Abstract
Dlx homeobox genes are mammalian homologs of the Drosophila Distal-less (Dll) gene. The Dlx/Dll gene family is of ancient origin and appears to play a role in appendage development in essentially all species in which it has been identified. In Drosophila, Dll is expressed in the distal portion of the developing appendages and is critical for the development of distal structures. In addition, human Dlx5 and Dlx6 homeobox genes have been identified as possible candidate genes for the autosomal dominant form of the split-hand/split-foot malformation (SHFM), a heterogeneous limb disorder characterized by missing central digits and claw-like distal extremities. Targeted inactivation of Dlx5 and Dlx6 genes in mice results in severe craniofacial, axial, and appendicular skeletal abnormalities, leading to perinatal lethality. For the first time, Dlx/Dll gene products are shown to be critical regulators of mammalian limb development, as combined loss-of-function mutations phenocopy SHFM. Furthermore, spatiotemporal-specific transgenic overexpression of Dlx5, in the apical ectodermal ridge of Dlx5/6 null mice can fully rescue Dlx/Dll function in limb outgrowth.
Keywords: Embryo, mouse, transgenic, knockout, cartilage, limb
Dlx homeobox genes are mammalian homologs of the Drosophila Distal-less (Dll) gene and consist of six genes that are organized into the Dlx1/2, Dlx5/6, and Dlx3/7 bigene clusters. The vertebrate Dlx genes are expressed in craniofacial primordia, developing brain, and limbs, including the apical ectodermal ridge (AER). The Dlx/Dll gene family is of ancient evolutionary origin and has been either implicated or demonstrated to play a critical function in limb/appendage outgrowth and development in essentially all animals in which it has been identified (for review, see Panganiban 2000). In Drosophila, Dll is expressed in the distal portion of the developing appendages (limbs) and is required for proper development of distal appendage structures. Partial loss-of-function Drosophila Dll alleles affect primarily distal leg structures, while more severe loss-of-function Dll alleles extend their effects to more medial structures such as the fly tibia and femur. During development, distal cells of the fly limb retain their requirement for Dll/Dlx until late stages, whereas more medial cells loose this requirement earlier during fly limb outgrowth (for review, see Panganiban 2000).
Split-hand/split-foot malformation (SHFM), also referred to as ectrodactyly or lobster-claw deformity (OMIM 183600) is a human limb defect that affects growth and patterning of the central digital rays (Temtamy and McKusick 1978). The malformation is characterized by missing digits, fusion of remaining digits (syndactyly), and median clefts. SHFM is typically inherited in an autosomal dominant fashion with incomplete penetrance, variable expressivity, and segregation distortion (Scherer et al. 1994). Four human SHFM disease loci genetically map to chromosomes 7q21.3-22.1 (SHFM1, OMIM 183600), Xq26-q26.1 (SHFM2, OMIM 313350), 10q24-q25 (SHFM3, OMIM 600095), and 3q27 (SHFM4, OMIM 603237) (Faiyaz ul Haque et al. 1993; Scherer et al. 1994; Nunes et al. 1995; Ianakiev et al. 2000). Moreover, SHFM can present by itself or in combination with craniofacial, urogenital, ectodermal, and/or hearing abnormalities (Scherer et al. 1994; O'Quinn et al. 1998; Tackels-Horne et al. 2001).
The naturally occurring Dactylaplasia (Dac) mutant mouse is a phenocopy model of SHFM (Chai 1981). Dac maps syntenically to human chromosome 10q23-q25 and is a model for SHFM3 (Johnson et al. 1995). Dac embryos initiate proper limb outgrowth up to embryonic day 10.5 (E10.5). However, by E11.5 the central portion of the AER degenerates, while the anterior and posterior portions of the AER remain intact (Seto et al. 1997; Crackower et al. 1998). The AER is critical for proper limb outgrowth and patterning by signaling to the underlying progress zone and by maintaining the zone of polarizing activity (for review, see Capdevila and Izpisua Belmonte 2001). While Dac may be a syntenic model for only SHFM3, it can be inferred that the degeneration of the AER may be a common pathogenic feature within all forms of SHFM.
In vertebrates, the Dlx genes are expressed in craniofacial primordia, developing brain, and limbs, including the AER. The paired Dlx5 and Dlx6 genes, which map to chromosome 7q22, are postulated as candidates for SHFM1 (Scherer et al. 1994; Crackower et al. 1996). Targeted disruption or ablation of Dlx1, Dlx2, Dlx1/2, or Dlx5 genes in mice results in craniofacial, bone, and vestibular defects (Qiu et al. 1997; Acampora et al. 1999; Depew et al. 1999). However, the limbs remain conspicuously absent of any abnormalities. The lack of limb defects in these mutants may be the result of genetic compensation or overlapping function with other members of the Dlx gene family. To explore this possibility, we performed a loss of Dlx5 and Dlx6 function study in mice. As hypothesized, we found that targeted disruption of both Dlx5 and Dlx6 results in bone, inner ear, and severe craniofacial defects. In addition, simultaneous loss of Dlx5 and Dlx6 gene products results in a phenocopy model of human SHFM1 and is the first demonstration of a role for mammalian Dlx genes in limb development.
Results
Targeted disruption of both Dlx5 and Dlx6 (Dlx5/6) in embryonic stem cells and mice
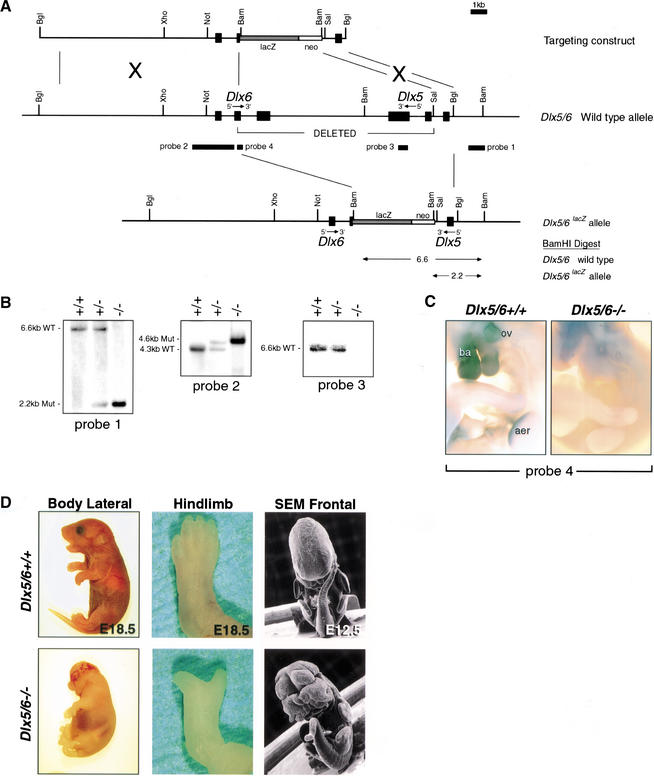
Due to their close proximity to one another on the chromosome, the Dlx5/6 null allele was achieved by making a single deletion inactivating both genes simultaneously. As outlined in Figure 1A, Dlx5 and Dlx6 are arranged in a transcriptionally convergent orientation. A correctly targeted Dlx5/6 null allele places lacZ under the transcriptional control of Dlx6 5′ regulatory elements and deletes ∼11 kb of DNA containing all the coding and noncoding sequence spanning the Dlx5 and Dlx6 homeoboxes. The absence of functional Dlx5 or Dlx6 homeodomains and associated C-terminal amino acids should thus result in null alleles for both Dlx5 and Dlx6. Correct targeting of Dlx5 and Dlx6 was confirmed by Southern blot analysis employing probes internal and external to the Dlx5/6 targeting construct (Fig. 1B). Correct Dlx5/6 targeting was further confirmed by employing an RNA in situ hybridization probe corresponding to Dlx6 sequences overlapping the homeobox and adjacent mRNA sequences which fall within the Dlx5/6 null allele deleted region. In Dlx5/6 heterozygotes, a clear signal was observed corresponding to expression in the branchial arches (ba), otic vesicle (ov), and AER from the wild-type allele. Analysis of Dlx5/6 null embryos, however, showed no corresponding signal in the same tissues or elsewhere (Fig. 1C), indicating that deletion of this region had been successfully achieved at the Dlx5/6 null locus.
Figure 1.
Simultaneous disruption of Dlx5 and Dlx6 by homologous recombination, genotyping, mRNA, and morphological analyses of embryos carrying a Dlx5/6 null allele. (A) Structure of the genomic Dlx5 and Dlx6 loci, ires.lacZ.neo targeting vector, and mutant allele following homologous recombination. Exons are depicted as black boxes. The transcriptional orientation of each gene is indicated. The probes employed for Southern and RNA in situ analyses are indicated with black rectangles. (B) Southern blot analysis of embryonic genomic DNA isolated from the yolk-sac was digested with BamHI and then hybridized with a 5′ (probe 1) in the undeleted region of Dlx5, 5′ (probe 2) in the undeleted region of Dlx6, or 3′ (probe 3) in the deleted region of Dlx5. The wild-type and mutant alleles were detected as 6.6 kb and 2.2 kb fragments, respectively, with probe 1 and 4.3 kb and 4.6 kb, respectively, with probe 2. The wild-type allele was detected as a 6.6 kb fragment and the mutant allele was undetected with probe 3. (C) Whole-mount in situ hybridization of E10.5 wild-type and Dlx5/6−/− embryos using probe 4. Dlx5/6−/− embryos lack probe 4 expression as probe 4 falls within the region deleted in the Dlx5/6−/− embryos. (D) Gross appearance of wild-type and Dlx5/6−/− embryos and hindlimbs at E18.5. Dlx5/6−/− embryos show an overall reduced size, exencephaly, kinked tail vertebrae, and split hindlimbs. Scanning electron microscopy (SEM) of whole-mount wild-type and Dlx5/6−/− embryos at E12.5, highlighting the craniofacial, tail, and limb malformations already clearly visible at this stage. Abbreviations: aer, apical ectodermal ridge; ba, branchial arches; ov, otic vesicle.
Dlx5/6+/− mutant mice were viable, fertile, and did not have any overt physical abnormalities. In contrast, Dlx5/6−/− mutant embryos were viable up to the time of birth, and exhibited severe limb, craniofacial, and axial skeletal defects (Fig. 1D). Obvious craniofacial defects included the complete absence of calvaria, resulting in exencephaly, reduction in the size of the eyes, and clefting and dysmorphogenesis of nasal, maxillary, and mandibular structures. Obvious axial skeletal defects included growth retardation and kinked tail vertebrae. The Dlx5/6 null limb defects phenocopied SHFM with variable penetrance in the forelimbs and complete penetrance in the hindlimbs. The cause of death in the Dlx5/6 nulls was likely related in part to the exencephaly, which results in cerebral trauma during fetal delivery and massive postnatal blood loss.
Dlx5/6-lacZ expression during craniofacial development
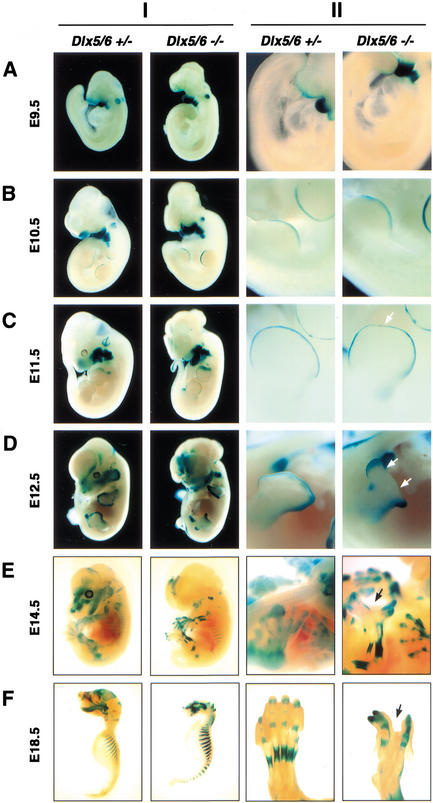
The Dlx5/6-lacZ expression pattern in heterozygous embryos was consistent with that previously revealed by use of RNA in situ hybridization or lacZ reporter expression (Simeone et al. 1994; Chen et al. 1996; Acampora et al. 1999; Depew et al. 1999). However, forebrain expression domains were reduced, likely owing to the deletion of a CNS-specific intergenic enhancer (Zerucha et al. 2000). Dlx5/6-lacZ expression was apparent by E8.5 in the ventral cephalic epithelium, nasal and otic placodes, and tail bud. At this stage of development, there was no apparent difference in lacZ expression or morphological abnormality in Dlx5/6−/− mutant embryos (not shown). At E9.5, strong Dlx5/6-lacZ expression was observed in the first and second branchial arches and the otic vesicle (Fig. 2A). A phenotypic difference was first observed at E9.5, with failure of the anterior neuropore to close in Dlx5/6−/− embryos (Fig. 2A). By E10.5, Dlx5/6-lacZ expression was observed in the third and fourth branchial arches and nasal placodes (Fig. 2B).
Figure 2.
Embryonic Dlx5 and Dlx6 expression revealed by whole-mount β-galactosidase staining of heterozygous and homozygous Dlx5/6 mutant embryos (Column I), with higher magnifications of limbs from the same embryos (Column II). (A) Dlx5/6 is strongly expressed in the first and second branchial arches and the otic pit, with less prominent expression in the tail and forelimb buds at E9.5. (B) Expression becomes apparent in the AER of both fore- and hindlimb buds by E10.5. (C) At E11.5, reduction in expression in the frontonasal prominence and medial AER (arrow) initiates. (D) Expression in the medial AER deteriorates by E12.5 in Dlx5/6−/− embryos. Arrows in the right-hand column indicate the loss of medial tissue and digits in the Dlx5/6−/− embryos. (E) Expression within all developing bones becomes apparent by E14.5, at which time the SHFM phenotype becomes recognizable in null mutants. (F) At E18.5, craniofacial and skeletal expression remains strong. Calvarial and mandibular expression is noticeably lost in Dlx5/6−/− embryos owing to the absence of these tissues.
Dlx5/6-lacZ expression was strong in the epithelium of the semicircular canals and endolymphatic duct of the inner ear of Dlx5/6+/− embryos at E11.5 (Fig. 2C). Subsequent phenotypic differences were observed in Dlx5/6−/− embryos at E11.5 with unrecognizable inner ear structures, absence of developing eyes, dysmorphic branchial arch derivatives, and reduced lacZ expression in the frontonasal prominence (Fig. 2C). Severe clefting of the entire nasal cavity in Dlx5/6−/− embryos became readily apparent by E14.5 (Fig. 2E). Dlx5/6-lacZ expression at this stage was severely reduced in frontonasal, maxillary, and mandibular tissues of Dlx5/6−/− embryos. Expression in tissues giving rise to facial and calvarial structures was noticeably lost. In addition, external and middle ear expression appeared consolidated, while inner ear expression was absent.
Dlx5/6-lacZ expression during axial skeletal development
Dlx5/6-lacZ expression was observed in all sites of developing endochondral and membranous bone formation. The first abnormality in axial skeletal development was apparent by E11.5, with the formation of a kinked tail in Dlx5/6−/− embryos (Fig. 2C). In heterozygous embryos, Dlx5/6-lacZ expression in clavicles, axial skeleton, and the leading edge of calvaria was first observed at E12.5 (Fig. 2D). By E14.5, expression was observed in the perichondral region of developing long bones, ribs, and digits (Fig. 2E). The pattern of Dlx5/6-lacZ expression in the axial skeleton was similar between Dlx5/6+/− and Dlx5/6−/− embryos, suggesting that there was no significant loss of Dlx5/6−/− cells in this domain, and furthermore, that the expression of Dlx5/6-lacZ was independent of any critical Dlx5/6 cross- or autoregulatory interactions.
Dlx5/6-lacZ expression during limb development
Dlx5/6-lacZ was expressed in all developing appendages including the external ears, genital tubercle, and limb buds (Fig. 2A–F). In the developing limbs, strong expression in the AER of both the hind- and forelimb buds was observed by E10.5 and extended into the anteroproximal margins of the forelimbs by E11.5. Expression in the genital tubercle was also evident at E11.5. The first phenotypic difference in limb development was observed in the hindlimb AER of Dlx5/6−/− embryos at E11.5. The medial portion at the AER had become thinner and the distal edge of the hindlimb had flattened. At E12.5, Dlx5/6-lacZ expression remained strong in the AER of Dlx5/6+/− embryos, and digital buds were apparent. However, in Dlx5/6−/− embryos, the medial hindlimb AER expression had degenerated, resulting in a distal flattening while still maintaining Dlx5/6-lacZ expression in AER lateral margins (Fig. 2D). By E14.5, Dlx5/6-lacZ was expressed in the epithelium and mesenchyme of digital tips and the perichondrium of the digits. In contrast, from one to three of the central hindlimb digits were absent in the hindlimbs of any given Dlx5/6−/− embryo with normal Dlx5/6-lacZ expression in the remaining digits. In addition, outgrowth of the remaining digits was diverted laterally away from the midline towards the remaining distal Dlx5/6-lacZ expression domains persisting in the lateral edges of the AER remnant.
Cartilage and bone defects in Dlx5/6−/− mice
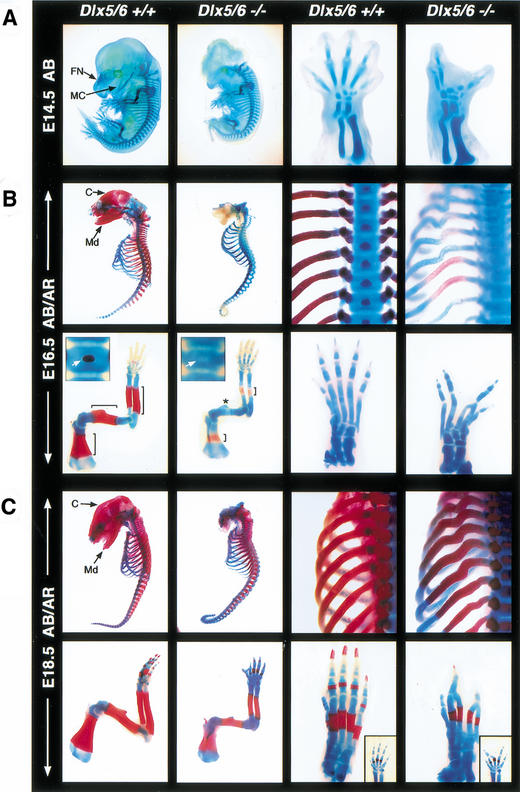
Severe craniofacial cartilage defects in Dlx5/6−/− embryos were apparent at E14.5. Exencephaly, with the absence of Alcian blue staining of frontonasal, supraoccipital, and rostral temporal areas, and the complete loss of Meckel's cartilage were the most striking defects (Fig. 3A). The inner ear capsule and middle ear cartilages were fused and severely dysmorphic, while external ear cartilage was absent. Moreover, cartilage giving rise to the cranial floor (basioccipital, basisphenoid, and sphenoid) and frontonasal prominence were present, but with severe patterning defects. Cartilage from the exoccipital and ventral temporal bone primordia extending to the distal nasal capsule were distinctly condensed and fused. It could not be determined as to whether middle and inner ear cartilages were included within the fused structure. External ear cartilage was absent and outgrowth of the dysmorphic nasal prominence was curved caudally. At E16.5, the dysmorphic cranial cartilage structure remained unossified, and maxillary and mandibular bones were absent. However, soft tissues (skin, whisker pads, tongue, and muscle) were present in a manner that defined where maxillary and mandibular structures should have developed. Endochondral ossification of the exoccipital bone primordia of Dlx5/6−/− embryos was present at E18.5. In addition, areas of membranous ossification had developed that suggested an attempt to form maxilla, premaxilla, and nasal bones.
Figure 3.
Craniofacial, axial, and appendicular skeletal defects of Dlx5/6−/− embryos. (A) Whole-mount Alcian blue (AB) staining reveals the absence of Meckel's, nasal prominence and central digit cartilages of E14.5 mutant embryos. (B) Alcian blue and alizarin red (AB/AR) staining shows the absence of calvaria, maxillary, and mandibular bones of E16.5 mutant embryos. In addition, the axial skeleton, including vertebral bodies (inset), has little or no ossification and the ribs are malformed. The forelimbs (left columns) also have a delay in ossification (bracketed region), or a complete absence of ossification (star) and hindlimb digits (right columns) are absent and/or fused. (C) The delay in ossification becomes less severe in the axial and appendicular skeleton by E18.5. However, the ribs remain disfigured and the ratio of forelimb ossification to cartilage remains retarded. Hindlimbs and forelimbs (inset) have clefting due to missing and fused digits. Abbreviations: C, calvaria; FN, frontonasal cartilage; MC, Meckel's cartilage; Md; Mandible.
Defects in limb cartilage were most striking in the hindlimbs of Dlx5/6−/− embryos. At E14.5, all combinations of the central digits were absent, similar to SHFM. Missing cartilage included phalanges, metatarsals, and tarsals (Fig. 3A–C). The most prevalent defect was the absence of the central digit. In addition, remaining adjacent digits tended to be either misshapen or fused at phalanges or metatarsals. Similar phenotypic defects were occasionally observed in the forelimbs (Fig. 3C, inset).
Endochondral bone defects were first observed at E16.5 with shorter or completely absent ossification centers in Dlx5/6−/− embryos compared to Dlx5/6+/+ or Dlx5/6+/− embryos (Fig. 3B). In addition, the ribs of Dlx5/6−/− embryos were severely dysmorphic, with the greatest aberrations in the region proximal to the vertebral column. An absence or delay in endochondral ossification was observed in all bones of the appendicular and axial skeleton of Dlx5/6−/− embryos at E16.5. By E18.5, affected endochondral ossification centers had achieved some minor developmental progression; however in all bones in the Dlx5/6−/− embryos, the transition from a cartilaginous to ossified skeleton remained severely retarded (Fig. 3C).
Dlx5/6 can regulate endochondral ossification
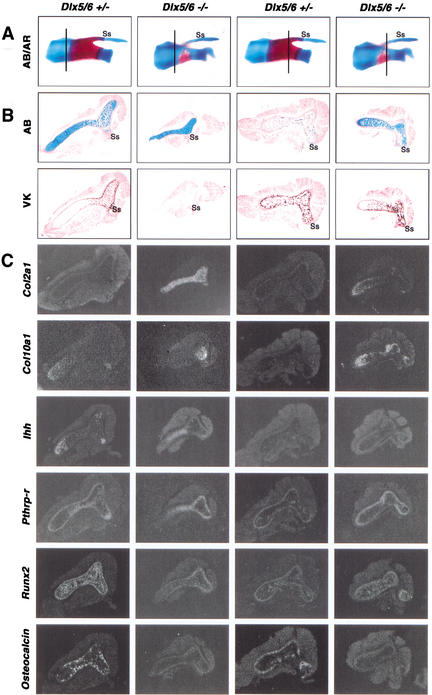
Histological analysis of the delayed endochondral ossification observed in Dlx5/6−/− embryos was performed on skeletal sections from E16.5 embryos. The scapula was specifically chosen because the projection of the spine of scapula (Ss) can be used as an anatomical point of reference to match corresponding sections from control and Dlx5/6 null embryos (Fig. 4A). In an area consisting of prehypertrophic, hypertrophic, and calcified (von Kossa-positive) chondrocytes in Dlx5/6+/− sections, comparable Dlx5/6−/− sections were composed of only prehypertrophic chondrocytes (Fig. 4B), indicating a retardation of the normal chondrocyte developmental program. In progressive serial sections, Dlx5/6+/− skeletal sections showed the presence of vascularization and a mineralized bone matrix. In contrast, comparable regions in Dlx5/6−/− embryos consisted of hypertrophic and calcified chondrocytes, minimal vascular invasion, and a predominantly cartilage matrix (Fig. 4B).
Figure 4.
Histological and molecular examination of the ossification of the scapula of heterozygous and Dlx5/6 null embryos at E16.5. (A) AB/AR staining of whole-mount scapulas shows the absence of ossification of the spine of scapula (Ss) and a minimal ossification within the center of the scapula, compared to heterozygous embryos. Vertical lines indicate the approximate area from which comparable serial sections were taken for each column of results. (B) In more proximal sections, AB-stained Dlx5/6−/− scapulas appear to lack both hypertrophic chondrocytes and a von Kossa (VK)-positive calcium matrix. (C) In contrast to control embryos, Col2a1 expression remains strong and Col10a1 appears to be increasing in the Dlx5/6 null sections. In addition, Ihh downregulation appears to be delayed in Dlx5/6 null sections, whereas Pthrp-r expression is unaffected. Runx2 expression appears normal in the perichondrium, but reduced in the chondrium of the Dlx5/6 null sections. Osteocalcin expression is completely absent in the Dlx5/6 null sections. In more distal sections, calcified hypertrophic chondrocytes are now apparent. Col2a1 expression is now almost absent in the Dlx5/6 null sections, and Col10a1 expression is strong. At this level, Runx2 appears normal in the Dlx5/6 null sections and Osteocalcin expression remains absent.
Molecular analysis of comparable E16.5 Dlx5/6−/− embryos also suggested a delay in the onset of the osteogenic pathway. Relative to wild-type embryos, Dlx5/6−/− embryos showed normal expression of Sox9, a regulator of precartilaginous condensation (data not shown). In contrast, Col2a1 and Col10a1 expression, indicators of prehypertrophic and hypertrophic chondrocytes, were still actively transcribed in Dlx5/6−/− sections, whereas in the same skeletal region in Dlx5/6+/− embryos, both genes had already been significantly downregulated (Fig. 4C). Fgf-Fgfr3 and Ihh-Pthrp-Pthrp-r signaling pathways are known to be important regulators of chondrocyte proliferation and differentiation that couples chondrogenesis to osteogenesis during endochondral ossification (Naski et al. 1998; Chung et al. 2001). Similar to what was observed for Col2a1 and Col10a1, there was a temporal delay in Fgfr3 and Ihh expression in the Dlx5/6−/− scapula (data not shown; Fig. 4C) such that comparable regions between control and Dlx5/6−/− null embryos showed opposing patterns of Fgfr3 and Ihh expression. Interestingly, Pthrp-r expression was unaffected in the Dlx5/6−/− null embryos (Fig. 4C).
The Runx2 (Osf2/Cbfa1) transcription factor has been shown to be a regulator of chondrocyte and osteoblast differentiation and a direct inducer of osteoblast-specific Osteocalcin expression (Ducy et al. 1997; Takeda et al. 2001). Similar Runx2 expression patterns were observed within the chondrium and perichondrium of comparable Dlx5/6+/− and Dlx5/6−/− skeletal sections (Fig. 4C). However, there appears to be a decrease in the number of cells expressing Runx2 within the chondrium. In contrast, Osteocalcin gene expression was dramatically absent in Dlx5/6−/− skeletal sections (Fig. 4A,C), suggesting that Dlx5/6 have a critical positive role in the osteoblast maturation pathway, and in their absence, the accumulation of mature osteoblasts is severely retarded or absent.
Dlx5/6 are essential for hindlimb AER maintenance
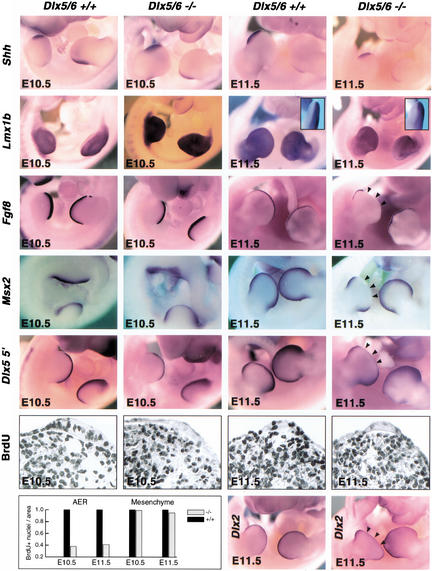
The earliest morphological limb abnormalities in Dlx5/6−/− embryos were observed at E11.5 (Fig. 2C). Therefore, molecular analysis of genes known to regulate limb development (for review, see Capdevila and Izpisua Belmonte 2001) was performed at this and earlier stages. Shh expression in the zone of polarizing activity has been shown to be a critical mediator of anterior-posterior limb patterning. Its expression was identical between wild-type and Dlx5/6−/− embryos at E10.5 and E11.5 (Fig. 5). Similarly, there were no alterations in the expression pattern of Lmx1b, a regulator of limb dorsal-ventral patterning (Fig. 5). Fgf8 expression in the AER, which is required for proximal-distal patterning and outgrowth, was normal for Dlx5/6−/− embryos at E10.5 (Fig. 5). However, Fgf8 expression in Dlx5/6−/− embryos was lost in the medial portion of the hindlimb AER by E11.5 (Fig. 5). Furthermore, Msx2 expression in the hindlimb AER and progress zone (PZ) of Dlx5/6−/− embryos was normal at E10.5 and by E11.5 was clearly absent in the medial AER and underlying PZ (Fig. 5). Dlx2, another member of the mammalian Dlx family, which shows strong similarity to Dlx5 and Dlx6 in expression and sequence, is coexpressed with Dlx5 and Dlx6 in the AER. No alterations of Dlx2 expression were observed prior to E11.5, suggesting that there were no critical cross-regulatory interactions between Dlx5/6 and Dlx2. At E11.5, expression of Dlx2 like that of the other AER markers, was missing from the medial region of the distal limb (Fig. 5). To follow the specific fate of cells normally expressing Dlx5 and Dlx6, we used a probe corresponding to the 5′ end of Dlx5, which is still present in the Dlx5/6 null mutation and should thus be transcribed. The expression of the Dlx5 5′ probe paralleled what was observed for other markers of the AER, namely normal expression was observed prior to E11.5, but following this stage there was a progressive loss of expression from the distal medial region of the limb. As normal levels and spatiotemporal expression of Dlx5 5′ was observed in the Dlx5/6−/− embryos, we can conclude that neither the position nor the migration of Dlx5/6 null cells was severely affected in the limbs of Dlx5/6−/− embryos prior to E11.5. Dss1 (deleted in split-hand/split-foot 1) is a gene encoding a unique protein located near Dlx5 and Dlx6 on the chromosome and is also expressed in the developing distal limb, including the AER (Crackower et al. 1996). Owing to its proximity to the SHFM1 critical region and its expression in the developing limb, Dss1 has been considered a candidate gene for SHFM1. Interestingly, no alteration of Dss1 expression in the developing limb of the Dlx5/6−/− embryos was observed prior to E11.5. From this stage on, Dss1 expression, like all other markers for the distal limb, was decreasing or missing from the distal medial portion of the Dlx5/6−/− limb (not shown).
Figure 5.
Molecular analysis of altered limb development of Dlx5/6−/− embryos. Whole-mount in situ hybridization shows that Shh and Lmx1b expression is normal at E10.5 and E11.5 in the Dlx5/6 null embryos. Fgf8 and Msx2 expression in the hindlimb AER is normal at E10.5, but is absent in the medial AER by E11.5. Arrowheads in the right-hand column indicate the loss of expression and missing tissue in the medial portion of the distal limb in the Dlx5/6−/− embryos. Analysis with a Dlx5 5′ riboprobe for the Dlx5 exon still present in the mutant allele reveals diminished Dlx5 expression in the medial AER by E11.5. Immunohistochemical staining and quantitation of BrdU incorporation reveals decreased cellular proliferation in the AER at E10.5 and E11.5, while proliferation in the underlying mesenchyme is unaffected. A compensatory increase of Dlx2 expression in the AER was not observed.
Morphological and molecular examination of the Dlx5/6−/− limbs suggested a progressive loss of distal medial limb cells with an onset immediately prior to E11.5. Possible explanations for cell loss are a decreased rate of cell proliferation or an increase in the number of cells locally undergoing programmed cell death. Programmed cell death in the AER and underlying mesenchyme was therefore measured by analyzing DNA fragmentation with a TUNEL assay; however, no significant difference was observed between wild-type and Dlx5/6−/− embryos in either of these cell types (not shown), suggesting that the loss of limb tissue observed in the Dlx5/6−/− embryos was unrelated to the specific cell death-mediated elimination of Dlx5/6−/− cells. In contrast, BrdU incorporation to mark cycling cells indicated a significant decrease in the number of proliferating cells within the hindlimb AER of Dlx5/6−/− embryos at both E10.5 and E11.5 (Fig. 5). However, cell proliferation in the underlying mesenchyme was normal at both stages for null embryos. These data indicate that a principal role for Dlx5/6 is maintenance of normal proliferation rates within the cells of the medial AER at E10.5 and E11.5. Loss of Dlx5/6 resulted in a localized decrease in cell proliferation within the medial AER and subsequent loss of limb structures (medial digits) arising from the underlying mesenchyme, which is regulated by the overlying AER.
Msx2 AER-specific transgene-driven Dlx5 expression rescues SHFM in Dlx5/6−/− mice
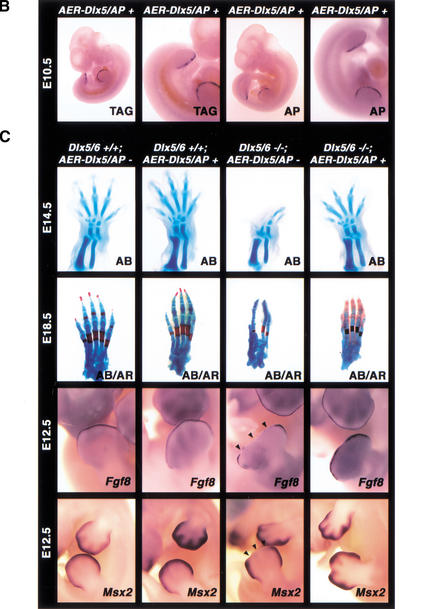
Transgenic mice were generated that overexpress Dlx5 in the AER under the control of the Msx2 AER-specific enhancer (Fig. 6A; Liu et al. 1994). The transgenic mice have no conspicuous phenotypic abnormalities even though transgene-specific expression by RNA in situ hybridization or of a coinjected alkaline phosphatase reporter confirmed that the AER-Dlx5 transgene was being expressed in the AER (Fig. 6B). Transgenic mice were subsequently bred onto the Dlx5/6 mutant background to determine whether AER-specific Dlx5 overexpression could rescue the SHFM defect in Dlx5/6−/− embryos. AER-Dlx5 transgenic Dlx5/6−/− embryos were stillborn and presented craniofacial and vertebral defects identical to those described for nontransgenic Dlx5/6−/− embryos. In addition, AER-Dlx5 expression was capable of fully rescuing the SHFM phenotype (Fig. 6C). Cartilage staining of E14.5 embryos showed normal hindlimb structures in both Dlx5/6+/+; AER-Dlx5+ and Dlx5/6−/−; AER-Dlx5+ mice (Fig. 6C). Proximal-distal outgrowth, patterning, and ossification within developing hindlimbs also proceeded in a normal manner (Fig. 6C). Furthermore, molecular analysis revealed that Fgf8 and Msx2 were expressed in the hindlimbs of Dlx5/6−/−; AER-Dlx5+ mice at E12.5 (Fig. 6C). As described above, Fgf8 and Msx2 expression were lost in the central hindlimb AER from E11.5 onward in the Dlx5/6 null embryos (Fig. 5).
Figure 6.
SHFM phenotypic rescue of Dlx5/6−/− embryos by Dlx5 transgene expression in the developing AER. (A) Structure of the Dlx5 and alkaline phosphatase (AP) transgenes that employ the Msx2 AER-specific enhancer. (B) Whole-mount in situ hybridization of AER-Dlx5/AP embryos showing AER expression of the tagged (TAG) Dlx5 transgene and transgene-specific alkaline phosphatase staining of the AER at E10.5. (C) AB staining at E14.5 shows restored hindlimb cartilage outgrowth and patterning of Dlx5/6−/− embryos expressing the AER-Dlx5/AP transgene. AB/AR cartilage and bone staining of the hindlimbs remains rescued at E18.5. Whole-mount in situ hybridization at E12.5 shows that AER expression of both Fgf8 and Msx2 can be restored by AER-Dlx5/AP expression in mutant mice. Arrowheads at E12.5 indicate the loss of marker expression and missing tissue in the medial portion of the distal limb in the Dlx5/6−/− embryos lacking the AER-Dlx5/AP transgene.
Discussion
Role of Dlx5/6 in control of craniofacial development
Dlx5/6−/− mice have a multitude of craniofacial and ear defects including the failure of Meckel's cartilage, mandible, and calvaria formation. The craniofacial and ear defects reported here are dramatically more severe than those observed in Dlx5-deficient mice (Acampora et al. 1999; Depew et al. 1999), which suggests that Dlx5 and Dlx6 have unique and redundant functions. Numerous genes are required for proper ear and craniofacial development, including Prx1, Prx2, Msx1, Msx2, Endothelin-1 (Edn1), and Endothelin-A receptor (ETA). Mice null for Prx1/2, Msx2, Edn1, or ETA have craniofacial and ear defects that have some similarity to a portion of the defects in Dlx5/6−/−mice (ten Berge et al. 1998; Thomas et al. 1998; Clouthier et al. 2000; Satokata et al. 2000). However, the defects in all of the aforementioned studies are to a lesser extent and severity. Furthermore, recent observations suggest that Dlx6 and not Dlx5 acts as a transducer of Edn1-dependent expression of dHAND in the first branchial arch of mice (Charite et al. 2001). The mechanisms by which Dlx5 and Dlx6 regulate craniofacial and ear development are obviously complex and are the focus of ongoing studies. We propose that they function directly and indirectly as global coordinators of a number of signaling pathways that are critical for craniofacial development.
Role of Dlx5/6 in control of endochondral ossification
Dlx5 and Dlx6 are essential regulators of endochondral ossification, as Dlx5/6−/− mice have normal early cartilage formation of long bones at E14.5, but axial and appendicular skeleton show severe retardation in chondrocyte/cartilage and osteoblast differentiation and subsequent mineralization. Prechondrocyte condensation appears unaffected in mutant mice, as Sox9 expression patterns and levels are normal. The requirement for Dlx5/6 appears during the transition from prehypertrophic to hypertrophic chondrocytes. The onset of Col2a1 expression is normal in the Dlx5/6−/− embryos, but the differentiation to Col10a1- and Runx2-expressing cells in the chondrium is either blocked or severely retarded. In addition, the gene expression patterns for Fgf-Fgfr3 and Ihh-Pthrp-Pthrp-r signaling pathways, key regulators of chondrogenesis, are likewise blocked or severely retarded. Disruption of these pathways can result in a delay in osteogenesis that is secondary to cartilage maturation, as Ihh has recently been shown to couple the transition from chondrogenesis to osteogenesis (Chung et al. 2001).
Dlx5 and Dlx6 could also be direct regulators of osteogenesis or osteoblast differentiation. Dlx5-deficient mice exhibit a mild delay in ossification of long bones, without effecting Runx2 expression (Acampora et al. 1999). It has been proposed that Dlx5 may be a downstream target of Runx2 or act independently, but in a similar role. Dlx5 and Dlx6 do not appear to be direct targets of Runx2, because Runx2-deficient mice almost completely fail to form bone (Komori et al. 1997; Otto et al. 1997). They do appear to act independently in the activation of Osteocalcin, as Osteocalcin expression is absent in skeletal sections of Dlx5/6−/− mice, while perichondral Runx2 expression is normal in adjacent sections. However, the present data suggest that Dlx5 and Dlx6 regulate cartilage maturation, which in turn regulates the onset of osteogenesis.
In vitro overexpression of Dlx5 can induce Osteocalcin production in murine osteoblasts (Miyama et al. 1999). Furthermore, there is increasing evidence that Dlx5 promotes activation of Osteocalcin by forming heterodimers with Msx2, which antagonizes the Msx2-mediated repression of Osteocalcin (Newberry et al. 1998; Shirakabe et al. 2001). Overexpression of Msx2 prevents osteoblast differentiation, whereas inhibition of Msx2 translation accelerates differentiation (Dodig et al. 1999; Liu et al. 1999). In addition, Msx2-deficient mice have alterations in both chondrogenesis and osteogenesis that cause defective endochondral bone formation (Satokata et al. 2000). Therefore, delayed endochondral ossification in Dlx5/6−/− mice could be the result of increased Msx2 activity, in the absence of altered Msx2 or Runx2 gene expression. Another possibility is that Dlx5/6 act along parallel pathways for the promotion and linkage of chondrocyte and osteoblast maturation. Dlx5/6 could possibly act as potentiators of Sox5 and Sox6 function. Sox5 and Sox6 are possible downstream targets of Sox9 and are essential for the transition from prehypertrophic to hypertrophic chondrocytes (Smits et al. 2001). This could be the basis for the reduced number of Runx2-expressing hypertrophic chondrocytes within the chondrium of Dlx5/6 null embryos. A reduction in the number of Runx2-expressing chondrocytes could subsequently result in a retardation of bone formation, as Runx2 is critical for hypertrophic differentiation and vascular invasion (Zelzer et al. 2001). This assumes that the total number or concentration of Runx2-expressing cells is critical. Alternately, Dlx5/6 could promote the differentiation of a mesenchymal progenitor to a mature osteoblast within the perichondrium. Perichondrial Runx2 expression or other factors might partially compensate for the loss of Dlx5/6; however, the compensation is incomplete and results in delayed endochondral ossification and stunted bone growth.
Role of Dlx5/6 in control of limb development
Dlx5/6 control proximal-distal patterning in the murine hindlimb by maintaining the medial portion of the AER. Degeneration of the AER in Dlx5/6−/− mice results in a phenocopy of SHFM1. The SHFM in Dlx5/6 null mice has similarities to the Dac mouse model of SHFM3; however, in Dac mice, the AER degenerates owing to aberrant cell death within the AER (Seto et al. 1997; Crackower et al. 1998). In Dlx5/6−/− embryos, no alterations in cell death were observed in the AER or adjacent mesenchyme. In the Dlx5/6−/− embryos, expression of Fgf8 and Msx2 was lost in the medial portion of the AER by E11.5, while Dlx5/6-lacZ was still being expressed. Since Dlx5/6 are normally expressed in the AER with expansion into the PZ by E12.5, they appear to be essential for the survival of at least one cell population in the AER. Observations from analyses of Dac mice have implicated Fgf8 as a regulator of AER maintenance and the cause of SHFM. The present results rule out Fgf8 as the cause of Dlx5/6-mediated SHFM because Dlx5/6−/− mice have decreased cell proliferation in the AER by E10.5 at a time when Fgf8 expression is normal. In addition, conditional loss of Fgf8 expression in the mouse limb bud does not result in any form of SHFM (Lewandoski et al. 2000; Moon and Capecchi 2000). In fact, the limb defects reported in conditional Fgf8 null mice are less severe than those observed in the Dlx5/6 null embryos.
Dlx5 and Dlx6 show redundant control of AER maintenance, as mice lacking Dlx5 alone have no obvious limb defects (Acampora et al. 1999; Depew et al. 1999), and overexpression of Dlx5 in the AER of Dlx5/6−/− mice rescues the SHFM in the absence of any Dlx6 expression. The targets of Dlx5 and/or Dlx6 protein function appear to be at maximal levels in wild-type mice, as overexpression of Dlx5 on a wild-type genetic background has no effect on limb development. One possible model consistent with these results would have wild-type Dlx5 and Dlx6 functioning as transcriptional repressors of an overlapping set of downstream target genes. When the levels of Dlx5- and Dlx6-mediated repression fall below a critical threshold owing to loss of Dlx5 and Dlx6 alleles, certain genes within the AER may become active and affect either directly or indirectly the proliferation of cells within the AER. On a wild-type background, the overexpression of Dlx5 via an introduced transgene may have no effect, as the normal targets of Dlx5 and Dlx6 are already fully repressed. Naturally, converse models with Dlx5 and Dlx6 functioning as “activators” of a repressor would also be consistent with the observed results.
In humans, SHFM can be inherited in a regular dominant manner over multiple generations, yet also show lack of penetrance with skipped generations or irregular ‘dominant’ inheritance. In mice, the loss of a single Dlx5/6 locus (gene pair) is fully recessive with regards to the SHFM phenotype. These results might indicate that the human genome contains certain combinations of genetic modifiers which make limb development more sensitive to reduced levels of Dlx5/6 protein or possibly that certain types of SHFM mutations in human function in a dominant-negative manner, possibly by negatively interfering with the normal function of a shared cofactor.
In conclusion, Dlx5 and Dlx6 are dynamic regulators of mammalian development, which are absolutely required for proper craniofacial and skeletal development and which display overlapping genetic functions in all tissues in which they are expressed. In addition, they appear to act as essential regulators of chondrogenesis and osteogenesis. Finally, they are essential for maintenance of the medial portion of the hindlimb AER, as their loss leads to AER degeneration that results in a phenocopy of SHFM1 with craniofacial defects. These results are also the first example of an evolutionary conserved role for mammalian Dlx genes in limb outgrowth and development, a function the Dlx/Dll gene family appears to share among all species with appendages.
Materials and methods
Generation of Dlx5/6 null mutant and AER-Dlx5 transgenic mice
Three overlapping lambda clones containing Dlx5 and Dlx6 from a 129/Sv genomic library were obtained following a low-stringency screen performed as described (Chen et al. 1996). The inserts from the phage were subcloned into plasmid pTZ18 for restriction mapping and DNA sequencing, which confirmed that they contained the genes for Dlx5 and Dlx6. An artificial NruI site was subsequently introduced into the homeobox sequence of Dlx6 by site-directed mutagenesis. The Dlx5/6 targeting construct pSL99 was generated using a 12-kb BglII-NruI homology arm spanning the 5′ end of the Dlx6 gene up to the artificial NruI site in the Dlx6 homeobox. The second homology arm of the targeting construct corresponded to a 1.5 kb SalI-BglII genomic fragment overlapping the 5′ end of Dlx5 and containing exon I (exon II and exon III encode the homeodomain). The ires-lacZ-neo cassette (Wang et al. 2001) was inserted with the ires sequence abutting the Dlx6 homeobox sequence; thus, a correctly targeted clone would lack a functional Dlx5 and Dlx6 homeodomain and associated C-terminal amino acids, which should result in null alleles for both genes. The ires-lacZ sequences therefore fall under the control of the Dlx6 5′ transcriptional regulatory regions, and any regulatory sequences that lay between Dlx5 and Dlx6 would thus be removed. ES cell electroporation, selection, Southern blotting, chimera production, and testing were performed essentially as described (Wang et al. 2001).
The AER-specific transgenes were constructed using the Msx2 AER-specific enhancer/promoter (Liu et al. 1994) kindly provided by Robert Maxson (University of Southern California Medical School, Los Angeles). To generate the AER-Dlx5 transgene, the 0.5 kb Msx2 AER-specific enhancer/promoter fragment was ligated to the 0.9 kb Dlx5 ORF (kindly provided by Cory Abate-Shen, UMDNJ, Piscataway, NJ), followed by a 0.96 kb BglII-NotI fragment derived from the 3′ untranslated region of Drosophila Hmx (DHmx) cDNA (Wang et al. 2000) inserted in the inverted orientation, which serves as a noncoding transgene-specific “tag” sequence employed specifically for RNA in situ hybridization purposes. Finally, a 0.25 kb SV40-derived polyadenylation sequence was ligated to the 3′ end (Frasch et al. 1995). To generate the AER-AP transgene, the above-mentioned Msx2 AER-specific fragment was ligated to the ires-alkaline phosphatase reporter gene (Li et al. 1997), which places the alkaline phosphatase cDNA under the control of the Msx2 AER-specific enhancer/promoter sequences. The above-mentioned 0.25 kb SV40-derived polyadenylation sequence was ligated to the 3′ end as well. Both transgenes were gel purified, mixed in equal molar ratios, and microinjected into 1-cell mouse embryos as described (Frasch et al. 1995). Southern blotting of tail tip DNA identified ten stable transgenic founder lines carrying either one or both transgene constructs. Transgenic lines having cointegrated both transgenes were identified by continual cosegregation of the two transgene alleles after several sequential generations of outbreeding (transgenic X wild-type) and were employed for further analysis.
Embryos ranging in age from E8.5 to 18.5 were collected for whole-mount β-galactosidase staining, as described (Wang et al. 2001). Noon of the vaginal plug date was designated as E0.5. AER-Dlx5/AER-AP transgenic embryos, ranging in age from E8.5 to E14.5, were collected for whole-mount alkaline phosphatase staining (Li et al. 1997) and RNA in situ hybridization (Wang et al. 2001) as described.
Skeletal, histological, and RNA in situ analyses
Whole-mount cartilage and ossified skeletal elements were analyzed by staining collected embryos with Alcian blue or combined Alcian blue/alizarin red. For cartilage staining, E12.5 to E14.5 embryos were fixed overnight in Bouin's solution, stained in Alcian blue, and cleared as described (Tribioli and Lufkin 1999). For combined cartilage/bone analysis, E15.5 to E18.5 embryos were collected, and skin and internal organs were removed and fixed overnight in 95% ethanol. Skeletal preparations were then stained with Alcian blue followed by alizarin red staining and then cleared, as described (Tribioli and Lufkin 1999).
For histological analysis of scapulas, embryos were collected at E16.5 and the forelimbs were isolated, oriented, and embedded in paraffin for serial transverse sectioning (7 μm) using standard histological techniques. The spine of scapula (Ss) was used as an anatomical landmark to match control and mutant sections for staining or RNA in situ hybridization (described below). Matched scapula sections were stained with Alcian blue or von Kossa's techniques as described (Sheehan and Hrapchak 1987; Tribioli and Lufkin 1999).
In situ hybridization experiments on transverse paraffin embedded scapula sections (described above) were performed as described (Wang et al. 2001). The following cDNAs were used to synthesize antisense [35S]UTP-RNA probes: 500 bp Col2a1, 500 bp Col10a1, 270 bp Runx2, 470 bp Osteocalcin, 400 bp Fgfr3, 1800 bp Ihh, 500 bp Pthrp-r, and 500 bp Sox9. Nonradioactive whole-mount in situ hybridization experiments were performed on embryos collected between E9.5 and E14.5, as described (Wang et al. 2001). The following cDNAs were used to synthesize antisense digoxigenin-11-UTP-RNA probes: 400 bp Fgf8, 1300 bp Lmx1b, 1000 bp Msx2, 960 bp DHmx, 330 bp Dlx5 5', 560 bp Dlx2, and 2600 bp Shh.
Cell proliferation and apoptosis
Cell proliferation was determined by measuring the incorporation of 5-bromo-2'-deoxyuridine (BrdU) into the cells of E10.5 to E13.5 embryos. Embryos were collected from pregnant mice one hour following a 50 μg/g intraperitoneal BrdU injection and processed as described (Tribioli and Lufkin 1999; Wang et al. 2001). Apoptotic cell death was determined by measuring cells containing fragmented DNA utilizing the ApopTag kit (Intergen), as described (Ahuja et al. 1997). Briefly, tissue sections were postfixed with ethanol:acetic acid (2:1) followed by quenching of endogenous peroxidases with 3% hydrogen peroxide. Sections were then treated with digoxigenin-11-dUTP in the presence of terminal transferase for 90 min at 37°C. The digoxigenin-11-dUTP reaction to the free end of DNA was detected by using diaminobenzidine as substrate for a peroxidase conjugated antidigoxigenin antibody. At least three embryos for each genotype at each embryonic stage were examined for either cell proliferation or apoptosis.
Acknowledgments
We thank Robert Maxson for the Msx2 AER-specific enhancer, Cory Abate-Shen for the Dlx5 ORF cDNA, Edoardo Boncinelli, Mitch Goldfarb, Jill Helms, Randy Johnson, Gerard Karsenty, Peter Koopman, Gail Martin, Andy McMahon, Shintaro Nomura, Bjorn Olsen, Francesco Ramirez, John Rubenstein, David Sassoon, and Motohiko Sato for providing RNA in situ probes. The NIH (AR43449, DE013076, and DE013741) has supported different parts of this work.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Thomas.Lufkin@mssm.edu; FAX 212-860-9279.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.988402.
References
- Acampora D, Merlo GR, Paleari L, Zerega B, Postiglione MP, Mantero S, Bober E, Barbieri O, Simeone A, Levi G. Craniofacial, vestibular and bone defects in mice lacking the Distal-less-related gene Dlx5. Development. 1999;126:3795–3809. doi: 10.1242/dev.126.17.3795. [DOI] [PubMed] [Google Scholar]
- Ahuja HS, Zhu Y, Zakeri Z. Association of cyclin-dependent kinase 5 and its activator p35 with apoptotic cell death. Dev Genet. 1997;21:258–267. doi: 10.1002/(SICI)1520-6408(1997)21:4<258::AID-DVG3>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Capdevila J, Izpisua Belmonte JC. Patterning mechanisms controlling vertebrate limb development. Annu Rev Cell Dev Biol. 2001;17:87–132. doi: 10.1146/annurev.cellbio.17.1.87. [DOI] [PubMed] [Google Scholar]
- Chai CK. Dactylaplasia in mice a two-locus model for development anomalies. J Hered. 1981;72:234–237. doi: 10.1093/oxfordjournals.jhered.a109486. [DOI] [PubMed] [Google Scholar]
- Charite J, McFadden DG, Merlo G, Levi G, Clouthier DE, Yanagisawa M, Richardson JA, Olson EN. Role of Dlx6 in regulation of an endothelin-1-dependent, dHAND branchial arch enhancer. Genes Dev. 2001;15:3039–3049. doi: 10.1101/gad.931701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Li X, Wang W, Lufkin T. Dlx5 and Dlx6: An evolutionary conserved pair of murine homeobox genes expressed in the embryonic skeleton. Ann N Y Acad Sci. 1996;785:38–47. doi: 10.1111/j.1749-6632.1996.tb56242.x. [DOI] [PubMed] [Google Scholar]
- Chung UI, Schipani E, McMahon AP, Kronenberg HM. Indian hedgehog couples chondrogenesis to osteogenesis in endochondral bone development. J Clin Invest. 2001;107:295–304. doi: 10.1172/JCI11706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouthier DE, Williams SC, Yanagisawa H, Wieduwilt M, Richardson JA, Yanagisawa M. Signaling pathways crucial for craniofacial development revealed by endothelin-A receptor-deficient mice. Dev Biol. 2000;217:10–24. doi: 10.1006/dbio.1999.9527. [DOI] [PubMed] [Google Scholar]
- Crackower MA, Motoyama J, Tsui LC. Defect in the maintenance of the apical ectodermal ridge in the Dactylaplasia mouse. Dev Biol. 1998;201:78–89. doi: 10.1006/dbio.1998.8938. [DOI] [PubMed] [Google Scholar]
- Crackower MA, Scherer SW, Rommens JM, Hui CC, Poorkaj P, Soder S, Cobben JM, Hudgins L, Evans JP, Tsui LC. Characterization of the split hand/split foot malformation locus SHFM1 at 7q21.3-q22.1 and analysis of a candidate gene for its expression during limb development. Hum Mol Gen. 1996;5:571–579. doi: 10.1093/hmg/5.5.571. [DOI] [PubMed] [Google Scholar]
- Depew MJ, Liu JK, Long JE, Presley JE, Meneses JJ, Pedersen RA, Rubenstein JL. Dlx5 regulates regional development of the branchial arches and sensory capsules. Development. 1999;126:3831–3846. doi: 10.1242/dev.126.17.3831. [DOI] [PubMed] [Google Scholar]
- Dodig M, Tadic T, Kronenberg MS, Dacic S, Liu YH, Maxson R, Rowe DW, Lichtler AC. Ectopic Msx2 overexpression inhibits and Msx2 antisense stimulates calvarial osteoblast differentiation. Dev Biol. 1999;209:298–307. doi: 10.1006/dbio.1999.9258. [DOI] [PubMed] [Google Scholar]
- Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: A transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- Faiyaz ul Haque M, Uhlhaas S, Knapp M, Schuler H, Friedl W, Ahmad M, Propping P. Mapping of the gene for X-chromosomal split-hand/split-foot anomaly to Xq26-q26.1. Hum Genet. 1993;91:17–19. doi: 10.1007/BF00230215. [DOI] [PubMed] [Google Scholar]
- Frasch M, Chen X, Lufkin T. Evolutionary-conserved enhancers direct region-specific expression of the murine Hoxa-1 and Hoxa-2 loci in both mice and Drosophila. Development. 1995;121:957–974. doi: 10.1242/dev.121.4.957. [DOI] [PubMed] [Google Scholar]
- Ianakiev P, Kilpatrick MW, Toudjarska I, Basel D, Beighton P, Tsipouras P. Split-hand/split-foot malformation is caused by mutations in the p63 gene on 3q27. Am J Hum Genet. 2000;67:59–66. doi: 10.1086/302972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KR, Lane PW, Ward-Bailey P, Davisson MT. Mapping the mouse dactylaplasia mutation, Dac, and a gene that controls its expression, mdac. Genomics. 1995;29:457–464. doi: 10.1006/geno.1995.9981. [DOI] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- Lewandoski M, Sun X, Martin GR. Fgf8 signalling from the AER is essential for normal limb development. Nat Genet. 2000;26:460–463. doi: 10.1038/82609. [DOI] [PubMed] [Google Scholar]
- Li X, Wang W, Lufkin T. Dicistronic LacZ and alkaline phosphatase reporter constructs permit simultaneous histological analysis of expression from multiple transgenes. Biotechniques. 1997;23:874–878. doi: 10.2144/97235st03. [DOI] [PubMed] [Google Scholar]
- Liu YH, Ma L, Wu LY, Luo W, Kundu R, Sangiorgi F, Snead ML, Maxson R. Regulation of the Msx2 homeobox gene during mouse embryogenesis: A transgene with 439 bp of 5′ flanking sequence is expressed exclusively in the apical ectodermal ridge of the developing limb. Mech Dev. 1994;48:187–197. doi: 10.1016/0925-4773(94)90059-0. [DOI] [PubMed] [Google Scholar]
- Liu YH, Tang Z, Kundu RK, Wu L, Luo W, Zhu D, Sangiorgi F, Snead ML, Maxson RE. Msx2 gene dosage influences the number of proliferative osteogenic cells in growth centers of the developing murine skull: A possible mechanism for MSX2-mediated craniosynostosis in humans. Dev Biol. 1999;205:260–274. doi: 10.1006/dbio.1998.9114. [DOI] [PubMed] [Google Scholar]
- Miyama K, Yamada G, Yamamoto TS, Takagi C, Miyado K, Sakai N, Ueno M, Shibuya H. A BMP-inducible gene, dlx5, regulates osteoblast differentiation and mesoderm induction. Dev Biol. 1999;208:123–133. doi: 10.1006/dbio.1998.9197. [DOI] [PubMed] [Google Scholar]
- Moon AM, Capecchi MR. Fgf8 is required for outgrowth and patterning of the limbs. Nat Genet. 2000;26:455–459. doi: 10.1038/82601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naski MC, Colvin JS, Coffin JD, Ornitz DM. Repression of hedgehog signaling and BMP4 expression in growth plate cartilage by fibroblast growth factor receptor 3. Development. 1998;125:4977–4988. doi: 10.1242/dev.125.24.4977. [DOI] [PubMed] [Google Scholar]
- Newberry EP, Latifi T, Towler DA. Reciprocal regulation of osteocalcin transcription by the homeodomain proteins Msx2 and Dlx5. Biochemistry. 1998;37:16360–16368. doi: 10.1021/bi981878u. [DOI] [PubMed] [Google Scholar]
- Nunes ME, Schutt G, Kapur RP, Luthardt F, Kukolich M, Byers P, Evans JP. A second autosomal split hand/split foot locus maps to chromosome 10q24-q25. Hum Mol Genet. 1995;4:2165–2170. doi: 10.1093/hmg/4.11.2165. [DOI] [PubMed] [Google Scholar]
- O'Quinn JR, Hennekam RC, Jorde LB, Bamshad M. Syndromic ectrodactyly with severe limb, ectodermal, urogenital, and palatal defects maps to chromosome 19. Am J Hum Genet. 1998;62:130–135. doi: 10.1086/301687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- Panganiban G. Distal-less function during Drosophila appendage and sense organ development. Dev Dyn. 2000;218:554–562. doi: 10.1002/1097-0177(200008)218:4<554::AID-DVDY1023>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Qiu M, Bulfone A, Ghattas I, Meneses JJ, Christensen L, Sharpe PT, Presley R, Pedersen RA, Rubenstein JL. Role of the Dlx homeobox genes in proximodistal patterning of the branchial arches: Mutations of Dlx-1, Dlx-2, and Dlx-1 and -2 alter morphogenesis of proximal skeletal and soft tissue structures derived from the first and second arches. Dev Biol. 1997;185:165–184. doi: 10.1006/dbio.1997.8556. [DOI] [PubMed] [Google Scholar]
- Satokata I, Ma L, Ohshima H, Bei M, Woo I, Nishizawa K, Maeda T, Takano Y, Uchiyama M, Heaney S, et al. Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nat Genet. 2000;24:391–395. doi: 10.1038/74231. [DOI] [PubMed] [Google Scholar]
- Scherer SW, Poorkaj P, Massa H, Soder S, Allen T, Nunes M, Geshuri D, Wong E, Belloni E, Little S. Physical mapping of the split hand/split foot locus on chromosome 7 and implication in syndromic ectrodactyly. Hum Mol Gen. 1994;3:1345–1354. doi: 10.1093/hmg/3.8.1345. [DOI] [PubMed] [Google Scholar]
- Seto ML, Nunes ME, MacArthur CA, Cunningham ML. Pathogenesis of ectrodactyly in the Dactylaplasia mouse: aberrant cell death of the apical ectodermal ridge. Teratology. 1997;56:262–270. doi: 10.1002/(SICI)1096-9926(199710)56:4<262::AID-TERA5>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Sheehan DC, Hrapchak BB. Theory and practice of histotechnology. Columbus, OH: Battelle Press; 1987. [Google Scholar]
- Shirakabe K, Terasawa K, Miyama K, Shibuya H, Nishida E. Regulation of the activity of the transcription factor Runx2 by two homeobox proteins, Msx2 and Dlx5. Genes Cells. 2001;6:851–856. doi: 10.1046/j.1365-2443.2001.00466.x. [DOI] [PubMed] [Google Scholar]
- Simeone A, Acampora D, Pannese M, D'Esposito M, Stornaiuolo A, Gulisano M, Mallamaci A, Kastury K, Druck T, Huebner K. Cloning and characterization of two members of the vertebrate Dlx gene family. Proc Natl Acad Sci. 1994;91:2250–2254. doi: 10.1073/pnas.91.6.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits P, Li P, Mandel J, Zhang Z, Deng JM, Behringer RR, de Croumbrugghe B, Lefebvre V. The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev Cell. 2001;1:277–290. doi: 10.1016/s1534-5807(01)00003-x. [DOI] [PubMed] [Google Scholar]
- Tackels-Horne D, Toburen A, Sangiorgi E, Gurrieri F, de Mollerat X, Fischetto R, Causio F, Clarkson K, Stevenson RE, Schwartz CE. Split hand/split foot malformation with hearing loss: First report of families linked to the SHFM1 locus in 7q21. Clin Genet. 2001;59:28–36. doi: 10.1034/j.1399-0004.2001.590105.x. [DOI] [PubMed] [Google Scholar]
- Takeda S, Bonnamy JP, Owen MJ, Ducy P, Karsenty G. Continuous expression of Cbfa1 in nonhypertrophic chondrocytes uncovers its ability to induce hypertrophic chondrocyte differentiation and partially rescues Cbfa1-deficient mice. Genes Dev. 2001;15:467–481. doi: 10.1101/gad.845101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temtamy SA, McKusick VA. The genetics of hand malformations. Birth Defects Orig Artic Ser. 1978;14:1–619. [PubMed] [Google Scholar]
- ten Berge D, Brouwer A, el Bahi S, Guenet JL, Robert B, Meijlink F. Mouse Alx3: An aristaless-like homeobox gene expressed during embryogenesis in ectomesenchyme and lateral plate mesoderm. Dev Biol. 1998;199:11–25. doi: 10.1006/dbio.1998.8921. [DOI] [PubMed] [Google Scholar]
- Thomas T, Kurihara H, Yamagishi H, Kurihara Y, Yazaki Y, Olson EN, Srivastava D. A signaling cascade involving endothelin-1, dHAND and msx1 regulates development of neural-crest-derived branchial arch mesenchyme. Development. 1998;125:3005–3014. doi: 10.1242/dev.125.16.3005. [DOI] [PubMed] [Google Scholar]
- Tribioli C, Lufkin T. The murine Bapx1 homeobox gene plays a critical role in embryonic development of the axial skeleton and spleen. Development. 1999;126:5699–5711. doi: 10.1242/dev.126.24.5699. [DOI] [PubMed] [Google Scholar]
- Wang W, Chan EK, Baron S, Van de Water T, Lufkin T. Hmx2 homeobox gene control of murine vestibular morphogenesis. Development. 2001;128:5017–5029. doi: 10.1242/dev.128.24.5017. [DOI] [PubMed] [Google Scholar]
- Wang W, Lo P, Frasch M, Lufkin T. Hmx: An evolutionary conserved homeobox gene family expressed in the developing nervous system in mice and Drosophila. Mech Dev. 2000;99:123–137. doi: 10.1016/s0925-4773(00)00488-3. [DOI] [PubMed] [Google Scholar]
- Zelzer E, Glotzer DJ, Hartmann C, Thomas D, Fukai N, Soker S, Olsen BR. Tissue specific regulation of VEGF expression during bone development requires Cbfa1/Runx2. Mech Dev. 2001;106:97–106. doi: 10.1016/s0925-4773(01)00428-2. [DOI] [PubMed] [Google Scholar]
- Zerucha T, Stuhmer T, Hatch G, Park BK, Long Q, Yu G, Gambarotta A, Schultz JR, Rubenstein JL, Ekker M. A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J Neuroscience. 2000;20:709–721. doi: 10.1523/JNEUROSCI.20-02-00709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]