguish the above possibilities, we performed the mutant analysis of SrcKD.
Figure 1.
Inhibition of Abl-induced Cbl phosphorylation by SrcKD. 293T cells were transiently transfected with the indicated expression plasmids. Proteins (50 μg) from total lysates (TL) of the transfected 293T cells were resolved by SDS/PAGE and Western-blotted with the antibodies indicated at the right. pY indicates an anti-phosphotyrosine antibody. Proteins also were immunoprecipitated from the lysates with anti-Cbl antibody (IPCbl) or with glutathione beads (IPGST). The arrow head indicates the position of Cbl. (A) SrcKD inhibits the tyrosine phosphorylation of Cbl by Abl. AblKD, kinase-deficient Abl. (B) Mutant analysis of SrcKD. Anti-Src antibody recognizes GSTSrcSH3 as well as Src. (C) The effect of the proline-rich region of Cbl.
The SH3 Domain of SrcKD Is Necessary but Not Sufficient for the Inhibition of the Abl Kinase.
Because the SH3 domain of Src has been shown to bind to Cbl (10), we examined the involvement of the SH3 domain of Src in this inhibitory activity. SrcKD/SH3 is a mutant in which both the SH3 domain and the kinase activity are deficient. We also used a small protein containing only the GST-fused SH3 domain of Src.
In this experiment, we coexpressed Abl, Cbl, and the Src mutants in 293T cells and examined the changes in the cellular tyrosine phosphorylation. We found that the SrcKD/SH3 mutant did not suppress the phosphorylation of Cbl (Fig. 1B, lane 4), indicating that a functional SH3 domain of Src is necessary for the inhibition. However, the Src SH3 domain alone did not inhibit the Cbl phosphorylation. Rather the SH3 domain further activated the cellular tyrosine phosphorylation although it bound to Cbl (Fig. 1B, lane 5). Because the SH3 domain of SrcKD is necessary for the inhibition, the GST-fused SH3 domain of Src should block the tyrosine phosphorylation of Cbl if SrcKD acts as a dominant negative for the endogenous Src. The result of these experiments, therefore, indicates that the full-length Src is required for the inhibitory activity of SrcKD and suggests that SrcKD is not acting as a dominant negative for the endogenous Src kinase. In addition, the SH2 domain was required for SrcKD to act as the suppressor (Fig. 1B, lane 6). One possibility is that the SH2 domain may bind to a phosphorylated tyrosine residue on Cbl. The other possibility is that because SrcKD is a major tyrosine-phosphorylated protein (Fig. 1B, lane 3), intramolecular interaction between the SH2 domain of SrcKD and phosphorylated tyrosine 527 (18) or tyrosine 416 of SrcKD could regulate the suppressor activity of SrcKD.
Overexpression of Cbl Proline-Rich Region Reduces the Inhibitory Effect of SrcKD.
The previous results suggested that the inhibitory activity of SrcKD is mediated by the direct interaction between the Src SH3 domain and Cbl. To determine whether the binding of the Src SH3 domain to the proline-rich region is essential for this inhibitory effect, we examined the effect of a peptide representing the proline-rich region of Cbl (Cbl437–647). This proline-rich fragment coprecipitated with Src (Fig. 1C, lane 4). Moreover its coexpression suppressed the effect of SrcKD and allowed the Cbl phosphorylation by Abl (Fig. 1C, lane 4), suggesting that the interaction between the proline-rich region of Cbl and the SH3 domain of SrcKD is essential for the inhibition.
The SH3 Domain of Abl Also Binds to Cbl.
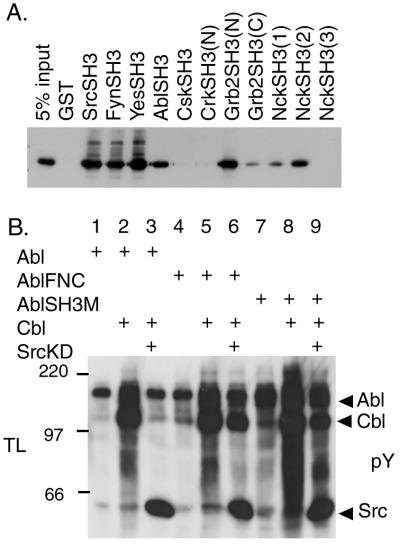
The proline-rich region of Cbl contains many possible binding sites for the SH3 domains of various proteins (19). To address the specificity of the binding between the SH3 domain of Src and the Cbl proline-rich region, we examined the in vitro binding affinity between various SH3 domains and Cbl. Many SH3 domains including those of Src, Fyn, Yes, Abl, Grb2, and Nck bound to Cbl (Fig. 2A). No binding was observed with the N-terminal SH3 domain of Crk, the SH3 domain of Csk, or the third SH3 domain of Nck (Fig. 2A). These results suggested that the interaction between Cbl and proteins containing SH3 domains is not restricted to the Src family kinases but includes the Abl kinase.
Figure 2.
Effect of Abl SH3 domain on Cbl phosphorylation. (A) The binding of various SH3 domains to Cbl. Various GST-fused SH3 domains, as indicated at the top, were prepared from bacteria and incubated with lysates from 293T cells expressing Cbl. GST-fusion proteins were immunoprecipitated with glutathione beads and Western-blotted with anti-Cbl antibody. (B) Role of the SH3 domain of Abl for the Cbl phosphorylation. Proteins (50 μg) from total lysates (TL) of 293T cells transfected with the indicated plasmids were resolved by SDS/PAGE and Western blotting with anti-phosphotyrosine (pY) antibody. Arrows indicate the position of Abl, Cbl, and Src.
The SH3 Domain of Abl Is Dispensable for Cbl Phosphorylation but Necessary for the Inhibitory Effect of SrcKD.
Because the SH3 domain of Abl can bind to Cbl (Fig. 2A) and the SH3 domain of Abl has been shown to interact functionally with a putative trans-acting inhibitor molecule (4, 5), we examined whether the SH3 domain of Abl is involved in the inhibition of Abl activity by SrcKD. We used the AblFNC mutant in which the SH3 domain of Abl is replaced with the SH3 domain of Crk as well as the Abl SH3-deficient mutant (AblSH3M). The expression of AblFNC or AblSH3M alone resulted in increased phosphorylation of cellular proteins (Fig. 2B, lanes 4 and 7), indicating that the kinase activity of AblFNC or AblSH3M is up-regulated compared with WT Abl. Coexpression of AblSH3M and Cbl in 293T cells induced Cbl phosphorylation (Fig. 2B, lane 8), showing that the Abl SH3 domain is not essential for the phosphorylation of Cbl by Abl. In addition, the phosphorylation of Cbl by AblFNC or AblSH3M was not inhibited by SrcKD (Fig. 2B, lanes 6 and 9). Because AblFNC or AblSH3M can not interact with Cbl by the SH3 domain of its mutants, these results suggest that the binding of the SH3 domain of Abl to Cbl is a step required for the inhibition by SrcKD.
The C-Terminal Region of Abl Interacts with the C-Terminal Region of Cbl.
The fact that the SH3 domain of Abl is not necessary for the phosphorylation of Cbl suggests that other regions of Abl might interact with Cbl, thus mediating the phosphorylation. We examined the C-terminal region of Abl that was reported to be essential for the in vivo function of Abl (20).
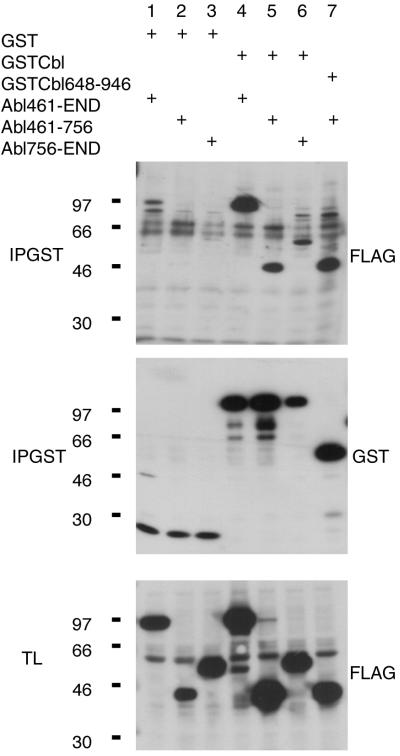
FLAG-tagged Abl C-terminal mutants, 461-End, 461–756, and 756-End, were expressed in 293T cells together with GST-tagged Cbl mutants (Fig. 3). The results showed that the C-terminal region of Abl amino acid 461-end (Fig. 3, lane 4) and amino acids 461–756 (Fig. 3, lane 5) had a high degree of affinity to Cbl. Further analysis demonstrated that the region amino acids 648–946 of Cbl was involved in the interaction with the region amino acids 461–756 of Abl (Fig. 3, lane 7). We also observed a limited binding of the mutant Abl amino acids 756-end to Cbl (Fig. 3, lane 6). Hence, we concluded that the binding between Abl and Cbl takes place between the C-terminal regions of both molecules.
Figure 3.
Abl and Cbl interact through their C-terminal regions. Proteins (50 μg) from total lysates (TL) of 293T cells transfected with the indicated plasmids were resolved by SDS/PAGE and Western-blotted with anti-FLAG tag (FLAG) antibody recognizing the FLAG-tagged Abl proteins. GST-Cbl fusion proteins were immunoprecipitated (IPGST) with glutathione beads and Western-blotted with the indicated antibodies.
The Binding of Abl to Monomeric Cbl Allows the Binding of Src to Cbl.
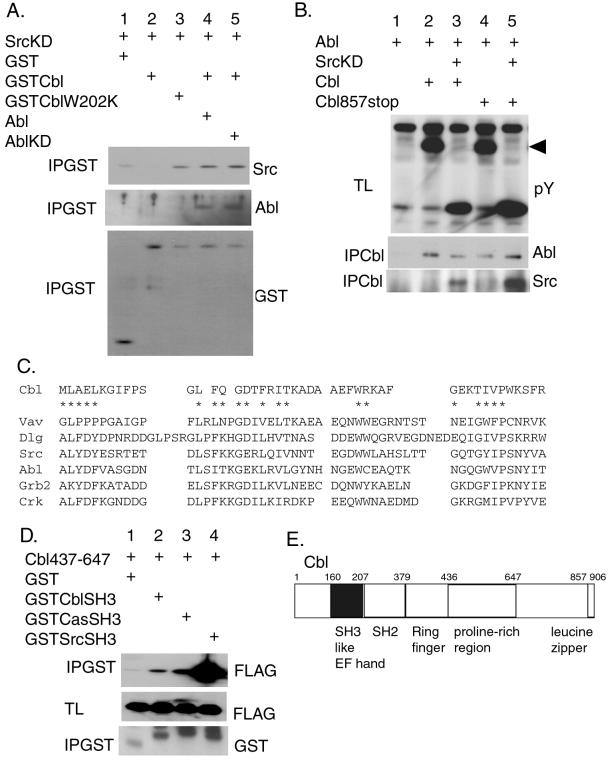
We observed that the Src SH3 domain binds to the Cbl proline-rich region and that Abl interacts with the Cbl C-terminal region. It is possible that these molecules form a ternary complex. To examine the possibility, 293T cells were cotransfected with vectors encoding Abl, SrcKD, and GST-tagged Cbl. GST-Cbl was precipitated, and the binding of Src and Abl was analyzed by Western blotting using anti-Src and anti-Abl antibodies (Fig. 4A). In the presence of Abl, both SrcKD and Abl were coprecipitated with Cbl (Fig. 4A, lane 4), indicating that a ternary complex was indeed formed. However, Cbl did not bind to SrcKD in the absence of Abl (Fig. 4A, lane 2) although the GST–SH3 domain of Src was able to interact with Cbl (Fig. 2A). The kinase activity of Abl was not necessary for the formation of ternary complex among Abl, SrcKD, and Cbl (Fig. 4A, lane 5). Because Src does not bind to Abl (not shown), its binding to Cbl in the presence of Abl could be explained by a modification of Cbl conformation induced by its binding to Abl. Because Cbl is reported to form a dimer through the leucine zipper region located at the very end of Cbl (16), we examined the suppression of tyrosine phosphorylation of Cbl by SrcKD as well as the binding of Abl to Cbl in the monomeric Cbl mutant (Cbl857stop) in which the leucine zipper region is deleted (16). The monomeric Cbl mutant was phosphorylated and coprecipitated with Abl to the same extent to WT Cbl (Fig. 4B, lanes 2 and 4), indicating that the leucine zipper region of Cbl was not necessary for the binding and the phosphorylation of Cbl by Abl. The SrcKD was coprecipitated with Cbl and inhibited the phosphorylation of the monomeric Cbl mutant by Abl (Fig. 4B, lane 5), demonstrating that the suppression of the Abl-mediated phosphorylation of Cbl by SrcKD occurs on a Cbl monomer.
Figure 4.
The binding between SrcKD and Cbl is regulated by Abl. (A) 293T cells were transiently transfected with the indicated expression plasmids. Proteins (500 μg) from cell lysates were precipitated with glutathione beads (IPGST) and Western-blotted with the antibodies indicated at the right. (B) The monomeric Cbl mutant (Cbl857stop) was coprecipitated with Abl and Src. 293T cells were transiently transfected with the indicated plasmids. Proteins from cell lysates were immunoprecipitated by using the anti-hemagglutinin antibody Y11 to precipitate Cbl (IPCbl) and Western-blotted with the anti-Abl and anti-Src antibodies. The total lysates (TL) also were Western-blotted with anti-phosphotyrosine antibody (pY). The arrow indicates the position of Cbl. (C) Alignment of Cbl SH3-like sequence with related SH3 domains from various proteins: Vav, Drosophila disclarge protein (Dlg), Src, Abl, Grb2, and Crk. The conserved residues of the SH3 domain are indicated by *. (D) The binding of the Cbl SH3-like sequence to the proline-rich region of Cbl. Proteins (50 μg) from total lysates (TL) of the transfected 293T cells were resolved by SDS/PAGE and Western-blotted with anti-FLAG tag (FLAG) antibody recognizing the Cbl437–647 FLAG-tagged fragment. GST-fusion proteins were precipitated with glutathione beads (IPGST) and Western-blotted with the indicated antibodies. (E) The schematic representation of Cbl indicating the position of the SH3-like sequence.
The Closed Structure of Cbl Can Be Maintained by the Interaction Between the N-Terminal Region and the C-Terminal Region of Cbl.
To investigate the mechanism of how a modification of Cbl conformation is induced by its binding to Abl, we examined the amino acid sequence of Cbl and found that Cbl contains an SH3-like sequence in its N terminus between amino acids 161 and 206 (Fig. 4C). The crystal structure of Cbl revealed that this region forms part of an alpha-helical EF hand domain but not an SH3 domain (21). However, by introducing the inactivating mutation in this putative SH3-like sequence of Cbl (W202K), we observed that the CblW202K mutant and SrcKD interact directly in the absence of Abl (Fig. 4A, lane 3). This finding suggested that this N-terminal region of Cbl might interact intramolecularly with its own proline-rich region at the C terminus and that the binding of Abl would abolish this interaction. As a consequence it would be possible for the SH3 domain of Src to interact with the unmasked proline-rich region of Cbl. We were able to show that the GST-tagged SH3-like sequence of Cbl coprecipitated with the proline-rich region of Cbl (Fig. 4D, lane 2). The affinity of the Cbl SH3-like sequence was much lower than that of the Src SH3 domain to the Cbl proline-rich sequence but was comparable to that of the p130Cas SH3 domain (Fig. 4D), supporting the above possibility.
Discussion
SrcKD Creates Steric Hindrance.
In this study, we demonstrated that SrcKD acts as a trans-acting inhibitor of the Abl kinase when we used Cbl as a substrate. We also found that Abl can open the binding site of Cbl to SrcKD (Fig. 4). Although the major tyrosine phosphorylation sites of Cbl by Abl are localized at tyrosine 700 and 774 (22), which are distinct from the Src binding region (Cbl437–647), the ternary complex of Abl, Cbl, and SrcKD could form a structure in which the access of the Abl kinase to the tyrosine residues of Cbl is blocked. Thus, one of the plausible models would be that SrcKD prevents the substrate presentation to the Abl kinase by creating steric hindrance.
The Mechanisms of Action of the Dominant Negative Proline-Rich Region of Cbl.
We showed that the proline-rich region of Cbl acts as a dominant negative molecule for the inhibition of the Abl kinase by SrcKD. This result is consistent with the idea that the binding of SrcKD is the major mechanism for its inhibitory function. The abundantly overexpressed proline-rich region may occupy SrcKD.
The effects of the proline-rich fragment of Cbl on the inhibitory activity of SrcKD raise one possible explanation for the activation of the c-Abl kinase by Cbl and its suppression by SrcKD. We found that the proline-rich region of Cbl also binds to the SH3 domain of Abl (Fig. 2A). This region then may activate the Abl kinase directly by blocking the action of endogenous putative trans-acting inhibitor molecules that interact with the SH3 domain of Abl. SrcKD may function as an inhibitor of the Abl kinase by binding to the same proline-rich motif of Cbl interacting with the SH3 domain of Abl, thereby preventing the removal of the endogenous inhibitors from the c-Abl kinase. However, because in our assay system we overexpressed Abl, Cbl, and SrcKD, it is unlikely that the endogenous Abl inhibitors account for all of the inhibitory activity. In addition, in accordance with the above theory, the Src SH3 domain alone was not effective in suppressing the Abl kinase.
The role of the Abl SH3 domain in the SrcKD inhibition of Cbl phosphorylation is still unclear at the moment. The result of the SH3 mutants of Abl raised the possibility that both the Src SH3 and Abl SH3 domains simultaneously bind to Cbl when SrcKD inhibits the Cbl phosphorylation by Abl. The identification of the proline-rich motif specific for each of the Src SH3 and Abl SH3 domains will be necessary for further analysis. Because the proline-rich region also interacts with Fyn, Yes, Grb2, and Nck, these molecules might regulate the Abl kinase by binding to Cbl. Indeed, FynKD is able to inhibit Cbl phosphorylation by Abl (unpublished result). Mayer and others also independently showed that Nck negatively regulates the Cbl phosphorylation by Abl (B. Mayer, personal communication).
The Closed Structure of Cbl.
We described that Cbl can form a closed structure that prevents the binding of Src to Cbl. The binding of Abl to Cbl is considered to trigger the structural changes in Cbl. In the case of other proteins that have a closed structure such as Src (2, 3) or Crk (23), the deletions of the C-terminal regions give rise to the formation of oncogenic proteins v-Src and v-Crk. Similarly, v-Cbl has a deletion of the C-terminal region of Cbl, including the proline-rich region of Cbl (19). Our results suggest that the oncogenic activity residing in the N-terminal region of Cbl can be released because of a disruption of a closed structure of Cbl. In addition, Cbl has another oncogenic form, 70Z Cbl, which contains a 17-aa deletion between the N-terminal and the C-terminal regions (19). We found that 70Z Cbl constitutively binds to Src although WT Cbl does not (unpublished result). This result also suggests that the constitutive conformational changes that open the structure of Cbl contribute to its oncogenic activation. Recent reports showed that the Cbl ring finger domain exhibits the ubiquitin ligase activity (24). The structural transition of Cbl induced by Abl might regulate the ligase activity because the ring finger domain is localized right next to the Src binding proline-rich region (Fig. 4E).
In summary, we showed that SrcKD directly inhibits Cbl phosphorylation by the Abl kinase. The analysis of mutants suggests that the complex of SrcKD and Cbl acts as an inhibitor of Abl. Although the physiological consequence of the interaction and the regulation of Abl, Cbl, and kinase-inactive form of WT Src is still unclear at the moment, it is possible that these interactions are important for the cytoskeleton reorganization (25–27). Plattner et al. (25) demonstrated that the kinase-activated Src activates Abl and the kinase-deficient Src reduces this activation in response to platelet-derived growth factor-regulating membrane ruffling. Kaplan et al. (27) showed that SrcKD regulates the cytoskeleton reorganization in osteoclasts, and Cbl has been found to act downstream of Src in these cells (10). The mechanism of Abl inhibition by SrcKD would not be limited to the inhibition of the WT Src kinase but would rather ensue from direct inhibition of the Abl kinase by SrcKD. Thus, this study suggests caution in interpreting data using a kinase-deficient molecule as a dominant negative to study the kinase activity.
Acknowledgments
We thank Makiko Maeda for technical assistance. We acknowledge D. Baltimore, B. Mayer, M. Sudol, S. Feller, K. Kirsh, H. Sabe, A. Wong, M. Chou, and J. Brugge for their gifts of reagents. We thank B. Mayer for communicating results before its publication. We also acknowledge P. Model and J. Kuriyan for helpful suggestions. M.-M.G. was supported by National Cancer Institute Training Grant CA09673–23. This work was supported by National Institutes of Health Grant CA44356 and a grant in aid from the Ministry of Education in Japan.
Abbreviations
- SH
Src homology
- SrcKD
kinase-deficient Src
- WT
wild type
- GST
glutathione S-transferase
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.060030697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.060030697
References
- 1.Cooper J A, Gould K L, Cartwright C A, Hunter T. Science. 1986;231:1431–1434. doi: 10.1126/science.2420005. [DOI] [PubMed] [Google Scholar]
- 2.Sicheri F, Moarefi I, Kuriyan J. Nature (London) 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 3.Xu W, Harrison S C, Eck M J. Nature (London) 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 4.Pendergast A M, Muller A J, Havlik M H, Clark R, McCormick F, Witte O N. Proc Natl Acad Sci USA. 1991;88:5927–5931. doi: 10.1073/pnas.88.13.5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayer B J, Baltimore D. Mol Cell Biol. 1994;14:2883–2894. doi: 10.1128/mcb.14.5.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wen S T, Van Etten R A. Genes Dev. 1997;11:2456–2467. doi: 10.1101/gad.11.19.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andoniou C E, Thien C B, Langdon W Y. EMBO J. 1994;13:4515–4523. doi: 10.1002/j.1460-2075.1994.tb06773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoon C H, Lee J, Jongeward G D, Sternberg P W. Science. 1995;269:1102–1105. doi: 10.1126/science.7652556. [DOI] [PubMed] [Google Scholar]
- 9.Murphy M A, Schnall R G, Venter D J, Barnett L, Bertoncello I, Thien C B F, Langdon W Y, Bowtell D D L. Mol Cell Biol. 1998;18:4872–4882. doi: 10.1128/mcb.18.8.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka S, Amling M, Neff L, Peyman A, Uhlmann E, Levy J B, Baron R. Nature (London) 1996;383:528–531. doi: 10.1038/383528a0. [DOI] [PubMed] [Google Scholar]
- 11.Takeya T, Hanafusa H. Cell. 1983;32:881–890. doi: 10.1016/0092-8674(83)90073-9. [DOI] [PubMed] [Google Scholar]
- 12.Soriano P, Montgomery C, Geske R, Bradley A. Cell. 1991;64:693–702. doi: 10.1016/0092-8674(91)90499-o. [DOI] [PubMed] [Google Scholar]
- 13.Schwartzberg P L, Xing L, Hoffmann O, Lowell C A, Garrett L, Boyce B F, Varmus H E. Genes Dev. 1997;11:2835–2844. doi: 10.1101/gad.11.21.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka M, Gupta R, Mayer B J. Mol Cell Biol. 1995;15:6829–6837. doi: 10.1128/mcb.15.12.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Brien M C, Fukui Y, Hanafusa H. Mol Cell Biol. 1990;10:2855–2862. doi: 10.1128/mcb.10.6.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartkiewicz M, Houghton A, Baron R. J Biol Chem. 1999;274:30887–30895. doi: 10.1074/jbc.274.43.30887. [DOI] [PubMed] [Google Scholar]
- 17.Georgescu M M, Kirsch K H, Shishido T, Zong C, Hanafusa H. Mol Cell Biol. 1999;19:1171–1181. doi: 10.1128/mcb.19.2.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jove R, Kornbluth S, Hanafusa H. Cell. 1987;50:937–943. doi: 10.1016/0092-8674(87)90520-4. [DOI] [PubMed] [Google Scholar]
- 19.Blake T J, Shapiro M, Morse H C, Langdon W Y. Oncogene. 1991;6:653–657. [PubMed] [Google Scholar]
- 20.Schwartzberg P L, Stall A M, Hardin J D, Bowdish K S, Humaran T, Boast S, Harbison M L, Robertson E J, Goff S P. Cell. 1991;65:1165–1175. doi: 10.1016/0092-8674(91)90012-n. [DOI] [PubMed] [Google Scholar]
- 21.Meng W, Sawasdikosol S, Burakoff S J, Eck M J. Nature (London) 1999;398:84–90. doi: 10.1038/18050. [DOI] [PubMed] [Google Scholar]
- 22.Andoniou C E, Thien C B, Langdon W Y. Oncogene. 1996;12:1981–1989. [PubMed] [Google Scholar]
- 23.Rosen M K, Yamazaki T, Gish G D, Kay C M, Pawson T, Kay L E. Nature (London) 1995;374:477–479. doi: 10.1038/374477a0. [DOI] [PubMed] [Google Scholar]
- 24.Joazeiro C A, Wing S S, Huang H, Leverson J D, Hunter T, Liu Y C. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 25.Plattner R, Kadlec L, DeMali K A, Kazlauskas A, Pendergast A M. Genes Dev. 1999;13:2400–2411. doi: 10.1101/gad.13.18.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonita D P, Miyake S, Lupher M L, Jr, Langdon W Y, Band H. Mol Cell Biol. 1997;17:4597–4610. doi: 10.1128/mcb.17.8.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaplan K B, Swedlow J R, Morgan D O, Varmus H E. Genes Dev. 1995;9:1505–1517. doi: 10.1101/gad.9.12.1505. [DOI] [PubMed] [Google Scholar]