Abstract
Coordinating the behaviors of different cell populations is essential for multicellular development. One important example for this can be found in ovule development in higher plants. Ovules give rise to the gametophyte in the distal nucellus and form protective sporophytic organs from the underlying chalaza. We show that the WUSCHEL (WUS) homeobox gene provides a mechanism to coordinate these events. WUS is expressed in the nucellus and our loss- and gain-of-function analyses show that WUS is not only necessary but also sufficient for integument formation from the chalaza. WUS protein is retained in the nucellus, indicating that WUS activity in the nucellus generates a downstream signal that non-cell-autonomously regulates integument initiation in the chalaza. This signal appears to act locally, thus determining the position of organ formation from chalazal cells adjacent to the nucellus. Analysis of WUS and AINTEGUMENTA functions indicates that integument initiation requires inputs from different ovule regions. Together with previous findings for shoot and floral meristems, where WUS signaling establishes a stem cell niche, our results indicate that WUS defines a signaling mechanism that is used repeatedly during plant development in coordinating the behavior of adjacent cell groups.
Keywords: WUSCHEL, Arabidopsis, signaling, ovule development
In higher plants, egg cell formation takes place in specialized somatic structures, the ovules. During their development, two neighboring cell groups, the nucellus and the chalaza, coordinately produce structures that eventually form an intimate physical and functional unit (Fig. 1): The nucellus, which is located at the distal end of the initially finger-like protruding ovule primordium, harbors the megaspore mother cell (mmc) from which the female gametophyte will form (Schneitz et al. 1995). After meiosis of the mmc one of the four haploid daughter cells survives and in turn undergoes three rounds of mitotic divisions. The resulting eight daughters constitute the mature female gametophyte (embryo sac), with one of them becoming the egg cell (Webb and Gunning 1990). The central chalaza initiates the integuments, protective sporophytic organs that grow around the nucellus and that after fertilization form the seed coat that protects the growing embryo (Esau 1977). The third, proximal region of the ovule, called funiculus, serves as a connection to the mother tissue, which can provide nutrients to the gametophyte.
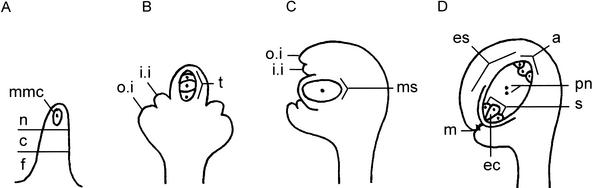
Figure 1.
Schematic diagram of wild-type ovule development. (A) Along the proximal–distal axis of ovule primordia three domains can be distinguished: the distal nucellus that harbors the megaspore mother cell (mmc), the central chalaza, and the proximal funiculus. (B) While the mmc divides meiotically to give rise to a tetrad of haploid cells, integuments are initiated from the chalaza that subsequently grow to enclose the nucellus. (C) The three distal cells of the tetrad die and the functional megaspore undergoes three rounds of mitotic division. (D) The mature embryo sac is composed of three antipodal cells at the chalazal end, two synergids at the micropylar end, one egg cell, and two polar nuclei, which eventually fuse to give the diploid central cell. (a) Antipodal cell; (c) chalaza; (ec) egg cell; (es) embryo sac; (f) funiculus; (i.i) inner integument; (m) micropyle; (mmc) megaspore mother cell; (ms) megaspore; (n) nucellus; (o.i) outer integument; (pn) polar nuclei; (s) synergid; (t) tetrad.
Not only does the survival of the prospective embryo require the protection offered by the seed coat, but already the early development of the female gametophyte appears to depend on proper integument formation: In several mutants (see below), defective integument formation coincides with a subsequent failure to progress through embryo sac development (e.g., discussed in Klucher et al. 1996), suggesting a tight linkage of these processes to ensure coordinated development and, ultimately, reproductive success. How this coordination is achieved is presently unknown.
Mutational analysis has revealed several genes that influence various aspects of ovule development (Chevalier et al. 2002). In nozzle mutants the most distal cells express chalazal markers and the nucellus is disorganized or absent, indicating that NOZZLE plays a central role in the formation of the nucellus (Schiefthaler et al. 1999; Balasubramanian and Schneitz 2000). Mutations in the AINTEGUMENTA (ANT) gene result in a failure of the ovule to initiate integument formation (Elliott et al. 1996; Klucher et al. 1996). The BELL1 (BEL1), INNER NO OUTER (INO), and HUELLENLOS (HLL) genes are involved in various aspects of integument outgrowth (Robinson-Beers et al. 1992; Modrusan et al. 1994; Gaiser et al. 1995; Reiser et al. 1995; Baker et al. 1997; Schneitz et al. 1997, 1998; Villanueva et al. 1999). ANT and HLL additionally play a role in the specification of funicular cells (Schneitz et al. 1998).
Here we present evidence that the homeobox gene WUSCHEL (WUS) regulates one important step in ovule development. WUS has been identified because of its central role in stem cell regulation in shoot and floral meristems. wus shoot meristems terminate prematurely after a few leaves are formed, and wus floral meristems terminate without forming a gynoecium (Laux et al. 1996). The WUS expression domain defines an organizing center in the shoot apex that specifies its overlying neighbors as pluripotent stem cells (Mayer et al. 1998). In shoot apices, WUS is sufficient to induce the expression of the CLV3 gene, which encodes a putative ligand of the CLV signaling pathway by which the stem cells signal back and restrict the WUS expression domain (Brand et al. 2000; Schoof et al. 2000). This feedback loop between the WUS and CLV3 genes appears to regulate size homeostasis of the stem cell population. In determinate flower primordia, initially the same self regulatory circuitry is established (Schoof et al. 2000). However, at the end of flower development, WUS additionally appears to contribute to the expression of its own repressor, the AGAMOUS (AG) gene, which in turn is required to terminate WUS expression and therefore floral meristem activity (Lenhard et al. 2001; Lohmann et al. 2001).
In addition to its role in shoot and floral meristems, we have found that WUS is also expressed in the nucellus of ovule primordia. We show that local WUS dependent signaling is required and sufficient to induce integument formation from the underlying chalazal domain. These results provide evidence for a novel pathway for interregional communication during ovule development and for determining the position of organ formation and suggest that similar short-range signaling modules are employed repeatedly during plant development to tune the behavior of neighboring cell groups.
Results
WUS is expressed in the nucellus of ovules
We first analyzed WUS mRNA expression in ovules by in situ hybridization. WUS mRNA was detected specifically in the distal part of ovules, the nucellus, from early stages on (Fig. 2A–D). No expression was observed in the chalaza or the funiculus. The intensity of the WUS expression signal was highest in stages 2-II to 2-III (stages after Schneitz et al. 1995) when integument primordia arise (Fig. 2B). In subsequent stages the WUS expression signal was less intense (Fig. 2C,D) and could not be detected after stage 3, which is when the embryo sac developed. In addition to the nucellar expression, a weak hybridization signal was observed in the epithelium of mature ovules (data not shown). Because we frequently observed a high background signal in the epithelium with various probes, this signal probably does not reflect specific WUS expression. In addition to its expression in ovules, WUS mRNA was also detected in developing anthers (data not shown).
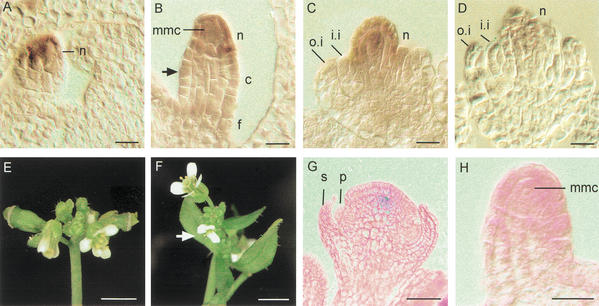
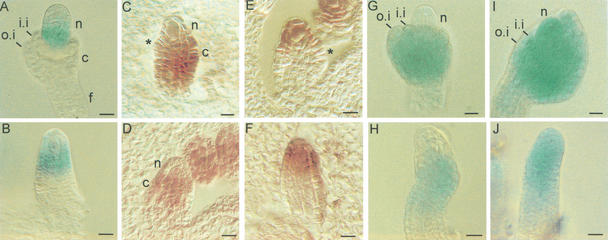
Figure 2.
WUS mRNA expression in ovules and characterization of CLV1::WUS plants. (A–D) In situ hybridization to tissue sections of various stages of wild type ovules. Signal is detected as brown color. (A) WUS mRNA is detected in young ovules in the nucellus. (B) At stage 2-II, when the inner integuments are initiated (arrow) a strong signal is detected in the nucellus of the ovule while no mRNA expression is observed in the chalaza and the funiculus. (C,D) WUS mRNA expression becomes weaker during the outgrowth of integuments at stages 2-IV (C) to 2-V (D). (E,F) Phenotype of CLV1::WUS (E) and CLV1::WUS; wus (F) inflorescences. In F, the arrow points at a flower resembling those found in wus mutants, whereas the top flower contains a gynoecium. (G,H) Expression of the linked CLV1::GUS reporter gene in a floral meristem (G) and a young ovule primordium of CLV1::WUS; wus plants. GUS activity is detected as blue colour in a central cell population of the floral meristem (G). No signal is obtained in the ovule primordium (H). (c) Chalaza; (f) funiculus; (i.i.) inner integument; (mmc) megaspore mother cell; (n) nucellus; (o.i.) outer integument; (p) petal primordium; (s) sepal primordium. Bars, 10 μm (A–D,H), 3 mm (E,F), and 30 μm (G).
In shoot meristems, WUS can induce expression of the CLV3 gene (Schoof et al. 2000). We therefore asked whether CLV3 is also expressed in ovules. We could not detect expression at the time when WUS is expressed either by a CLV3::GUS reporter gene or by in situ hybridization experiments using a CLV3 antisense probe (data not shown).
Generation of ovules lacking WUS activity
An analysis of a possible role of WUS in ovule development is hampered by the fact that wus floral meristems terminate before a gynoecium is formed. Therefore, we aimed to rescue the wus meristem defect and obtain gynoecia with wus mutant ovules. For this purpose we expressed WUS under the control of the CLV1 promoter in a wus mutant background. The CLV1 promoter provides expression in the meristem but is not expressed in ovules (see below).
CLV1::WUS control plants in a wild-type background displayed an enlarged meristem, a fasciated stem and flowers that formed gynoecia with supernumerary carpels (Fig. 2E) (Schoof et al. 2000). This phenotype resembles that of clv mutants and is caused by the relatively large expression domain of the CLV1::WUS transgene in comparison to the size of the endogenous WUS expression domain (Schoof et al. 2000).
We crossed the wus mutation into the CLV1::WUS plants and identified F2 plants homozygous for the wus mutation by PCR. The wus mutation did not affect the vegetative phenotype of the CLV1::WUS plants, indicating that the wus mutant defects in the shoot meristem were completely suppressed by the transgene. Each CLV1::WUS; wus plant generated two types of flowers (Fig. 2F): Approximately 40% of the flowers resembled the flowers of non-transgenic wus mutants in that they formed no more than four stamens and no gynoecium (Laux et al. 1996). In contrast, ∼60% (n = 117) of the flowers formed gynoecia with supernumerary carpels indistinguishable from the flowers of the CLV1::WUS control in a wild-type background (Table 1), indicating that the effects of the wus mutation in the respective floral meristems were suppressed by the transgene.
Table 1.
Floral organ numbers of CLV1::WUS; wus, CLV1::WUS, and wild-type plants
| Genotype
|
Organ numbers
|
|||
|---|---|---|---|---|
| Sepals
|
Petals
|
Anthers
|
Carpels
|
|
| CLV1::WUS; wus | 3.9 (0.4) | 3.5 (0.6) | 5.6 (0.8) | 4.1 (0.9) |
| CLV1::WUS | 4.1 (0.5) | 3.8 (0.5) | 5.3 (0.5) | 4.1 (0.8) |
| Wild type | 4.0 (0.0) | 4.0 (0.0) | 5.9 (0.3) | 2.0 (0.0) |
For each case the organ numbers of the three youngest flowers of seven individual plants were counted. These flowers always had developed a gynoecium in CLV1::WUS; wus plants. The average organ numbers and the standard deviation (parenthesis) are given.
WUS is required for integument initiation
Gynoecia of both CLV1::WUS control and CLV1::WUS; wus plants formed ovules. We confirmed that the CLV1::WUS transgene was not expressed in ovules using a linked CLV1::GUS reporter gene (see Materials and Methods): GUS staining was detected in the expected CLV1 expression domain in shoot and floral meristems (Fig. 2G) but not in ovules of CLV1::WUS controls or CLV1::WUS; wus plants (Fig. 2H). This confirmed that we had generated wus mutant ovules. For simplicity we will refer to ovules from CLV1::WUS; wus plants as wus ovules.
Ovule development in CLV1::WUS control plants (n = 29) was indistinguishable from wild type, indicating that the presence of the transgenes had no effect on ovule development (Fig. 3). In CLV1::WUS; wus plants (n = 22), ovule development was indistinguishable from control plants only until stage 2-I (Fig. 3A,B). At stage 2-I, control plants initiated integuments: First, the epidermal cells enlarged at the prospective position of the integuments (Fig. 3C); subsequently the inner and the outer integument primordia arose (Fig. 3E) and grew around the nucellus (Fig. 3G,I). In contrast, wus ovules did not show epidermal cell enlargement or any signs of integument initiation (Fig. 3D,F). The ovules rather developed into finger-like protrusions (Fig. 3 H,J) of a length comparable to the size of ovules in control plants (Fig. 3I,J). Eventually wus ovules appeared to degenerate (data not shown).
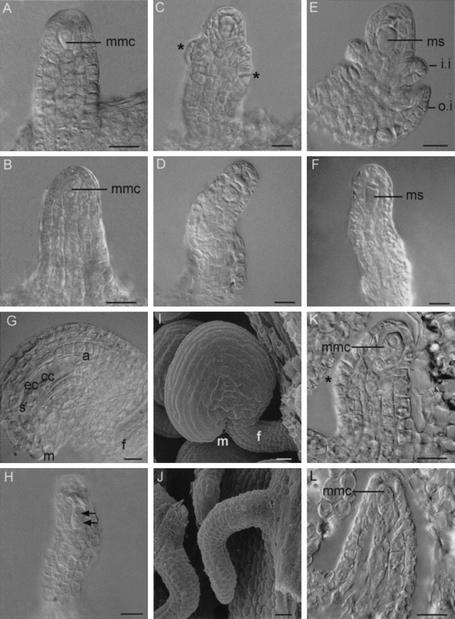
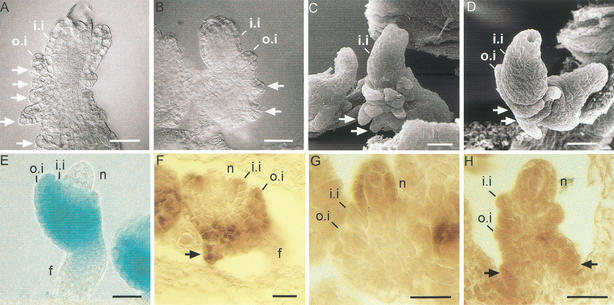
Figure 3.
Ovule development in CLV1::WUS and CLV1::WUS; wus plants. (A–H) DIC microscopy images of ovule development in CLV1::WUS plants (A,C,E,G) and CLV1::WUS; wus (B,D,F,H) plants. (A,B) Stage 1-II, ovule primordia have grown out. The megaspore mother cell is apparent in the subepidermal layer. (C,D) In CLV1::WUS ovules the inner integument initiates (asterisks in C). At the comparable stage of wus ovules no integument formation is evident (D). (E,F) In both CLV1::WUS (E) and wus ovules (F) the functional megaspore is evident.(G,H) At stage 3-V in CLV1::WUS ovules the two nuclear central cell, the egg cell, nuclei of the synergids, and the antipodal cells are visible (G). Corresponding ovules of CLV1::WUS; wus plants arrest at the two nuclear embryo sac stage (H). Arrows point at the embryo sac nuclei. (I,J) SEM image of mature CLV1::WUS (I) and wus (J) ovules: In CLV1::WUS the integuments enclose the nucellus and the ovule is bent. The corresponding wus ovule is naked because of the lack of integuments. (K,L) Histological sections of CLV1::WUS wild-type (K) and wus (L) ovules at stage 2-II. The asterisk indicates the site of inner integument formation as observed by epidermal cell enlargement (K). No such cell enlargement is evident in the corresponding wus ovule (L). (a) Antipodal cells; (cc) central cell; (ec) egg cell; (es) embryo sac; (f) funiculus; (i.i.) inner integument; (m) micropyle; (mmc) megaspore mother cell; (ms) functional megaspore; (o.i.) outer integument; (s) synergids. Bars, 10 μm.
Because the wus mutation results in partial differentiation of cells of the shoot apex, we analyzed whether cell differentiation in ovules was altered by the wus mutation. In histological paraffin sections, we did not observe differences in either cell size or vacuolization between ovules of control plants and wus ovules (Fig. 3K,L).
In summary, these results show that in ovules the WUS gene is required for integument initiation.
Megagametogenesis is incomplete in wus ovules
A hallmark of ovule development is meiosis and the development of the female gametophyte in the nucellus. Indistinguishable from wild type, megagametogenesis in CLV1::WUS control plants started with the enlargement of the megaspore mother cell and meiosis at stage 2-IV (Fig. 4A) that results in most cases in a linear tetrad of meiotic daughter cells (Fig. 4C). At stage 3-I, three tetrad cells degenerate, whereas the cell closest to the chalaza, the functional megaspore, undergoes three rounds of mitotic divisions to give the embryo sac with the egg cell and two accompanying synergids at the micropylar end, one diploid central cell and three antipodal cells at the chalazal end (Fig. 3G).
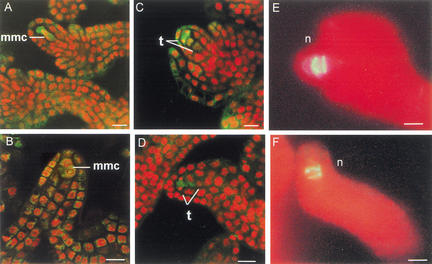
Figure 4.
Meiosis in CLV1::WUS and wus ovules. (A–D) Confocal microscopy of CLV1::WUS and wus ovules. Nuclei are detected in red, cytoplasm appears green. (A,B) In CLV1::WUS (A) and wus (B) ovules two nuclei are visible in the megaspore mother cell. (C,D) Tetrad formation is evident in CLV1::WUS (C) and wus (D) ovules. (E,F) Dark-field microscopy of CLV1::WUS and wus ovules. Ovules were stained with aniline blue to detect callose accumulation, which is indicative of meiotically dividing cells. Autofluorescence of the tissue is detected in red. Callose accumulation is shown as bright color. In both CLV1::WUS (E) and wus (F) ovules, two bands of callose accumulation are detected. (mmc) Megaspore mother cell; (n) nucellus; (t) tetrad. Bars, 10 μm.
In wus ovules the enlargement of the megaspore mother cell, division of the nuclei (Fig. 4B) and tetrad formation (Fig. 4D) were indistinguishable from control plants. We confirmed that the division of the megaspore mother cell in wus ovules was meiotic by staining with aniline blue. Aniline blue stains callose, which specifically marks the cell plate of the meiotically dividing megaspore mother cell (Rodkiewitz 1970). We found 72% (n = 59) of wus ovules staining for callose (Fig. 4F), which was comparable to the 79% (n = 74) of stained ovules in control plants (Fig. 4E), indicating that meiosis takes place in wus ovules. Subsequently, wus mutant ovules frequently formed a two-nuclear embryo sac (Fig. 3H), but mature embryo sacs were never observed. Instead, the ovules started to degenerate (data not shown).
Therefore, wus ovules were unable to complete embryo sac development. Because a similar arrest in embryo sac development has been described for other mutants defective in integument formation (Klucher et al. 1996), this defect could be a secondary effect caused by the lack of integuments in wus ovules.
Integument initiation requires independent inputs from the nucellus and the chalaza
We next asked whether WUS function could be integrated into known genetically defined pathways of integument formation. ant ovules fail to initiate integuments very similar to wus ovules (Klucher et al. 1996). To address whether ANT expression requires WUS activity, we performed in situ hybridization with an ANT probe. In control ovules, we found ANT expression predominantly in the chalaza at the time of integument initiation (Fig. 5C). The ANT expression pattern in wus ovules was indistinguishable from that in control plants (Fig. 5D). To address whether vice versa, WUS expression requires ANT activity, we analyzed WUS expression in ant ovules, using a WUS::GUS reporter gene that reflects the wild-type WUS mRNA expression pattern in ovules, but, in contrast to the mRNA expression, often showed a more intense staining in the basal part of the nucellus (cf. Fig. 5A to 2C). We found that the WUS:: GUS reporter was expressed in the nucellus of ant ovules indistinguishably from the expression in control plants (Fig. 5B).
Figure 5.
Gene expression in wus and ant ovules. (A,B) Expression of a WUS::GUS reporter gene. (A) In wild type, GUS activity is detected throughout the nucellus, but, in contrast to the WUS mRNA expression, is more pronounced in the basal portion of the nucellus. (B) In ant mutants, GUS activity was found in the same region as in wild type. (C,D) ANT mRNA expression in wild-type (C) and wus (D) ovules. At the time point of integument initiation (asterisk in C) ANT mRNA is found in the chalaza of wild-type ovules and the pattern obtained for wus ovules is indistinguishable from the wild-type expression. (E,F) WUS mRNA expression in wild type (E) and wus mutants (F). At the time point of integument initiation (asterisk in E) WUS mRNA is found exclusively in the nucellus of wild-type ovules and the same pattern is obtained in wus mutants. (G–J) Expression of an AG::GUS reportergene in wild-type and wus background. (G,H) Before the embryo sac develops, GUS activity is detected in the chalaza of wild-type ovules (G). Thereafter GUS activity is additionally evident in the nucellus (I). The GUS activity pattern obtained with wus ovules (H,J) is indistinguishable from that in wild type. (c) Chalaza; (f) funiculus; (i.i.) inner integument; (n) nucellus; (o.i.) outer integument. Bars, 10 μm.
Therefore, WUS and ANT do not regulate the promoter activity of the other gene, suggesting that both genes represent independent pathways and that inputs from both the chalaza and the nucellus are required for integument initiation.
The normal expression pattern of ANT in wus ovules suggested that the chalaza was properly established and that the defects observed were not caused by mis-specification of chalazal identity. To further confirm that wus ovules were correctly partitioned into domains, we used WUS and AG expression as additional regional markers. As described above, WUS is expressed exclusively in the nucellus of wild-type ovules (Fig. 5E) until stage 3-I. This expression pattern was not altered in wus ovules (Fig. 5F). The expression of an AG::GUS reporter (Sieburth and Meyerowitz 1997) specifically marks the chalaza of wild-type ovules until stage 3-I (Fig. 5G). Thereafter the expression extends into the nucellus (Fig. 5I). In wus ovules, the reporter was expressed in the same pattern as in wild type (Fig. 5H,J). Therefore, using these three regional markers, we could not detect any defects in the specification of nucellar and chalazal identities in the absence of WUS activity, suggesting that the effects of the wus mutation on integument initiation were not due to gross misspecification of ovule region identities.
WUS is sufficient to induce integument initiation
We next asked whether WUS is not only required for integument initiation, but is also sufficient to induce integuments. For this purpose, we expressed WUS ectopically from an ANT cis regulatory region in a wild-type background. This promoter provided expression in the chalaza and the integument primordia (Fig. 6E), indistinguishable from the ANT mRNA expression pattern. Because ANT::WUS plants show severe meristem defects and do not develop beyond the seedling stage (Schoof et al. 2000), we used an inducible system based on a translational fusion between WUS and the ligand-binding domain of the glucocorticoid receptor (GR). In the absence of a ligand for GR, the fusion protein is retained in the cytoplasm by virtue of its interaction with the Hsp90 complex (Dalman et al. 1991) but can move into the nucleus after application of the GR-ligand dexamethasone. Without induction, ANT::WUS-GR plants identified by PCR developed indistinguishably from wild type. In contrast, 6 d after induction we detected ectopic outgrowths that resembled integuments in the basal region of the ovule (Fig. 6A–D). These structures expressed the ANT gene, which in wild type is expressed in integument primordia (Fig. 6F). It should be noted that the outgrowth of both the normal integuments and ectopic organs appeared disturbed, which is in line with our previous observation from studies in the shoot meristem showing that WUS expression inhibits organ differentiation (Schoof et al. 2000).
Figure 6.
Ectopic expression of WUS. (A–D) Ovules of dexamethasone induced ANT::WUS-GR plants. DIC microscopy images (A,B) and SEM images (C,D) showing the nucellus enclosed by the inner integument. The outer integument does not grow beyond initial stages. Integument-like structures (arrows) are visible below the normal inner and outer integuments. (E) Expression of an ANT::GUS reportergene in wild-type plants. GUS activity is shown as blue color marking the chalaza including the inner and outer integument. (F) ANT mRNA expression in ovules of induced ANT::WUS-GR plants. ANT mRNA is detected in the chalaza and additionally in the newly formed ectopic structure (arrow). (G,H) WUS mRNA expression. (G) Control plant. WUS mRNA signal is restricted to the nucellus. (H) Ovule of induced ANT::WUS-GR plant. WUS mRNA expression is visible throughout the ovule primordium, including the region that forms ectopic organs (arrows). (f) Funiculus; (i.i.) inner integument; (n) nucellus; (o.i.) outer integument. Bars 20 μm.
In control experiments, no ectopic organs were formed in ANT::WUS-GR plants sprayed with a mock solution lacking dexamethasone or in plants that did not harbor the ANT::WUS-GR transgenes sprayed with dexamethasone (data not shown). To confirm that WUS was ectopically expressed, we performed in situ hybridization with a WUS probe. In contrast to wild-type ovules where WUS is expressed exclusively in the nucellus (Fig. 6G or 2C), ovules of induced ANT::WUS-GR plants displayed ectopic WUS expression in the chalaza and even in the funiculus (Fig. 6H).
In summary, ectopic WUS expression in the ovule is sufficient to induce the initiation of integument-like organs.
A functional WUS-GUS fusion protein remains in the cells of the nucellus
Having shown that WUS expression in one cell population, the nucellus, initiates organ formation in a neighboring cell group, the chalaza, we pondered the molecular mechanism underlying the communication between these cell populations. There are several examples, where transcription factors move from cell to cell (Lucas et al. 1995; Nakajima et al. 2001), and we therefore addressed whether WUS protein itself migrates from the nucellus into the chalaza. For this purpose, we analyzed the localization of WUS protein by using a WUS–GUS translational fusion expressed under the control of the WUS promoter. This transgene was introduced into a wus mutant background and the functionality of the modified WUS protein was confirmed by its ability to rescue the wus mutant phenotype (Fig. 7B,C). GUS staining in ovules of WUS::WUS-GUS; wus plants was exclusively detected in the nucellus, but not in the chalaza, where the mutant defect is observed, or the funiculus (Fig. 7A–C). Although based on this experiment we cannot exclude that endogenous WUS protein can move into neighboring cells, this result indicates that WUS activity in nucellus cells is sufficient for normal integument development, suggesting that its function is not direct but is mediated by at least one yet unidentified downstream signal that migrates from the nucellus to the chalaza.
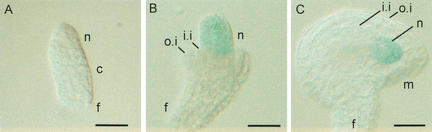
Figure 7.
WUS protein localization in ovules. (A–C) Expression of a WUS-GUS translational fusion in wus mutants. Ovules developed as wild type ovules indicating that the translational fusion is functional. (A) In young stages no GUS activity is detected. At the time of integument formation (B) GUS activity is observed exclusively in the nucellus. GUS activity persists until early stages of embryo sac development (C). (c) Chalaza; (f) funiculus; (i.i.) inner integument; (m) micropyle; (n) nucellus; (o.i.) outer integument. Bars 20 μm.
Discussion
Seed formation in higher plants exemplifies the more general requirement for coordinating cell behavior in the development of multicellular organisms, because neighboring ovule domains must act together to allow for the formation of a functional reproductive unit: The distal nucellus produces the female gametophyte and later on the embryo, while the underlying chalaza forms the integuments, protective organs that grow around the nucellus and later give rise to the seed coat. Here we address how the development of the individual ovule domains is coordinated. We show that the nucellus instructs neighboring chalazal cells to form integuments by WUS-dependent local signaling.
WUS signaling in the ovule
Our loss- and gain-of-function analyses indicate that WUS signaling plays a central role in ovule development: WUS activity is not only required, but also sufficient for the initiation of integuments from the chalaza. However, WUS mRNA and WUS protein are restricted to the nucellus. This indicates a novel signaling mechanism by which the nucellus governs organ formation in the neighboring chalaza: expression of WUS in the nucellus activates a downstream signal that emanates from the nucellus and induces organ initiation in the directly neighboring chalazal cells.
Our misexpression experiment shows that the entire chalaza and at least part of the funiculus can initiate organ formation. This raises the question of how the position of integuments is determined in wild-type ovules. A conceiveable explanation is that the WUS-dependent signal emanating from the nucellus acts only at a short range, thereby instructing specifically the chalazal cells adjacent to the nucellus to initiate integuments.
In contrast to chalazal cells, the nucellus itself does not form integuments. This suggests that nucellus cells are not competent to respond to the signal they produce, whereas their neighboring cells are. What causes this different competence? Our data show that integument initiation requires independent inputs from WUS and ANT pathways. Because ANT is only expressed in the chalaza but is not expressed in the nucellus, this difference could account for the inability of nucellar cells to respond to WUS signaling.
Therefore, the position of organ formation in ovules appears to be determined by the range of the WUS signal from the nucellus and the responsiveness of cells.
WUS function in shoot meristems and ovules
WUS is expressed in the apical part of shoot and floral meristems and in the distal domain of ovules and in both cases, WUS signaling plays a central role in establishing developmental differences between neighboring cell populations. These similarities raise the question of whether apical shoot meristems and ovules evolved from a common precursor or whether they may have recruited WUS function independently. Shoot meristems and ovules both are apical structures that initiate organs at some distance from the apex. Paleobotanical evidence suggests that the nucellus of the ovule is derived from an apical meristem: A widely accepted hypothesis for seed plant evolution, the telome theory, proposes that the nucellus originated from one sporangium-bearing shoot axis (fertile telome) out of several telomes in a dichotomously branching system (Kenrick and Crane 1997).
On the other hand, WUS signaling appears to have different developmental consequences that reflect the differences in the workings of the shoot meristem and the ovule. The shoot meristem is an indeterminate dynamic stem-cell system, which continuously produces cells at the apex that are consumed by organ formation at the periphery. WUS signaling from a small cell group, the organizing center, confers stem-cell identity on its overlying neighbors. One important readout of WUS signaling is the expression of the CLV3 gene in the stem cell region. CLV3 encodes a ligand for the CLV signaling pathway by which the stem cell region signals back to the organizing center and restricts the size of the WUS expression domain. The WUS/CLV3 feedback loop appears to keep the size of the stem-cell pool constant.
In contrast, ovules are determinate structures, which form only a few organs. Here, WUS signaling from the nucellus initiates organ formation in the underlying chalaza. In contrast to the shoot meristem, WUS does not activate CLV3 expression in ovules. There is no evidence for the presence of stem cells or an apparent supply of cells from the nucellus to the chalaza, which may explain why regulators of stem cell homeostasis, such as the CLV pathway, are not expressed there.
Therefore, the presence of WUS signaling in ovules and shoot meristems could reflect a conserved signaling module that has become embedded into different developmental programs and therefore supports the view that the nucellus is derived from an apical meristem.
WUS defines a repeatedly employed signaling module
Cell identity in plants is determined by position. This implies extensive intercellular communication to allow cells to assess their position and developmental task. Our data suggest that WUS defines a short-range signaling module, which is employed both in shoot meristems and in ovules to coordinate the development of neighboring cell groups.
What is the biological significance of this signaling mechanism? In the shoot meristem, WUS signaling establishes a stem cell niche and, by interacting with CLV3, ensures stem cell homeostasis. In ovules, it appears to coordinate two critical events for seed development, formation of the female gametophyte and generation of protective structures to ensure seed survival. Therefore, the induction of integument formation by WUS-dependent signals from the nucellus provides a mechanism by which the tissue that gives rise to the female gametophyte with the egg cell and eventually the embryo ensures the formation of its own protective structures, which in turn allows for the progression of gametophyte development.
Materials and methods
The wus-1 mutant and plant growth conditions have been described previously (Laux et al. 1996). The ant72F5 mutant was kindly provided by Kai Schneitz (University of Zürich, Switzerland). This allele displays a very strong ovule phenotype similar to the putative null allele ant-1 (Klucher et al. 1996).
Construction of transgenes and plant transformation
We used the pOpL two-component expression system (Moore et al. 1998) for all transgenic experiments with the exception of the analysis of AG::GUS activity in wus mutant ovules (see below). For simplicity we refer to, for example, plants of the genotype CLV1::LhG4, pOp::WUS as CLV1::WUS. Generation of the CLV1::LhG4, pOp::WUS-pOp::NLSGUS and ANT::LhG4 transgenic lines has been described (Schoof et al. 2000).
For the analysis of AG::GUS expression in wus mutant ovules, we employed a direct CLV1::WUS construct, which has been described previously (Schoof et al. 2000), and an AG::GUS reporter line kindly provided by L. Sieburth (Sieburth and Meyerowitz 1997).
To create pOp::WUS-GR transgenic plants, the WUS open reading frame was amplified using primers WUS5BAM (5′-AGTCGGGATCCACACACATGG-3′) and WUS3BAM+2 (5′-GAGCGGATCCAGACGTAGCTCAAGAG-3′), digested with BamHI and inserted into pRS020 (kindly provided by R. Sablowski; Sablowski and Meyerowitz 1998), which harbors the C-terminal ligand-binding domain of the rat glucocorticoid receptor (GR), to generate a translational fusion of WUS to GR. The WUS fragment was sequenced to exclude amplification errors. The resulting WUS-GR fusion was then inserted into pBarMOp, a modification of pGPTV-BAR (Becker et al. 1992), which contains the pOp-promoter to yield plasmid MT175.
For the WUS::GUS reporter construct, the WUS coding sequence was removed from the genomic region extending 4.4 kb upstream and 1.5 kb downstream of the WUS start and stop codons, respectively, and replaced by a unique SacI site using a PCR-based approach (MT61). NLSGUS (van der Krol and Chua 1991) was inserted into this SacI site and the resulting WUS:: GUS fusion was transferred to pBarA, a derivative of pGPTV-BAR, to yield MT87.
To express a translational fusion of WUS and GUS in wus-1 mutants, we used the pOpL two-component system. To generate the WUS::LhG4 construct, the LhG4 coding region was excised from pBin-LhG4 (provided by I. Moore, Department of Plant Science, Oxford University, UK) and subcloned into the unique SacI site of MT61 (see above). The resulting WUS::LhG4 fragment was then subcloned into pBarA to yield MT95. For the pOp::WUS-GUS construct, the WUS coding region was fused in-frame to the 5′ end of the GUS coding region preceded by an oligonucleotide that encodes five repeats of the dipeptide Gly–Ala as a spacer (MT225). The WUS–GUS fragment was excised from MT225 and ligated to pBarMOp to yield MT226.
To generate a CLV3::NLSGUS construct, the 5′ genomic region preceding the CLV3 ORF was amplified from Ler genomic DNA using primers CLV3ECORV5LEFT (5′-CTTTGATATCG CGGTTTGTGTAAATGG-3′) and CLV3BAM5RIGHT (5′-ATG GATCCTTAGAGAGAAAGTGACTGAGTG-3′), digested with EcoRV and BamHI and subcloned into pVIP35, which had been digested with NotI, blunt-ended with T4-DNA polymerase, and digested with BamHI to yield MT189. pVIP35 is a derivative of pBluescript harboring the NLSGUS coding region and a nos transcription terminator (van der Krol and Chua 1991) between the BamHI and EcoRI sites. The 3′ genomic region downstream of the CLV3 ORF was amplified from Ler genomic DNA using primers CLV3SAC3LEFT (5′-TTGAGCTCCCTTGACCTAAT CTCTTGTTGC-3′) and CLV3SAC3RIGHT (5′-CCGAGCTC TAGTGTTTCACCAAAGTCC-3′), digested with SacI and subcloned into MT189, which had been partially digested with SacI to generate MT190. The resulting CLV3::NLSGUS fragment was excised from MT190 by partial digestion with SacI and ligated to pBarM, a derivative of pGPTV-BAR (Becker et al. 1992), which had been digested with SacI, to yield plasmid MT194. Details are available on request.
Constructs were introduced into Agrobacterium strain GV3101(pMP90) (Koncz and Schell 1986) by electroporation. Plant transformation was done using the “floral dip” method (Clough and Bent 1998).
Microscopy
Fixation, clearing, and preparation of the ovules for light microscopy was performed as described (Siddiqi et al. 2000). For confocal laser scanning microscopy inflorescences were fixed and afterwards stained with arginine and propidium iodide as described (Clark et al. 1993). Optical sections were generated using a Leica TCS-NT confocal laser scanning microscope (Leica Microsystems, Heidelberg, Germany)
PCR-based genotyping
Plants were genotyped at the WUS locus by dCAPS (Neff et al. 1998) using primers wus1A (5′-TTGAATTAATGAATTATAG TTTGATACG-3′) and wus1S (5′-TTGAAGTTATGGATCTTG ATTGG-3′) at an annealing temperature of 55°C. The PCR product was digested with RsaI, which gives fragments of 246, 65, and 28 bp for the wild-type allele and 246 and 93 bp for the wus-1 allele.
Expression analysis
GUS staining of whole-mount ovules was performed largely as described (Schoof et al. 2000). However, ovules were not cleared in ethanol, but were immediately mounted in 60% glycerol. For sections, FAA-fixed and dehydrated material was embedded in Paraplast Plus (Oxford Labware, St. Louis). The sections were dewaxed by immersion in Histoclear, stained with 0.01% ruthenium red (3 min), and mounted in 60% glycerol. In situ hybridization using WUS as a probe was performed as described previously (Mayer et al. 1998). To generate the ANT probe, plasmid p5delta4 (Elliott et al. 1996) was digested with BamHI and transcribed with T7 RNA Polymerase.
Acknowledgments
We thank Jiri Friml, Nico Geldner, Gerd Jürgens, Swen Schellmann, Kathrin Schrick, and the members of our laboratory for comments on the manuscript; Eike Rademacher, Jürgen Berger, and Oliver Grandjean for technical help; and James Doyle, Veronique Pautot, and Jan Traas for helpful discussions. We are grateful to Robert Sablowski, Leslie Sieburth, and Kay Schneitz for providing seeds and other materials. This work was supported by a Landesgraduiertenförderung stipend to R.G.-H., a stipend of the Boehringer Ingelheim Fonds to M.L., and by grants of the Deutsche Forschungsgemeinschaft to T.L.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL laux@biologie.uni-freiburg.de; FAX 49-761-203-2745.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.225202.
References
- Baker S, Robinson-Beers K, Villanueva J, Gaiser J, Gasser C. Interactions among genes regulating ovule development in Arabidopsis thaliana. Genetics. 1997;145:1109–1124. doi: 10.1093/genetics/145.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, Schneitz K. NOZZLE regulates proximal–distal pattern formation, cell proliferation, and early sporogenesis during ovule devlopment in Arabidopsis thaliana. Development. 2000;127:4227–4238. doi: 10.1242/dev.127.19.4227. [DOI] [PubMed] [Google Scholar]
- Becker D, Kemper E, Schell J, Masterson R. New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol. 1992;20:1195–1197. doi: 10.1007/BF00028908. [DOI] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science. 2000;289:617–619. doi: 10.1126/science.289.5479.617. [DOI] [PubMed] [Google Scholar]
- Chevalier D, Sieber P, Schneitz K. The genetic and molecular control of ovule development. In: O'Neill S, Roberts J, editors. Annual plant reviews: Plant reproduction. Sheffield, UK: Sheffield Academic Press; 2002. pp. 61–65. [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development. 1993;119:397–418. doi: 10.1242/dev.119.2.397. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Dalman F, Scherrer L, Taylor L, Akil H, Pratt W. Localisation of the 90 kDa heat shock protein-binding site within the hormone-binding domain of the glucocorticoid receptor by peptide competition. J Biol Chem. 1991;266:3482–3490. [PubMed] [Google Scholar]
- Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQJ, Gerentes D, Perez P, Smyth DR. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell. 1996;8:155–168. doi: 10.1105/tpc.8.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau K. Anatomy of seed plants. New York, NY: John Wiley and Sons; 1977. [Google Scholar]
- Gaiser JC, Robinson-Beers K, Gasser CS. The Arabidopsis SUPERMAN gene mediates asymetric growth of the outer integuments of ovules. Plant Cell. 1995;7:333–345. doi: 10.1105/tpc.7.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenrick P, Crane P. The origin and early diversification of land plants: A cladistic study. Washington, DC.: Smithsonian Institution Press; 1997. [Google Scholar]
- Klucher KM, Chow H, Reiser L, Fischer RL. The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell. 1996;8:137–153. doi: 10.1105/tpc.8.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel Agrobacterium binary vector. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- Laux T, Mayer KFX, Berger J, Jürgens G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development. 1996;122:87–96. doi: 10.1242/dev.122.1.87. [DOI] [PubMed] [Google Scholar]
- Lenhard M, Bohnert A, Jürgens G, Laux T. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell. 2001;105:805–814. doi: 10.1016/s0092-8674(01)00390-7. [DOI] [PubMed] [Google Scholar]
- Lohmann J, Huong R, Hobe M, Busch M, Parcy F, Simon R, Weigel D. A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell. 2001;105:793–803. doi: 10.1016/s0092-8674(01)00384-1. [DOI] [PubMed] [Google Scholar]
- Lucas WJ, Bouché-Pillon S, Jackson DP, Nguyen L, Baker L, Ding B, Hake S. Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science. 1995;270:1980–1983. doi: 10.1126/science.270.5244.1980. [DOI] [PubMed] [Google Scholar]
- Mayer KFX, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- Modrusan Z, Reiser L, Feldmann KA, Fischer RL, Haughn GW. Homeotic transformation of ovules into carpel-like structures in Arabidopsis. Plant Cell. 1994;6:333–349. doi: 10.1105/tpc.6.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore I, Galweiler L, Grosskopf D, Schell J, Palme K. A transcription activation system for regulated gene expression in transgenic plants. Proc Natl Acad Sci. 1998;95:376–381. doi: 10.1073/pnas.95.1.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Sena G, Nawy T, Benfey P. Intercellular movement of the putative transcription factor SHR in root patterning. Nature. 2001;413:307–311. doi: 10.1038/35095061. [DOI] [PubMed] [Google Scholar]
- Neff M, Neff J, Chory J, Pepper A. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J. 1998;14:387–392. doi: 10.1046/j.1365-313x.1998.00124.x. [DOI] [PubMed] [Google Scholar]
- Reiser L, Modrusan Z, Margossian L, Samach A, Ohad N, Haughn GW, Fischer RL. The BELL1 gene encodes a homeodomain protein involved in pattern formation in the Arabidopsis ovule primordium. Cell. 1995;83:735–742. doi: 10.1016/0092-8674(95)90186-8. [DOI] [PubMed] [Google Scholar]
- Robinson-Beers K, Pruitt RE, Gasser CS. Ovule development in wild-type Arabidopsis and two female sterile mutants. Plant Cell. 1992;4:1237–1249. doi: 10.1105/tpc.4.10.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodkiewitz B. Callose in cell walls during megasporogenesis in angiosperms. Planta. 1970;93:39–47. doi: 10.1007/BF00387650. [DOI] [PubMed] [Google Scholar]
- Sablowski RW, Meyerowitz EM. A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell. 1998;92:93–103. doi: 10.1016/s0092-8674(00)80902-2. [DOI] [PubMed] [Google Scholar]
- Schiefthaler U, Balasubramanian S, Sieber P, Chevalier D, Wisman E, Schneitz K. Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc Natl Acad Sci. 1999;96:11664–11669. doi: 10.1073/pnas.96.20.11664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneitz K, Hülskamp M, Pruitt RE. Wild-type ovule development in Arabidopsis thaliana: A light microscopy study of cleared whole-mount tissue. Plant J. 1995;7:731–749. [Google Scholar]
- Schneitz K, Hülskamp M, Kopczak SD, Pruitt RE. Dissection of sexual organ ontogenesis: A genetic analysis of ovule development in Arabidopsis thaliana. Development. 1997;124:1367–1376. doi: 10.1242/dev.124.7.1367. [DOI] [PubMed] [Google Scholar]
- Schneitz K, Baker S, Gasser C, Redweik A. Pattern formation and growth during floral organogenesis: HUELLENLOS and AINTEGUMENTA are required for the formation of the proximal region of the ovule primordium in Arabidopsis thaliana. Development. 1998;125:2555–2563. doi: 10.1242/dev.125.14.2555. [DOI] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KFX, Jürgens G, Laux T. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- Siddiqi I, Ganesh G, Grossniklaus U, Subbiah V. The DYAD gene is required for progression through female meiosis in Arabidopsis. Development. 2000;127:197–200. doi: 10.1242/dev.127.1.197. [DOI] [PubMed] [Google Scholar]
- Sieburth L, Meyerowitz EM. Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenetically. Plant Cell. 1997;9:355–365. doi: 10.1105/tpc.9.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Krol AR, Chua N-H. The basic domain of plant B-ZIP proteins facilitates import of a reporter protein into plant nuclei. Plant Cell. 1991;3:667–675. doi: 10.1105/tpc.3.7.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva J, Broadhvest J, Hauser B, Meister R, Schneitz K, Gasser C. INNER NO OUTER regulates abaxial-adaxial patterning in Arabidopsis ovules. Genes & Dev. 1999;13:3160–3169. doi: 10.1101/gad.13.23.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb MC, Gunning BES. Embryo sac development in Arabidopsis thaliana. I. Megasporogenesis, including the microtubular cytoskeleton. Sex Plant Reprod. 1990;3:244–256. [Google Scholar]