Abstract
In Arabidopsis, WRKY factors comprise a large gene family of plant-specific transcriptional regulators controlling several types of plant stress responses. To understand the regulatory role of WRKY proteins during such processes, we identified targets of the senescence- and defense-associated WRKY6 factor. WRKY6 was found to suppress its own promoter activity as well as that of a closely related WRKY family member, indicating negative autoregulation. On the other hand, WRKY6 positively influenced the senescence- and pathogen defense-associated PR1 promoter activity, most likely involving NPR1 function. One novel identified target gene, SIRK, encodes a receptor-like protein kinase, whose developmental expression is strongly induced specifically during leaf senescence. The transcriptional activation of SIRK is dependent on WRKY6 function. Senescing leaves of wrky6 knockout mutants showed a drastic reduction, and green leaves of WRKY6 overexpression lines showed clearly elevated SIRK transcript levels. Furthermore, the SIRK gene promoter was specifically activated by WRKY6 in vivo, functioning very likely through direct W-box interactions.
Keywords: cDNA-AFLP, WRKY transcription factor, receptor kinase, SIRK, autoregulation, PR1
In plants, as in other organisms, many developmental processes and responses to different stress stimuli underlay complex regulatory mechanisms operating at the level of gene expression (Lemon and Tjian 2000). Consistent with this regulatory complexity, nearly 6% of the total genes within the Arabidopsis genome code for transcription factors (Riechmann et al. 2000). One major family of plant-specific transcriptional regulators in Arabidopsis is represented by the WRKY gene family, comprising 74 members. WRKY factors belong to the zinc-finger-type class of proteins (Eulgem et al. 2000). Although still poorly studied, WRKY factors have been implicated in the regulation of certain plant processes, such as pathogen defense, wound response, and senescence (Eulgem et al. 2000). To understand the biological significance of WRKY factors during such processes, their in vivo target genes must be identified. Potential WRKY target genes have been suggested based on the general binding activity of WRKY factors to their recognized cis-element, TGACC/T, or W box (Eulgem et al. 2000; Yu et al. 2001). Almost nothing is known concerning trans-regulating activities of defined WRKY proteins on different target gene promoters, although transactivating capabilities of WRKY factors have been shown (de Pater et al. 1996; Eulgem et al. 1999; Hara et al. 2000).
Recently, we characterized one member of the Arabidopsis WRKY family, designated WRKY6, in more detail (Robatzek and Somssich 2001). The strongest WRKY6 expression was observed during leaf senescence but was also found in certain other tissues including floral organ abscission zones. In addition, expression of WRKY6 was influenced by several external and internal stimuli often associated with senescence and plant defense. Based on inhibitor studies, WRKY6 could be classified as an immediate-early-type gene not requiring de novo protein synthesis for its activation (F. Turck and I.E. Somssich, pers. comm.). Therefore, WRKY6 function is most likely involved in regulating certain early steps of these processes. Consistent with its function as a transcriptional regulator, the WRKY6 protein was found to be exclusively localized to the plant cell nucleus.
Here, we report the use of Arabidopsis wrky6 knockout mutants and a WRKY6 overexpression line to monitor WRKY6 trans-regulation activity on individual gene promoters and to screen for target genes. Several putative targets were identified. Our studies reveal that WRKY6 can function both as a positive and negative regulator of transcription, and in particular we identified one potential direct target gene very likely encoding an important signaling component of leaf senescence and defense response.
Results
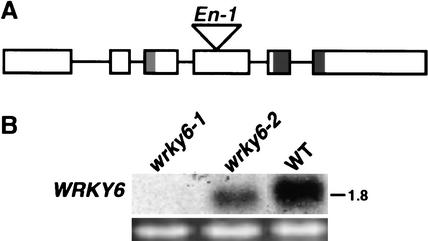
To study WRKY6 function and to isolate candidate target genes, we took advantage of a stable wrky6 knockout mutant, wrky6-2, which is derived from the En-1 insertion line wrky6-1 (Fig. 1A). The wrky6-1 line still carries an En-1 transposon inserted in the fourth exon of the WRKY6 gene, resulting in a total loss of WRKY6 transcript accumulation (Fig. 1B). In contrast, the wrky6-2 line carries a frame-shift mutation leading to a stop codon, owing to incorrect excision of the En-1 transposon, resulting in a deletion of 56 bp within the WRKY6 ORF. Although WRKY6 transcript was detectable in the wrky6-2 line, the translation product lacks 290 amino acids of the protein including its DNA-binding domain (data not shown).
Figure 1.
Identification of wrky6 knockout mutants. (A) Schematic representation of the Arabidopsis WRKY6 gene. Exons (boxes) and introns (lines) are indicated. The regions of the leucine zipper (light gray) and the WRKY domain (dark gray) as well as the position of the En-1 insertion (triangle) are shown. (B) Expression analysis of WRKY6 in senescent leaves of two knockout mutants, wrky6-1 and wrky6-2, compared with wild type (WT). The 28S rRNA band of the ethidium bromide-stained gel is shown for loading control.
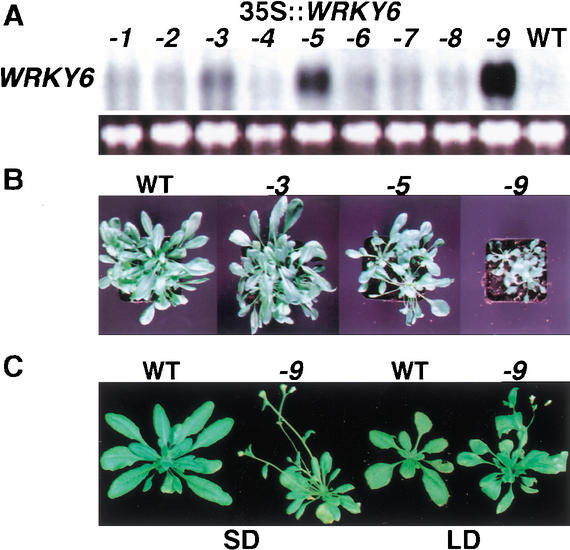
In addition, we used previously generated transgenic lines ectopically overexpressing WRKY6. Three lines, CaMV 35S::WRKY6-3, CaMV 35S::WRKY6-5, and CaMV 35S::WRKY6-9, showed clearly elevated levels of WRKY6 transcript in mature leaves, whereas no WRKY6 expression was observed in wild-type plants (Fig. 2A). The severity of the mutant phenotypes of the lines CaMV 35S::WRKY6-3, CaMV 35S::WRKY6-5, and CaMV 35S::WRKY6-9 strongly correlated with increasing expression levels of WRKY6 (Fig. 2B). The highest expressing line, CaMV 35S::WRKY6-9, was most strongly affected, showing a complex stress-related mutant phenotype (Fig. 2C). The plants were dwarfed with partly necrotic leaves, early flowering, and a reduction in their apical dominance.
Figure 2.
WRKY6 overexpression lines. (A) Expression of WRKY6 in nine independent T2 transgenic plants carrying a CaMV 35S::WRKY6 construct compared with wild type (WT). The ethidium bromide-stained 28S rRNA band is shown for loading control. (B) Dosage-dependence of the mutant phenotypes. Plants of the overexpressor lines CaMV 35S::WRKY6-3, -5, and -9 showing increasing levels of WRKY6 transcript are compared with wild type. (C) Comparison of the strongest overexpressor line CaMV 35S::WRKY6-9 with wild type. Plants were grown either under short-day (SD) or long-day (LD) conditions.
WRKY6 negatively influences its own promoter function
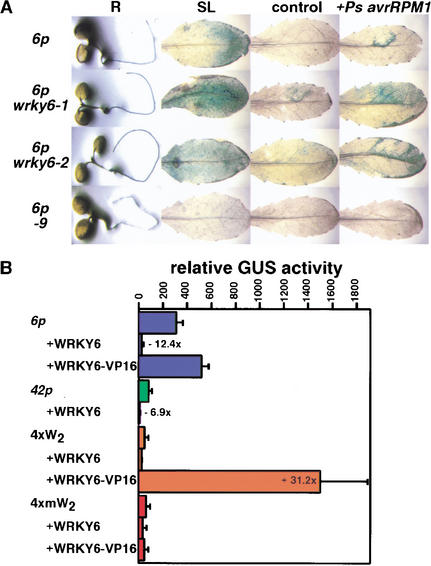
To follow the effects of WRKY6 on its own promoter activity, transgenic lines carrying a WRKY6 promoter–reporter fusion were crossed with wrky6-1 and wrky6-2 knockout mutants as well as with the CaMV 35S::WRKY6-9 line. The WRKY6 promoter function, monitored by GUS activity, was analyzed with respect to tissue-specific and pathogen-triggered expression. Whereas wild-type plants showed strong GUS activity in roots and senescing leaves, this effect was even more pronounced in the wrky6 knockout mutants (Fig. 3A). The opposite effect was observed in the WRKY6 overexpressor. Only very faint GUS signals could be detected in roots, and no signals were present in senescing leaves. This indicates that WRKY6 is negatively regulating its own promoter-mediated expression, which occurs in a broad spectrum of cell types.
Figure 3.
Repressor activity of WRKY6. (A) WRKY6 promoter-driven GUS reporter gene activity (6p) was monitored in wild-type plants, in the knockout mutants wrky6-1 and wrky6-2, as well as in the overexpressor line CaMV 35S::WRKY6-9 (-9). Shown are GUS-stained roots (R), senescing leaves (SL), and mature leaves 5 h postinoculation with 108-CFU bacterial solution (+Ps avrRPM1) or with 10 mM MgCl2 (control). (B) Transient cotransfection assays with different target gene promoters and WRKY6. Presented are relative activities of WRKY6 promoter (6p), WRKY42 promoter (42p), tetramerized W2-box (4xW2), and mutated tetramerized W2-box (4xmW2) driven GUS reporter gene constructs after transfection of cell culture-derived Arabidopsis protoplasts. Transient transfections were done either with reporter constructs alone or combined with an effector construct containing a CaMV 35S-driven WRKY6 cDNA (WRKY6) or a CaMV 35S-driven fusion of the WRKY6 cDNA to the VP16 activation domain (WRKY6–VP16). Each bar represents the median of four independent transfections. Normalized GUS values were obtained using a control luciferase plasmid for standardization. Relative fold induction or repression values ⩾twofold are depicted.
The repression effect was also seen under inducing conditions, namely, upon inoculation with the avirulent bacterial strain Pseudomonas syringae pv. tomato DC3000 (Ps avrRPM1). In contrast to wild-type plants, infected leaves of WRKY6 overexpression lines showed no inducible WRKY6 promoter-dependent GUS activity (Fig. 3A). On the other hand, loss of function of WRKY6 caused a clear enhancement of the WRKY6 promoter-mediated reporter gene activity. In these mutants, bacterial challenge as well as control treatments with MgCl2 resulted in increased GUS signal. In addition, the observed local restriction of GUS activity to infection sites in wild-type plants was clearly relaxed. The spread of GUS activity into noninoculated leaf areas of the wrky6 knockout mutants suggests that WRKY6 may be required for down-regulating its own expression once a certain threshold level has been achieved. Thus, WRKY6 may directly or indirectly function in limiting certain plant responses to a specific cell layer surrounding the site of pathogen ingress.
The enhancement of WRKY6 promoter-mediated GUS activity in the wrky6 knockout mutants points in the direction of WRKY6 showing repressor activity. To further address this question, we performed transient transfections in Arabidopsis protoplasts. Cotransfections of the WRKY6 promoter–reporter fusion with CaMV 35S::WRKY6 resulted in a drastic 12-fold repression activity of WRKY6 on its own transcription (Fig. 3B). Similarly, a sevenfold repression activity was seen using a WRKY42 promoter–reporter fusion. WRKY42, the WRKY6 homolog (Eulgem et al. 2000), also shares a similar promoter architecture of certain regulatory modules with WRKY6 (Robatzek and Somssich 2001).
The WRKY6 repression activity may be due to competition with other transcriptional activators or interference with coactivators. To clarify this point, we modulated the activity of WRKY6 by fusion to the strong activation domain of VP16, and observed diminishment of the repressor activity of WRKY6. The WRKY6–VP16 protein negated the negative effect on WRKY6 promoter activity, showing, instead, a slight induction above background values (Fig. 3B). The VP16 fusion to WRKY6 was not affecting nuclear targeting nor specific promoter-binding capability, because strong activation is observed for WRKY6–VP16 when a tetramerized W2-box element was used to drive expression of the reporter gene, resulting in a 31-fold induction of GUS activity. No such increase was seen when a block mutation was introduced into the W-box motif. This strongly suggests that WRKY6 binds to W-box elements, which is in perfect agreement with all previous reports about cognate binding sites of WRKY factors (Eulgem et al. 2000). Whether W boxes are the only recognized cis-acting element of WRKY6 needs further elucidation.
Positive WRKY6 activity on PR1 promoter function
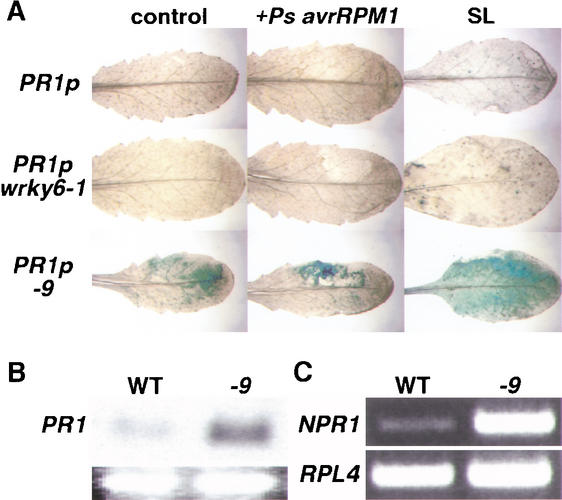
PR-type genes were previously described to be potential WRKY target genes (Eulgem et al. 2000). Given that PR1 contains several W boxes within its promoter (Maleck et al. 2000), including one involved in negatively regulating expression during systemic acquired resistance (SAR; Lebel et al. 1998), we tested whether PR1 gene expression is influenced by WRKY6. For this, we crossed PR1 promoter–reporter transgenic lines with the wrky6-1 knockout mutant and the CaMV 35S::WRKY6-9 line (Fig. 4A). Upon local infiltration of mature leaves with avirulent bacteria, the PR1-promoter-mediated level of GUS activity was strikingly high in the WRKY6 overexpression line, whereas in the wrky6 knockout mutant background, GUS activities similar to wild type were found. This pathogen-inducibility was detectable at much earlier time points (3–5 h) than in wild-type plants (24–48 h, Fig. 4A; data not shown). Clear GUS activities were also present in control inoculations with MgCl2, whereas only low GUS activities were observed in completely untreated control leaves. Leaf senescence slightly induces PR1 gene expression (Robatzek and Somssich 2001), which is drastically increased in the WRKY6 overexpression line (Fig. 4A; SL). In contrast, no other plant tissue showed such an up-regulation of the PR1 promoter activity (data not shown). Increased basal PR1 gene expression was confirmed by RNA blot analysis (Fig. 4B). Together these data indicate that WRKY6 overexpression causes a general up-regulation of PR1, but more importantly, mediates a stronger and faster response under stress inducing conditions (Fig. 4A). Recently, Yu et al. (2001) showed that WRKY factors can activate NPR1 via W boxes present within its promoter. NPR1 is a key regulator of the SAR-dependent signal pathway leading to PR1 expression (Cao et al. 1997). As shown in Figure 4C, overexpression of WRKY6 also results in elevated NPR1 transcript levels. This would suggest that WRKY6 action on PR1 seems to be indirect and likely involves NPR1 function.
Figure 4.
Effect of WRKY6 overexpression on PR1 and NPR1. (A) Activation of PR1-promoter-driven reporter gene activity by WRKY6. GUS activity observed in representative leaves of transgenic plants harboring the PR1 promoter-driven reporter gene in wild type (PR1p), wrky6-1 mutant, and WRKY6-9 (-9) overexpressor. GUS-stained mature leaves are shown either 5 h postinoculation with 108-CFU bacterial solution (+Ps avrRPM1) or with 10 mM MgCl2 (control), as well as senescing leaves (SL). (B) RNA blot analysis of PR1 in mature leaves of wild-type (WT) and WRKY6 overexpression lines (-9). The 28S rRNA ethidium bromide-stained band is shown as loading control. (C) RT–PCR analysis of NPR1 in mature leaves of wild-type (WT) and WRKY6 overexpression lines (-9). For control RPL4 (ribosomal protein L4) transcript was amplified.
Potential genes regulated by WRKY6
To isolate additional candidate target genes, we applied a cDNA-AFLP-based differential display approach (Durrant et al. 2000). We compared transcript populations either derived from roots of wild-type plants and wrky6-2 mutants, because roots are tissues of high WRKY6 expression (Robatzek and Somssich 2001), or derived from all aerial parts of wild-type and CaMV 35S::WRKY6-9 plants. Screening of ⩾12,000 different cDNA fragments resulted in the identification of 154 differentially expressed clones from root transcripts, designated R1–R154, and 63 clones from aerial part transcripts, designated P1–P63. The expression of ∼44% of the R-clones and ∼59% of the P-clones was up-regulated in the wrky6-2 and in the CaMV 35S::WRKY6-9 mutants, respectively.
Sequence analysis of the cDNA-AFLP fragments revealed, in 33% of the cases, homologies to only hypothetical ORFs. A number of candidates showed strong similarities to Ca2+-, defense-, and senescence-related genes, as well as different types of kinases, including receptor-like protein kinases (Table 1). To confirm the cDNA-AFLP results, we selected clones based on their sequence homologies and differential expression pattern. RT–PCR studies using independent RNA preparations verified 70% of the tested R-clones and 50% of the P-clones (data not shown).
Table 1.
cDNA-AFLP fragments and homologies to sequences in the Arabidopsis database
| Clonea
|
Expressionb
|
Accession no.c
|
Similarity
|
W
|
as1d
|
|---|---|---|---|---|---|
| R11e | +++ | AC024081 | JA-regulatory protein NAC2 | 2 | 0 |
| R16 | ++ | AC011698 | NAM-like protein | 2 | 0 |
| R18 | −− | T06055 | kinesin domain containing protein | 4 | 2 |
| R40 | ++ | AB023034 | xylosidase | 2 | 1 |
| R41e | −− | AC010718 | putative calmodulin | 5 | 1 |
| R43 | −− | AB003590 | sulfate transporter | 2 | 1 |
| R48 | +++ | AJ270302 | putative β-galactosidase | 3 | 0 |
| R50 | −− | AC010926 | putative casein kinase | 3 | 1 |
| R52/53 | −− | AL163818 | Ca2+-transporting ATPase-like protein | 4 | 2 |
| R62e | +++ | S66346 | SEN1 | 5 | 1 |
| R64 | +++ | X89866 | glutathione peroxidase | 0 | 0 |
| R67e | −−− | AC006551 | alcohol dehydrogenase-like | 2 | 0 |
| R68 | −−− | T02156 | glucosidase homolog | 4 | 1 |
| R72 | −−− | AC007651 | putative glutathione transferase | 3 | 1 |
| R74e | −− | T05493 | thaumatin-like PR protein | 3 | 0 |
| R81 | −−− | AF058919 | putative calmodulin-binding heat shock protein | 3 | 1 |
| R92 | −− | AC000132 | receptor-like protein kinase | 2 | 2 |
| R96 | −− | AC011708 | putative pectin esterase | 4 | 1 |
| R102e | +++ | AC006931 | putative lipase | 0 | 1 |
| R105 | − | AF217546 | calmodulin-binding protein | 1 | 0 |
| R129 | ++ | AC009519 | MAP kinase-like protein | 1 | 2 |
| R140e | +++ | 2924653 | heat shock protein HSP81-2 | 4 | 0 |
| R143 | + | AC020579 | putative disease resistance protein | 5 | 3 |
| R144e | ++ | Y14590 | class IV chitinase | 6 | 1 |
| P2 | +++ | AC005275 | putative xyloglucan endotransglycosylase | 1 | 2 |
| P4 | +++ | AL162506 | fructose-bisphosphate aldolase-like protein | 5 | 1 |
| P7e | −− | AC002339 | zinc protease-like protein | 2 | 2 |
| P24e | +++ | T00540 | receptor-like protein kinase | 9 | 0 |
| P27e | +++ | AC002336 | putative expansin | 3 | 0 |
| P29 | − | S51478 | drought-induced protein Di19 | 1 | 0 |
| P34e | ++ | AC005724 | putative calmodulin-binding protein | 2 | 2 |
| P35 | ++ | AL163812 | fructosidase-like protein | 2 | 0 |
| P38 | −−− | T09930 | thioredoxin homolog | 0 | 0 |
| P40 | − | T04549 | AP2 domain protein homolog | 2 | 0 |
| P55e | −−− | AC010676 | putative porin | 2 | 0 |
| P57 | − | AC006592 | homeobox factor HB6 | 2 | 0 |
| P58 | − | T02644 | ABC-type transport protein homolog | 2 | 1 |
Designated cDNA-AFLP fragments.
Differential expression of the cDNA-AFLP fragments detected in the null-mutant or the overexpression line compared to wild type.
GenBank accession numbers of identified genes corresponding to the cDNA-AFLP fragments.
Number of W boxes and as1-like elements present within 1 kb putative promoter sequence of corresponding genes.
Confirmed by RNA blot or RT–PCR analysis (others not tested).
Because W boxes, TGACC/T, are the cognate binding sites of WRKY factors (Eulgem et al. 2000), we searched 1-kb putative promoter sequences of these candidate target genes for their presence. In addition, we checked for as1-like elements (Rushton and Somssich 1998), which also contain the highly conserved TGAC core motif. Although a single W box within a promoter is sometimes sufficient to mediate WRKY-dependent gene expression, a clustering of W boxes is often observed (Eulgem et al. 1999; Maleck et al. 2000). Indeed, some of the isolated potential WRKY6 target genes contained numerous W boxes within their promoters (Table 1). Based on these data, the most promising WRKY6 target gene was chosen for further investigations.
The receptor-like protein kinase SIRK is a WRKY6 target
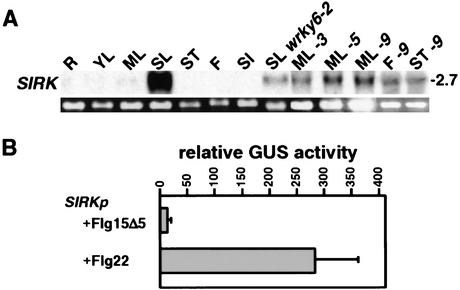
The gene (GenBank accession no. T00540) corresponding to the cDNA-AFLP fragment P24, showing induced expression in CaMV 35S::WRKY6-9 plants, encodes a typical leucine-rich repeat receptor-like protein kinase (Shiu and Bleecker 2001). Expression profiling using different plant tissues revealed a strong association of P24 with the process of senescence, it being highly induced in senescent leaves but not detectable in any of the other tested organs (Fig. 5A). Based on its expression pattern, we renamed P24 to AtSIRK for Arabidopsis thaliana senescence-induced receptor-like kinase. In contrast to the wild-type situation, the level of SIRK transcript detected in senescent leaves of the wrky6-2 knockout mutant was drastically reduced. Furthermore, elevated SIRK expression was also detected in mature leaves, stems, and flowers of WRKY6 overexpression lines. Taken together, these results strongly imply that high SIRK expression is dependent on WRKY6. Because the developmental expression patterns of WRKY6 and SIRK are only partly overlapping, transcriptional activation of SIRK by WRKY6 seems to be leaf senescence-specific.
Figure 5.
Dependence of SIRK gene expression on WRKY6. (A) Expression analysis of SIRK transcripts in different tissues, including roots (R), young leaves (YL), mature leaves (ML), senescing leaves (SL), stems (ST), flowers (F), or siliques (SI), derived from wild-type plants (WT), the wrky6-2 knockout mutant, or from the overexpressor lines -3, -5, and -9. The 28S rRNA band of the ethidium bromide-stained gel is shown as loading control. (B) Effect of the bacterial-derived elicitor flagellin 22 (Flg22) on SIRK promoter activity in transient transfection assays. Presented are relative activities of the SIRK promoter (SIRKp) driven GUS reporter gene after addition of the active (Flg22) and inactive (Flg15Δ5) forms of the elicitor. Each bar represents the median of four independent transfections. Normalized GUS values were obtained using a control luciferase plasmid.
In addition, WRKY6 expression is induced by bacterial pathogen infection (Fig. 3A). We therefore analyzed the responsiveness of the SIRK promoter to the bacterial elicitor flagellin (Felix et al. 1999). Transient transfection assays in protoplasts revealed an 18-fold increase of GUS reporter activity using the active versus the inactive elicitor (Fig. 5B). WRKY6 may therefore also play a role in this response.
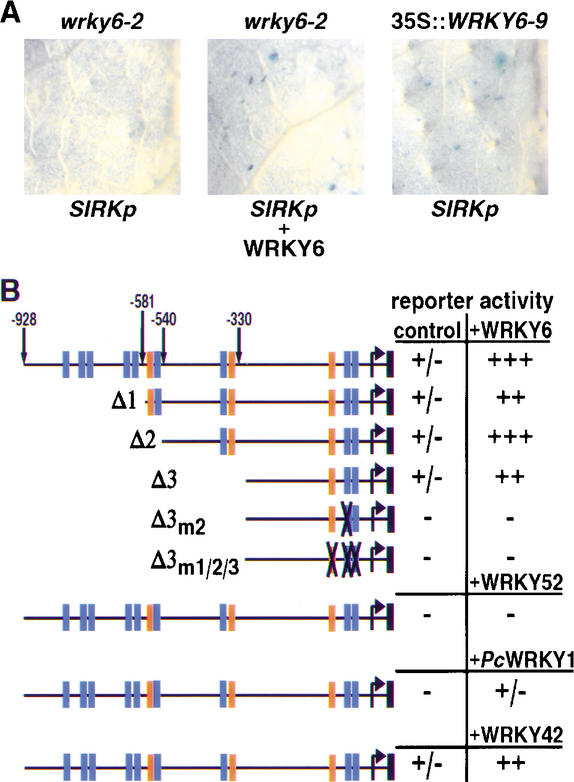
Transient transfections of green leaves were used to monitor WRKY6-dependent activation of the W-box-rich SIRK gene promoter, which in stable transgenic Arabidopsis SIRKp::GUS lines was shown to mediate leaf senescence-inducible expression (data not shown). Cobombardments of an SIRK promoter fusion to the GUS reporter gene with CaMV 35S::WRKY6 in either wrky6-2 knockout mutants or wild-type plants resulted in strong GUS activities, whereas the promoter–reporter construct on its own showed only faint background activities (Fig. 6A). Furthermore, bombardments of the promoter–reporter construct alone in WRKY6 overexpression lines showed strong GUS activities. Therefore, WRKY6 is able to transactivate SIRK gene expression in vivo.
Figure 6.
Specificity of the WRKY6–SIRK promoter interaction. (A) Representative leaves of four independent biolistic-mediated transient transfection assays using a 0.9-kb SIRK promoter-driven GUS reporter gene (SIRKp). Bombardments of detached mature leaves of wrky6-2 knockout mutants and the 35S::WRKY6-9 overexpressor line were done either with SIRKp alone or in combination with a CaMV 35S-driven WRKY6 cDNA construct (WRKY6). (B) Results of bombardment assays using various SIRK-promoter deletion/mutation GUS constructs as depicted schematically on the left. The respective sizes relative to the transcription start site (bent arrow) as determined by 5′RACE (O. Noubibou, pers. comm.; GenBank accession no. AF486619) are indicated by arrows. W boxes (TGACC/T) are marked by blue, TGACA motifs by orange rectangles, and mutated elements by crosses. SIRK promoter deletion constructs, Δ1, Δ2, Δ3, and mutation constructs, Δ3m2 and Δ3m1/2/3, were bombarded in detached mature wild-type leaves either alone (control) or in combinations with the effectors (+WRKY6, +WRKY52, +PcWRKY1, +WRKY42). Activation of the SIRK promoter constructs is indicated by relative values ranging from − (no GUS staining), +/− (1–5 GUS-positive cells in total), ++ (>20 GUS-positive cells per leave), to +++ (>50 GUS-positive cells per leave). Three independent experiments were performed.
Because the SIRK promoter contains nine W boxes, and WRKY6 could function through one or several of these, we analyzed a SIRK promoter–reporter deletion series (Fig. 6B). WRKY6 was still capable of activating the shortest deletion construct (Δ3) containing only two of the nine W boxes and one TGACA motif. Mutations within these three elements (Δ3m1/2/3) completely abolished its ability to activate the reporter gene. Interestingly, a single block mutation within the second W box between positions −43 and −49 bp (Δ3m2) equally led to total loss of function. This shows that, indeed, at least one W box is important for WRKY6 recognition. Furthermore, cobombardment with an SIRK promoter-derived construct spanning the four W boxes within region −581 to −736 bp did not lead to a significant increase of GUS activities above background values (data not shown). Therefore, WRKY6–SIRK-promoter interactions rely on more than just the presence of W-box motifs.
To show specificity on the protein side, we investigated cobombardements with two defense-associated WRKY factors, namely, WRKY52 and PcWRKY1 (Fig. 6B). In both cases, no obvious GUS activities were detected, indicating a specific requirement for WRKY6. However, WRKY42, the closest WRKY6 family homolog, was capable of activating the SIRK promoter (Fig. 6B). Regulation of SIRK promoter activity seems to involve functionally redundant members of the WRKY family.
Discussion
Functional redundancy within multigene families often complicates genetic attempts to define the role of individual members (Bouche and Bouchez 2001). This also appears to be the case for the wrky6 knockout mutation, which resulted in no obvious mutant phenotype. In certain cases, overexpression of the respective gene can give clues to its biological function. However, particularly with transcription factors like WRKY6, ectopic expression leading to nonphysiological concentrations of the protein can affect a plethora of regulatory networks and yield multiple mutant phenotypes, thereby negating conclusions derived from inference. Despite such problems, our results using cDNA-AFLP differential display indicate that the single WRKY6 knockout does result in altered gene expression profiles. This indicates that functional redundancy is not complete. Furthermore, several putative target genes identified in these comparative analyses (Table 1) corroborate our previous findings that WRKY6 is involved in controlling processes related to senescence and pathogen defense (Robatzek and Somssich 2001). These include genes encoding the senescence-associated protein 1, (SEN1), a protease; the jasmonic acid regulatory protein NAC2; a glutathione transferase (Nam 1997; Dong 1998); as well as several genes encoding defense-related proteins (R22, R74, R143, R144). The SEN1 gene promoter contains five W boxes within the first 1 kb of sequence. Its expression was strongly up-regulated in the wrky6 knockout mutant, indicating that WRKY6 may act as a negative regulator on this promoter. Additional genes identified in our study represent signaling components of calcium and kinase cascades, which also function during senescence and pathogen defense. Interestingly, similar sets of potential WRKY-regulated genes were identified in expression profiling experiments addressing SAR (Maleck et al. 2000; Petersen et al. 2000). Furthermore, chitinases and also receptor-like protein kinases have been proposed to be possible WRKY targets (Yang et al. 1999; Du and Chen 2000; Ohtake et al. 2000).
Both senescence and hypersensitive response, a successful defense strategy against numerous pathogens, are forms of programmed cell death (PCD). Because several defense-associated genes are expressed during leaf senescence, and defense-related mutants show alterations in senescence-associated gene expression, cross-talk between distinct PCD pathways do exist (Quirino et al. 1999; Morris et al. 2000).
WRKY6 activator and repressor function
All studied WRKY proteins have been shown to act as positive transcriptional regulators (de Pater et al. 1996; Eulgem et al. 1999; Hara et al. 2000). A negative function for WRKY factors was merely derived from inference (Lebel et al. 1998; Li et al. 1999). In this report, we showed that WRKY6 clearly acts as a negative regulator on its own and on WRKY42 expression, but the mechanism remains unknown. The WRKY6 protein does contain regions homologous to known trans-activation domains (Robatzek and Somssich 2001), but lacks obvious similarities to trans-repression domains (Hanna-Rose and Hansen 1996). Therefore, WRKY6 repressor activity may be direct, functioning via a novel type of repressor domain, or its action could be indirect through interaction/interference with other proteins.
Although we cannot exclude the possibility that the negative autoregulation of WRKY6 is mediated via W boxes, transcriptional repression of WRKY42 by WRKY6 points to another mechanism. The putative WRKY42 promoter sequence contains no W-box consensus motifs, indicating either an indirect WRKY6 effect or the involvement of other cis-acting elements. Such elements may be modifications of the W-box consensus, because several TGAC-core motifs of the TGACG (Rushton and Somssich 1998) or of the TGACA type (Desveaux et al. 2000) are present within the WRKY42 promoter.
WRKY6 acts as a positive regulator on PR1 expression. Most likely, this is because of an activation rather than a competition mechanism caused by ectopic WRKY6 expression, given that, apart from leaves, no such effect was observed in other tissues. Direct involvement of WRKY6 in PR1 transcription is supported by the presence of several W boxes within the PR1 promoter, and by the fact that elevated NPR1 levels alone are insufficient to induce PR1 (Cao et al. 1998). On the other hand, the further substantial increase of PR1 expression in the overexpressor line under stress conditions favors a more indirect role of WRKY6. The PR1 upstream regulator NPR1 has been shown to be a WRKY target gene (Yu et al. 2001), and WRKY6 may be one of its activators or alternatively impinge on the function of a specific WRKY factor. Despite these elevated levels for both NPR1 and PR1 in leaves of the WRKY6 overexpressor lines, we could not detect a significant enhancement of resistance or increased cell death toward compatible and incompatible strains of Pseudomonas syringae pv tomato DC3000 (lacking or carrying avrRPM1; data not shown). It should be noted that elevated levels of endogenous NPR1 and PR1 need not necessarily lead to resistance (Greenberg et al. 2000). One likely explanation is that the observed levels are insufficient, because it has been shown that NPR1 confers pathogen resistance in a dosage-dependent fashion (Cao et al. 1998).
Dual activities of transcription factors can be dependent on the cell environment and the type or level of signal input (Hoecker et al. 1995). Concentration-dependence is one mechanism of dual functionality by which transcription factors can act as activators or repressors (Ogbourne and Antalis 1998; Rushlow et al. 2001). Differing expression levels of WRKY6 may therefore determine whether target gene transcription is stimulated or repressed. Protein interactions and the abundance of interacting partners within different cell types or upon stress conditions contribute as well to the mechanism of dual functionality (Motohashi et al. 2000). This may also be valid for WRKY6, because it contains a leucine zipper capable of mediating dimerizations (S. Robatzek and I.E. Somssich, unpubl.).
The senescence-induced receptor kinase SIRK
Our data strongly imply that WRKY6 acts upstream of SIRK in the process of leaf senescence. This interaction appears to be direct, acting through at least one W box present within the SIRK promoter, and involving a specific requirement for WRKY6 function. We cannot, however, completely exclude alternative possibilities, for example, that WRKY6 induces other WRKY genes whose products interact with the W-box element. To date, SIRK is the only identified plant receptor kinase developmentally expressed solely during leaf senescence. One other receptor kinase, PvSARK from bean, has been associated with senescence, but is also detected in roots (Hajouj et al. 2000). The senescence-signaling pathway is often linked to pathogen defense (Quirino et al. 1999), and SIRK and WRKY6 are targets of both programs. Interestingly, the 1-kb SIRK promoter is capable of perceiving signals from these two cascades. Consistent with SIRK being a WRKY6 target gene, the temporal accumulation of SIRK mRNA upon Flg22 stimulation followed the rapid and transient increase of WRKY6 transcript in a slightly delayed manner (C.B. Zipfel and S. Robatzek, pers. comm.). Furthermore, preliminary results show that W-box elements are also required for flagellin responsiveness of this promoter (O. Noubibou, P. Rushton, and I.E. Somssich, pers. comm.). Whether common or distinct W boxes and WRKY factors mediate the signals from both pathways remains to be determined.
A connection between WRKY proteins and other receptor-like kinases as potential targets has been suggested based on the clustering of W boxes within their promoter regions and the ability of WRKY factors to bind to such elements in vitro (Du and Chen 2000; Ohtake et al. 2000). The expression of these receptor-like kinases was shown to be inducible upon treatment with salicylic acid (Ohtake et al. 2000), and the expression of one gene, RLK3, was also induced by pathogen attack (Czernic et al. 1999). Whether W boxes mediate these responses was not shown. Nevertheless, because WRKY6 expression is also up-regulated by salicylic acid and by bacterial infection (Fig. 3; Robatzek and Somssich 2001), WRKY6 may participate in their transcriptional control as well. Interestingly, the expression pattern of HAESA, another kinase gene with W-box clusters in its promoter (Ohtake et al. 2000), shows a strong overlap with that of WRKY6. Expression of both genes is highly activated in floral organ abscission zones (Jinn et al. 2000; Robatzek and Somssich 2001), suggesting another possible link between WRKY6 and a potential target gene in such cells. All of these receptor-like kinases show only ∼30% amino acid identity to SIRK; therefore, they most likely act in different signal perception/transduction pathways. WRKY6 could be a transcriptional regulator of distinct receptor kinase genes functioning in specific cells and during certain developmental stages in response to different external and internal signaling cues.
Database searches identified two additional receptor-like kinases with high homologies to SIRK. The proteins encoded by the genes F27F23.1 and F27F23.3 show 60.6% and 59.9% identity, respectively. The F27F23.3 gene contains six W boxes within its first 1 kb of promoter sequence, indicating that at least one other SIRK-related receptor kinase could be under the control of WRKY factors.
Receptor-like kinases serve as receivers and transducers of external and internal stimuli. Various input signals are transmitted through phosphorylation/dephosphorylation cascades, which lead to changes in gene expression patterns. To date, only a few receptor-like kinases have been linked to certain plant processes. These include CLV1 in meristem organization, ERECTA in organ shape, BRI1 in brassinolide signaling, FLS2 in flagellin signaling, HAESA in floral organ abscission, and BrSRK1 in self-incompatibility (Shiu and Bleecker 2001). WRKY proteins are expected to be substrates of kinases and/or phosphatases (Eulgem et al. 2000). This is consistent with recent identification of a set of specific potential WRKY effector genes being constitutively expressed in a MAP kinase mutant, mpk4, which negatively regulates SAR (Petersen et al. 2000). A hypothetical model derived from our results would suggest a dual function for WRKY6 during some stage of leaf senescence, which is initiated by binding of a senescence-triggered ligand to SIRK. Concomitant to this, expression of WRKY6 is induced. SIRK function activates a downstream kinase cascade resulting in modification of WRKY6 protein, thereby enabling it to activate the transcription of several genes including SIRK and to down-regulate WRKY6 expression.
As for other multigene families, unraveling the biological role of individual WRKY transcription factors and how they contribute to the establishment of the complex plant regulatory network remains a challenging endeavor. The identification of WRKY6 target genes, especially ones involved in the process of leaf senescence, is of particular importance given that, to date, nearly no regulatory components of leaf senescence are known (Nam 1997; Woo et al. 2001). Isolation of SIRK knockout mutants via reverse genetics or dsRNAi and their combination with other mutants affecting senescence and/or defense response will surely facilitate the molecular dissection of this important process as will the identification of additional components influencing or being influenced by WRKY6 function.
Materials and methods
Plant growth and treatments
Plant growth conditions for obtaining plant material, bacterial growth and infections, and histochemical GUS staining were performed as described by Robatzek and Somssich (2001).
Knockout mutants
A knockout mutation of the WRKY6 gene (GenBank accession no. AF331712) was identified by a PCR-based screen of an En-1 insertion population as described previously (Baumann et al. 1998). The combination of the WRKY6-specific primer 5′-ATC CCG TCG TGA CTA GAC ATT GAC-3′ and the En-1-specific primer 5′-GAG CGT CGG TCC CCA CAC TTC TAT AC-3′ led to the isolation of the line 6AAK67 as a wrky6 mutant. The En-1 insertion in the mutant (wrky6-1) was confirmed by Southern analysis, and its exact position following codon 263 determined by sequencing. The footprint within the wrky6-2 mutant was detected using WRKY6-specific primers flanking the original En-1 insertion site. Both mutants contain three additional En-1 insertions after twice back-crossing to wild-type plants. Homozygous plants for the wrky6 mutation were used for expression analysis.
Transgenic plants
WRKY6 cDNA was amplified by RT–PCR and introduced behind the CaMV 35S promoter into the XhoI and SacI sites in the pBT8 construct, a derivative of pBT2 (Weisshaar et al. 1991). Following digestion with ClaI and SacI, the CaMV 35S::WRKY6 fragment was introduced into the binary vector pGPTV (Koncz and Schell 1986). In addition to the WRKY6 coding region, the construct carries 37 bp of the 5′ untranslated region (UTR) and 64 bp of the 3′ UTR. The correctness of the constructs was verified by sequencing. Stable A. thaliana Col-0 transgenic lines were generated using the Agrobacterium tumefaciens-mediated gene-transfer procedure involving infiltration of inflorescences (Clough and Bent 1998). Independent transgenic lines were selected for kanamycin resistance and confirmed by Southern analysis. Plants of the T2 generation were used in detailed molecular and phenotypic studies.
Promoter reporter lines
6p::GUS (Robatzek and Somssich 2001) and PR1p::GUS (Lebel et al. 1998) were crossed into wrky6-1, wrky6-2 knockout mutants and the CaMV 35S::WRKY6-9 overexpressing line. Transgenic plants were selected for kanamycin resistance, and by Southern and PCR analysis. Expression studies were done using homozygous wrky6 mutants, and heterozygous CaMV 35S::WRKY6-9 lines.
Northern/RT–PCR analysis
Different tissues of A. thaliana plants ecotype Col-0 were used for total RNA extraction with the RNA/DNA-maxi kit (QIAGEN). In all cases, 10 μg of total RNA was loaded per lane, and the gels were blotted using standard molecular procedures (Sambrook et al. 1989). DNA probes were radioactively labeled by random priming using [α-32P]dCTP (Amersham) and the Ready-To-Go kit (Pharmacia).
RT–PCR was performed with 50 ng of total RNA, the NPR1-specific primers 5′-CTG TTG ATG GAC ACC ACC ATT GAT GG-3′ and 5′-GTC TGC GCA TTC AGA AAC TCC TTT AGG C-‘, or the RPL4-specific primers 5′-GTG ATA GGT CAG GTC AGG GAA CAA C3-‘ and 5′-CCA CCA CCA CGA ACT TCA CCG CGA GTC-‘, using the Ready-To-Go RT–PCR beads (Amersham) according to the manufacturer’s instructions.
cDNA-AFLP differential display
The method of cDNA-AFLP differential display was done as previously described (Durrant et al. 2000). For template construction, 1 μg of double-purified poly(A)+ mRNA derived from sterile-grown root tissue (wrky6-2 knockout mutants and wild type) and all aerial plant tissue (CaMV 35S::WRKY6-9 and NPTII gene transgenic wild type germinated under selective conditions) was used. Selective amplifications were done in primer combinations of Apo-WD10, 11, 12, 22, 58, and 63 with Mse-WD31 to WD46 (Durrant et al. 2000). Identified differential signals were re-amplified, cloned into the TOPO vector (Stratagene), and sequenced.
Transient transfections
For each reporter construct, the relevant promoter containing 5′-ATG upstream regions; 1315 bp (6p), 1132 bp (42p), and 928 bp (SIRKp) were amplified by PCR and introduced into the HindIII and BamHI sites of the pUC9-GUS reporter construct (van de Löcht et al. 1990). The promoter regions were fused translationally to the Escherichia coli uidA gene (Jefferson et al. 1987). These constructs therefore also contained 21 bp (6p), 21 bp (42p), and 9 bp (SIRKp) of the respective ORFs. The core TGAC motifs of the DNA elements were changed to ATTG within the SIRK-promoter deletion constructs (Δ3m2 and Δ3m1/2/3), as indicated in Figure 6B, using the megaprimer method (Landt et al. 1990). The pBT8 construct containing the CaMV 35S-driven WRKY6 cDNA was used as the WRKY6 effector. WRKY6 fusion to the transactivation domain of VP16 (derived from Herpes simplex virus protein 16) was achieved by cloning a PCR-amplified WRKY6 cDNA into the XhoI and PinAI sites, replacing the bZIP sequence of a pBT8 derivative (Feldbrügge et al. 1994). The 4xW2 GUS reporter contained a tetramer of the hexameric TTGACC W-box motif (Eulgem et al. 1999), and the 4xmW2 GUS construct contained the tetramer of CATTGT (Rushton et al. 2002).
Particle bombardments were done as previously described (Shirasu et al. 1999). For each combination 15–20 mature leaves were transfected 4 h after detachment with 3 μg of SIRKp::GUS reporter variants together with 3 μg of empty vector or effector plasmids WRKY6, WRKY42, WRKY52, (Deslandes et al. 2002), and the parsley PcWRKY1 (Eulgem et al. 1999). Bombardments were done at 900 psi with a 7× diffuser in a vacuum chamber (Bio-Rad). GUS staining was performed 16 h after incubation under long-day conditions. Efficiency of the bombardments was monitored using a strong constitutive 35S::GUS construct.
Protoplast isolation derived from cultured Arabidopsis cells, and transient cotransfection experiments were performed as previously described (van de Löcht et al. 1990; Hartmann et al. 1998; Jin et al. 2000). For each assay, 2 × 106 protoplasts were transfected with 10 μg of promoter–GUS reporter together with 5 μg of empty vector, WRKY6, or WRKY6-VP16 effectors along with 5 μg of 35S::LUC reference plasmids. Protein, LUC, and GUS activity measurements were carried out 20 h after incubation in the dark. LUC expression was used to normalize for specific GUS activities. For assays using the active and inactive forms of the bacterial elicitor flagellin (Felix et al. 1999), transfected protoplasts were incubated in the presence of 1 nM elicitor.
Acknowledgments
This work was partly supported by the DFG funded Graduiertenkolleg für Molekulare Analysen von Entwicklungsprozessen (IIIGK-GRK 306/1). We thank Klaus Hahlbrock, Paul Schulze-Lefert, and Thomas Boller for continuous support. We thank Catherine Kistner (Sainsbury Laboratory, Norwich) for help with the cDNA-AFLP differential display; Robert Dietrich (Syngenta, Raleigh, NC) for providing the PR1p::GUS transgenic lines; Laurant Deslandes for providing the 35S::WRKY52 and 35S::WRKY42 constructs; Isabell Hermann for providing the 42p::GUS construct; Octave Noubibou for providing the SIRKp::GUS deletion constructs; and Petra Köchner, Elke Logemann, Brigitte Schauf, and Anja Reinstädler for technical assistance.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL somssich@mpiz-koeln.mpg.de; FAX 49-221-5062313.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.222702.
References
- Baumann E, Lewald J, Saedler H, Schulz B, Wisman E. Successful PCR based reverse genetic screens using an En-1 mutagenised Arabidopsis thaliana population generated via single-seed descent. Theor Appl Genet. 1998;97:729–734. [Google Scholar]
- Bouche N, Bouchez D. Arabidopsis gene knockout: Phenotypes wanted. Curr Opin Plant Biol. 2001;4:111–117. doi: 10.1016/s1369-5266(00)00145-x. [DOI] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell. 1997;88:57–63. doi: 10.1016/s0092-8674(00)81858-9. [DOI] [PubMed] [Google Scholar]
- Cao H, Li X, Dong X. Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc Natl Acad Sci. 1998;95:6531–6536. doi: 10.1073/pnas.95.11.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Czernic P, Visser B, Sun W, Savoure A, Deslandes L, Marco Y, Van Montagu M, Verbruggen N. Characterization of an Arabidopsis thaliana receptor-like protein kinase gene activated by oxidative stress and pathogen attack. Plant J. 1999;18:321–327. doi: 10.1046/j.1365-313x.1999.00447.x. [DOI] [PubMed] [Google Scholar]
- de Pater S, Greco V, Pham K, Memelink J, Kijne J. Characterization of a zinc-dependent transcriptional activator from Arabidopsis. Nucleic Acids Res. 1996;24:4624–4631. doi: 10.1093/nar/24.23.4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Theulières T, Hirsch J, Feng DX, Bittner-Eddy P, Beynon J, Marco Y. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is confered by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc Natl Acad Sci. 2002;99:2404–2409. doi: 10.1073/pnas.032485099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desveaux D, Despres C, Joyeux A, Subramaniam R, Brisson N. PBF-2 is a novel single-stranded DNA binding factor implicated in PR-10a gene activation in potato. Plant Cell. 2000;12:1477–1489. doi: 10.1105/tpc.12.8.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X. SA, JA, ethylene, and disease resistance in plants. Curr Opin Plant Biol. 1998;1:316–323. doi: 10.1016/1369-5266(88)80053-0. [DOI] [PubMed] [Google Scholar]
- Du L, Chen Z. Identification of genes encoding receptor-like protein kinases as possible targets of pathogen- and salicylic acid-induced WRKY DNA-binding proteins in Arabidopsis. Plant J. 2000;24:837–847. doi: 10.1046/j.1365-313x.2000.00923.x. [DOI] [PubMed] [Google Scholar]
- Durrant WE, Rowland O, Piedras P, Hammond-Kosack KE, Jones JDG. cDNA-AFLP reveals a striking overlap in race-specific resistance and wound response gene expression profiles. Plant Cell. 2000;12:963–977. doi: 10.1105/tpc.12.6.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Schmelzer E, Hahlbrock K, Somssich IE. Early nuclear events in plant defence signalling: Rapid gene activation by WRKY transcription factors. EMBO J. 1999;18:4689–4699. doi: 10.1093/emboj/18.17.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/s1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- Feldbrügge M, Sprenger M, Dinkelbach M, Yazaki K, Harter K, Weisshaar B. Functional analysis of a light-responsive plant bZIP transcriptional regulator. Plant Cell. 1994;6:1607–1621. doi: 10.1105/tpc.6.11.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999;18:265–276. doi: 10.1046/j.1365-313x.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- Greenberg JT, Silverman PF, Liang H. Uncoupling salicyclic acid-dependent cell death and defense-related responses from disease resistance in the Arabidopsis mutant acd5. Genetics. 2000;156:341–350. doi: 10.1093/genetics/156.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajouj T, Michelis R, Gepstein S. Cloning and characterization of a receptor-like protein kinase gene associated with senescence. Plant Physiol. 2000;124:1305–1314. doi: 10.1104/pp.124.3.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna-Rose W, Hansen U. Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 1996;6:229–234. doi: 10.1016/0168-9525(96)10022-6. [DOI] [PubMed] [Google Scholar]
- Hara K, Yagi M, Kusano T, Sano H. Rapid systemic accumulation of transcripts encoding a tobacco WRKY transcription factor on wounding. Mol Gen Genet. 2000;263:30–37. doi: 10.1007/pl00008673. [DOI] [PubMed] [Google Scholar]
- Hartmann U, Valentine WJ, Christie JM, Hays J, Jenkins GI, Weisshaar B. Identification of UV/blue light-response elements in the Arabidopsis thaliana chalcone synthase promoter using a homologous protoplast transient expression system. Plant Mol Biol. 1998;36:741–754. doi: 10.1023/a:1005921914384. [DOI] [PubMed] [Google Scholar]
- Hoecker U, Vasil IK, McCarty DR. Integrated control of seed maturation and germination programs by activator and repressor functions of Viviparous-1 of maize. Genes & Dev. 1995;9:2459–2469. doi: 10.1101/gad.9.20.2459. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-Glucuronidase a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 2000;19:6150–6161. doi: 10.1093/emboj/19.22.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinn T-L, Stone JM, Walker JC. HAESA, an Arabidopsis leucine-rich repeat kinase, controls floral organ abscission. Genes & Dev. 2000;14:108–117. [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J. The promotor of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- Landt O, Grunert H-P, Hahn U. A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene. 1990;96:125–128. doi: 10.1016/0378-1119(90)90351-q. [DOI] [PubMed] [Google Scholar]
- Lebel E, Heifetz P, Thorne L, Uknes S, Ryals J, Ward E. Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J. 1998;16:223–233. doi: 10.1046/j.1365-313x.1998.00288.x. [DOI] [PubMed] [Google Scholar]
- Lemon B, Tjian R. Orchestrated response: A symphony of transcription factors for gene control. Genes & Dev. 2000;14:2551–2569. doi: 10.1101/gad.831000. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang Y, Clarke JD, Li Y, Dong XI. Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell. 1999;98:329–339. doi: 10.1016/s0092-8674(00)81962-5. [DOI] [PubMed] [Google Scholar]
- Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton KA, Dangl JL, Dietrich RA. The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat Genet. 2000;26:403–410. doi: 10.1038/82521. [DOI] [PubMed] [Google Scholar]
- Morris K, MacKerness S, Page T, John CF, Murphy AM, Carr JP, Buchanan-Wollaston V. Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J. 2000;23:677–685. doi: 10.1046/j.1365-313x.2000.00836.x. [DOI] [PubMed] [Google Scholar]
- Motohashi H, Katsuoka F, Shavit JA, Engel JD, Yamamoto M. Positive or negative MARE-dependent transcriptional regulation is determined by the abundance of small Maf proteins. Cell. 2000;103:865–875. doi: 10.1016/s0092-8674(00)00190-2. [DOI] [PubMed] [Google Scholar]
- Nam HG. The molecular genetic analysis of leaf senescence. Curr Opin Biotech. 1997;8:200–207. doi: 10.1016/s0958-1669(97)80103-6. [DOI] [PubMed] [Google Scholar]
- Ogbourne S, Antalis TM. Transcriptional control and the role of silencers in transcriptional regulation in eukaryotes. Biochem J. 1998;331:1–14. doi: 10.1042/bj3310001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtake Y, Takahashi T, Komeda Y. Salicylic acid induces the expression of a number of receptor-like kinase genes in Arabidopsis thaliana. Plant Cell Physiol. 2000;41:1038–1044. doi: 10.1093/pcp/pcd028. [DOI] [PubMed] [Google Scholar]
- Petersen M, Broderson P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE, et al. Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell. 2000;103:1111–1120. doi: 10.1016/s0092-8674(00)00213-0. [DOI] [PubMed] [Google Scholar]
- Quirino BF, Normanly J, Amasino RA. Diverse range of gene activity during Arabidopsis thaliana leaf senescence includes pathogen-independent induction of defense-related genes. Plant Mol Biol. 1999;40:267–278. doi: 10.1023/a:1006199932265. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C-Z, Keddie J, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Robatzek S, Somssich IE. A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defense-related processes. Plant J. 2001;28:123–133. doi: 10.1046/j.1365-313x.2001.01131.x. [DOI] [PubMed] [Google Scholar]
- Rushlow C, Colosimo PF, Lin M-C, Xu M, Kirov N. Transcriptional regulation of the Drosophila gene zen by competing Smad and Brinker inputs. Genes & Dev. 2001;15:340–351. doi: 10.1101/gad.861401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE. Transcriptional control of plant genes responsive to pathogens. Curr Opin Plant Biol. 1998;1:311–315. doi: 10.1016/1369-5266(88)80052-9. [DOI] [PubMed] [Google Scholar]
- Rushton, P.J., Reinstädler, A., Lipka, V., Lippok, B., and Somssich, I.E. 2002. Pathogen-inducible synthetic plant promoters. Plant Cell (in press). [DOI] [PMC free article] [PubMed]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shirasu K, Nielsen K, Piffanelli P, Oliver R, Schulze-Lefert P. Cell-autonomous complementation of mlo resistance using a biolistic transient expression system. Plant J. 1999;17:293–300. [Google Scholar]
- Shiu S-H, Bleecker AB. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci. 2001;98:10763–10768. doi: 10.1073/pnas.181141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Löcht U, Meier I, Hahlbrock K, Somssich IE. A 125 bp promoter fragment is sufficient for strong elicitor-mediated gene activation in parsley. EMBO J. 1990;9:2945–2950. doi: 10.1002/j.1460-2075.1990.tb07486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisshaar B, Armstrong GA, Block A, da Costa e Silva O, Hahlbrock K. Light-inducible and constitutively expressed DNA-binding proteins recognizing a plant promoter element with functional relevance in light responsiveness. EMBO J. 1991;10:1777–1786. doi: 10.1002/j.1460-2075.1991.tb07702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Chung KM, Park J-H, Oh SA, Ahn T, Hong SH, Jang SK, Nam H-G. ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell. 2001;13:1779–1790. doi: 10.1105/TPC.010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Wang Z, Fan B, Chen C, Chen Z. A pathogen- and salicylic acid-induced WRKY DNA-binding activity recognizes the elicitor response element of tobacco class I chitinase gene promoter. Plant J. 1999;18:141–149. [Google Scholar]
- Yu D, Chen C, Chen Z. Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell. 2001;13:1527–1539. doi: 10.1105/TPC.010115. [DOI] [PMC free article] [PubMed] [Google Scholar]