Abstract
Although the active digestion of mating-type minus (mt−) chloroplast DNA (cpDNA) in young zygotes is considered to be the basis for the uniparental inheritance of cpDNA in Chlamydomonas reinhardtii, little is known about the underlying molecular mechanism. One model of active digestion proposes that nucleases are either synthesized or activated to digest mt− cpDNA. We used a native-PAGE/in gelo assay to investigate nuclease activities in chloroplasts from young zygotes, and identified a novel Ca2+-dependent nuclease activity. The timing of activation (∼60–90 min after mating) and the localization of the nuclease activity (in mt− chloroplasts) coincided with the active digestion of mt− cpDNA. Furthermore, the activity of the nuclease was coregulated with the maturation of mating-type plus (mt+) gametes, which would enable the efficient digestion of mt− cpDNA. Based on these observations, we propose that the nuclease (designated as Mt+-specific DNase, MDN) is a developmentally controlled nuclease that is activated in mt+ gametes and participates in the destruction of mt− cpDNA in young zygotes, thereby ensuring uniparental inheritance of chloroplast traits.
Keywords: Non-Mendelian inheritance, chloroplast DNA, nuclease, native-PAGE/in gelo assay, chloroplast transformation
Chloroplasts and mitochondria contain their own genomes, which are transmitted to progeny in a non-Mendelian, maternal fashion in diverse taxa of sexual eukaryotes: higher plants, mosses, ferns, algae (Correns 1909; Kirk and Tilney-Basset 1978; Sears 1980; Whatley 1982; Hagemann and Schröder 1989; Kuroiwa 1991), fungi (Mitchell and Mitchell 1952; Kawano et al. 1987), and animals (Hutchison et al. 1974; Sutovsky and Schatten 2000), including humans (Giles et al. 1980). However, the molecular mechanism of uniparental inheritance remains unclear.
The genetics of uniparental inheritance have been extensively examined in the haploid unicellular green alga Chlamydomonas reinhardtii (Harris 1989; Gillham 1994). There are two mating types of C. reinhardtii, mating type plus (mt+) and mating type minus (mt−), which are controlled by a single complex locus on linkage group VI (Ebersold et al. 1962). C. reinhardtii undergoes a sexual life cycle, which includes defined stages of differentiation. Vegetative cells differentiate into gametes under conditions of nitrogen starvation and light irradiation (Beck and Haring 1996). Within minutes of their being mixed, gametes of opposite mating types adhere by their flagella and fuse to form zygotes. After a mandatory period of dormancy, the zygote undergoes meiosis and germination to produce four haploid progeny (Harris 1989). More than 90% of the meiotic progeny thus formed inherit chloroplast (cp) traits from the mt+ parent, a phenomenon that was first documented almost 50 years ago (Sager 1954).
In 1972, a biochemical study showed that the ratio of mt− chloroplast DNA (cpDNA) to mt+ cpDNA decreased 6 to 24 h after mating. Sager and colleagues proposed that mt− cpDNA was digested by restriction enzymes, whereas mt+ cpDNA was protected by methylation, in a process that resembled bacterial restriction-methylation (Sager and Lane 1972). Indeed, an increase in the methylation level of mt+ cpDNA was detected 7 h after mating (Burton et al. 1979; Royer and Sager 1979), and DNA methyltransferases with molecular weights of 60 kD and 200 kD were later purified (Sano et al. 1981). However, this hypothesis was weakened by data from experiments with a hypermethylation mutant (me-1; Bolen et al. 1982), and with methylation inhibitors, such as 5-aza Cyd and L-ethionine (Feng and Chiang 1984; Umen and Goodenough 2001).
The 203-kb chloroplast genome of C. reinhardtii (GenBank accession no. AF396929; J. Maul, J.W. Lilly, and D.B. Stern, unpublished results at http://www.biology.duke.edu/chlamy_genome/chloro.html) is present in ∼80–100 copies per cell (Gillham 1978) and is organized into 5–10 DNA–protein complexes, which are called chloroplast nucleoids (Kuroiwa et al. 1981). In 1982, Kuroiwa et al. found that DAPI-stained mt− cp nucleoids disappeared preferentially in young zygotes within 50 min of mating (Kuroiwa et al. 1982). The preferential disappearance of fluorescent mt− cp nucleoids occurred well before DNA digestion was detected (6–24 h after mating) by biochemical methods (Sager and Lane 1972). Single-cell analysis using optical tweezers was performed to investigate whether the mt− cpDNA was digested or simply diffused throughout the chloroplast (Nishimura et al. 1999). The individual fates of mt+ and mt− zygotic cpDNAs were followed by the insertion, using particle gun bombardment, of the aadA gene sequence into mt+ and mt− cpDNAs. Single zygotes with or without mt− cp nucleoids were selectively obtained by the optical tweezer and analyzed using nested PCR, which revealed that mt− cpDNA molecules were actively digested during the preferential disappearance of mt− cp nucleoids. This active digestion of mt− cpDNA is probably the basis for maternal inheritance of cpDNA.
Prevailing models suggest that the pathway through which mt− nucleoids are selectively destroyed has two distinct steps: protection of mt+ cpDNA and destruction of mt− cpDNA (Kuroiwa 1985; Gillham 1994), with the destruction step mediated by nucleases that degrade the unprotected mt− cpDNA within the first 60 min after mating. Kuroiwa et al. (1982) observed the pattern of digestion of cp nucleoids and found that each chloroplast nucleoid is digested within 10 min, without swelling. This pattern of digestion cannot be explained by the action of restriction endonucleases, because they simply make cuts in DNA to produce fragments that would be easily observed by fluorescent microscopy using a DNA-specific fluorochrome (Schwartz and Samad 1997). Therefore, they concluded that nonspecific nucleases, such as DNase I, were involved in the elimination of mt− cpDNA from the zygotes (Kuroiwa et al. 1982), rather than a sequence-specific restriction enzyme (Burton et al. 1977). Umen and Goodenough (2001) followed the fate of mt− cpDNA on Southern blots using the aadA tag, and found that the putative degradation intermediates were absent, thus indicating that cpDNAs were rapidly degraded into nucleotides. This finding supports the possibility that a sequence-nonspecific nuclease might be responsible for the active digestion of mt− cpDNA.
The SDS-PAGE/in gelo assay is a highly sensitive technique to detect sequence-nonspecific endo- and exonucleases. This method has been used to detect nucleases that are responsible for various physiological processes, such as nuclear DNA digestion during programmed cell death in animals (Urbano et al. 1998) and plants (Thelen and Northcote 1989; Mittler and Lam 1995; Xu and Hanson 2000). Using this technique, Ogawa and Kuroiwa (1985a) compared the nuclease activities of C. reinhardtii gametes and zygotes to identify nucleases that were specifically required for the uniparental inheritance of cpDNA. Six polypeptides were detected that had Ca2+-dependent nuclease activity, and they were collectively designated as Nuclease C.
Because Ca2+ is required for the preferential digestion of mt− cpDNA (Kuroiwa 1985), it was proposed that some of these nucleases might engage in preferential digestion. However, no differences in nuclease activity were detected between the gametes and zygotes, and it was not possible to identify the nucleases responsible for cpDNA digestion (Ogawa and Kuroiwa 1985a). On the other hand, an experiment with partially purified Nuclease C provided an important insight into the protection of mt+ cpDNA. Ogawa and Kuroiwa showed that the mt+ cp nucleoids of mature mt+ gametes were resistant to Nuclease C, whereas the cp nucleoids of mature mt− gametes and vegetative cells of both mating types were rapidly digested by treatment with Nuclease C (Ogawa and Kuroiwa 1985b). This result indicates that gametogenesis is linked to the protection of mt+ cp nucleoids, and that Ca2+-dependent nucleases may be responsible for preferential digestion.
One problem with the SDS-PAGE/in gelo assay is that the proteins must be denatured and reconstituted before nuclease activity can be detected. Thus, the processing steps might either inactivate or activate nucleases and lead to artifactual results. Indeed, nucleases that are irreversibly inactivated by SDS treatment include DNase II, which is composed of three protein subunits (α1, α2, and β) that are dissociated by SDS treatment (Wang et al. 1998). Conversely, certain nucleases are activated by dissociation from their partner components (Enari et al. 1998). The latter type of nuclease would therefore be artificially activated by SDS treatment. In this work, we investigated nuclease activities using a method that allows protein separation without denaturation (native-PAGE/in gelo assay).
First, we observed the precise timing of active digestion by quantifying the DAPI fluorescence of mt− cp nucleoids using an ultrasensitive microscopic photometry system, called VIMPCS (video-intensified microscopic photon counting system; Kuroiwa et al. 1986). Next, we used a tagged chloroplast transformant, AG8 mt+ (Ishikura et al. 1999), to investigate whether the preferential digestion of mt− cpDNA occurred before or after the fusion of mt+ and mt− chloroplast membranes. Our results revealed that preferential digestion was completed before chloroplast fusion. Then, we isolated chloroplasts from zygotes and analyzed changes in nuclease activity using a native-PAGE/in gelo assay. From these experiments, a Ca2+-dependent nuclease was identified as an important candidate for the preferential digestion of mt− cpDNA. The activity of this nuclease was rapidly assimilated or activated in mt− chloroplasts within 60–90 min after zygote formation, which precisely corresponds to the timing and pattern of mt− cpDNA digestion. Furthermore, the nuclease activity was regulated simultaneously with the maturation of mt+ gametes, which might represent a mechanism to ensure the efficient digestion of mt− cpDNA. From these results, we designated the nuclease as MDN (Mt+-specific DNase) and proposed it as a possible driving force for the uniparental inheritance of cpDNA in C. reinhardtii.
Results
Quantitative monitoring of mt− cpDNA destruction
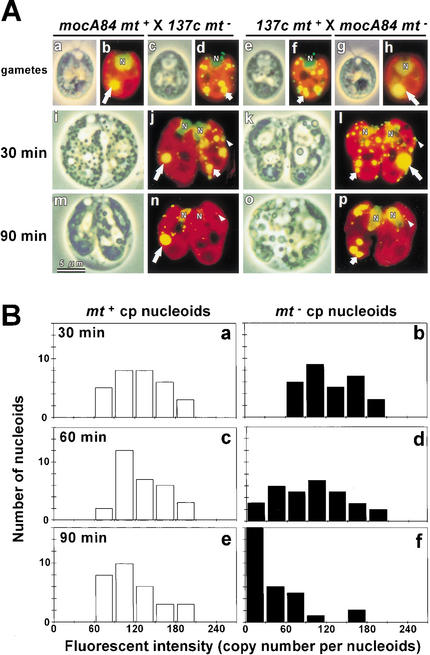
To monitor the precise timing of mt− cpDNA digestion, the fluorescence emitted from DAPI-stained mt− cp nucleoids was quantified using ultrasensitive microscopic photometry (VIMPICS; Kuroiwa et al. 1986). When wild-type gametes were used, mt+ cp nucleoids were indistinguishable from mt− cp nucleoids. In addition, wild-type cells normally contain five to eight cp nucleoids that vary in size and shape, which makes it difficult to assess their fluorescence accurately. To eliminate these difficulties, the moc mutant (Monokaryotic Chloroplast; Misumi et al. 1999) was used. The mocA84 mt+ mutant contains only one large cp nucleoid, which can be clearly distinguished from wild-type cp nucleoids and can easily be quantified by VIMPCS (Fig. 1A,B).
Figure 1.
(A) Phase-contrast (a,c,e,g,i,k,m,o) and fluorescent (b,d,f,h,j,l,n,p) images of living gametes (137c mt+ [e,f]; 137c mt− [c,d]; mocA84 mt+ [a,b]; mocA84 mt− [g,h]), and zygotes at 30 min (i–l) and 90 min (m–p) after mating for mocA84 mt+ × 137c mt− (i,j,m,n), and 137c mt+ × A84 mt− (k,l,o,p) crosses. The cells are stained with SYBR Green I. The yellow-green spheres (N) are cell nuclei. Chlorophyll autofluorescence is red. The yellow spots of the moc-type cp nucleoids, wild-type cp nucleoids, and the mitochondrial nucleoids are indicated by large arrows, small arrows, and white arrowheads, respectively. (B) Frequency distribution of the fluorescence of DAPI-stained moc-type nucleoids, quantified by VIMPCS at 30, 60, and 90 min after mating, and expressed in terms of cpDNA copy number. The fluorescence of mt+ (white) and mt− (black) cp nucleoids in the mocA84 mt+ × 137c mt−, and 137c mt+ × A84 mt− crosses are shown.
Figure 1A shows the fluorescent images of SYBR Green I-stained living zygotes from a cross between mocA84 mt+:wild-type 137c mt− and its reciprocal. SYBR Green I is a DNA-specific fluorochrome that can be used to visualize nuclear, mitochondrial, and chloroplast DNA molecules simultaneously in living cells (Nishimura et al. 1998). The moc-type cp nucleoids retained their condensed morphology, even after mating. Serial microscopic observations of the process of mt− cpDNA digestion revealed that the timing and pattern of mt− cpDNA digestion were almost the same in the wild-type and moc-type cp nucleoids, despite their morphological differences. The moc-type cp nucleoids were rapidly digested starting at the periphery and progressing to the center, in the absence of any swelling. The digestion process was completed within 10 min. These observations confirm the rapidity of mt− cpDNA digestion (Nishimura et al. 1999; Umen and Goodenough 2001). On the other hand, the mt+ and mt− mitochondrial nucleoids remained unchanged during the active digestion of mt− cp nucleoids, indicating that digestion was selective for mt− cpDNA.
The histograms in Figure 1B show the frequency distribution of fluorescence intensity per mt+ or mt− moc-type cp nucleoid. The distribution shows that the average copy number of cpDNA per moc-type nucleoid was ∼60–90, which was similar to the copy number of cpDNA per chloroplast in wild-type cells (Misumi et al. 1999). The distribution of mt+ cp nucleoids was unchanged during the 90-min period after mating, and no change was detected in mt+ cpDNA content. Although the distribution of mt− cp nucleoids at 30 min after mating was similar to that of mt+, with a peak at 60–90 copies, the distribution peak shifted to a slightly lower value at 60 min and shifted dramatically to 0 at 90 min. These results for mt− cpDNA digestion can be interpreted as follows: mt− cpDNA is unchanged until 30 min after mating, and mt− cpDNA digestion begins at 45–60 min after mating. Once initiated, digestion occurs rapidly and is completed within ∼10 min. On the other hand, mt+ cpDNA molecules are neither digested nor replicated during the maturation of young zygotes.
Chloroplast fusion and active digestion of mt− cpDNA
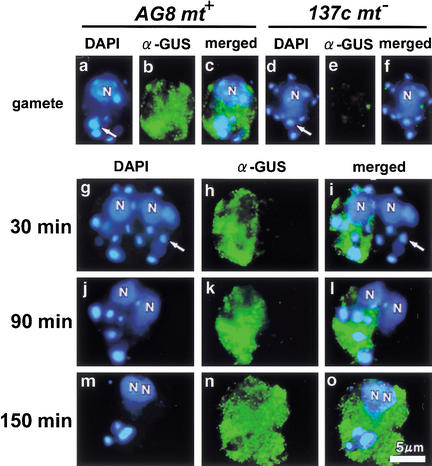
Previous electron-microscopic observations showed that the mt+ and mt− chloroplasts fused into a single chloroplast during the maturation of young zygotes (Cavalier-Smith 1976). Whether the active digestion of mt− cpDNA occurs before or after chloroplast fusion is an important consideration in understanding the molecular mechanism that controls the preferential digestion and protection of cpDNA in young zygotes. To address this question, a chloroplast transformant (AG8 mt+) was used (Ishikura et al. 1999). The cpDNA of this strain was transformed by particle gun bombardment with the uidA gene, resulting in the accumulation of GUS (β-glucuronidase) proteins in mt+ chloroplasts. AG8 mt+ cells were crossed with 137c mt− cells, and the timing of chloroplast fusion was monitored by observing the diffusion of GUS protein into mt− chloroplasts. The cells were simultaneously stained with DAPI to compare the timing of mt− cpDNA digestion with that of chloroplast fusion (Fig. 2).
Figure 2.
Fluorescent images of gametes (AG8 mt+ [a–c]; 137c mt− [d–f]) and zygotes (AG8 mt+ × 137c mt−) at 30 (g–i), 90 (j–l), and 150 (m–o) min after mating. Each of the cells is double stained with DAPI (a,d,g,j,m) and FITC-conjugated antibody against GUS protein (b,e,h,k,n). Merged images of DAPI- and FITC-labeled cells are also shown (c,f,i,l,o). The Cp nucleoids are indicated by large arrows. (N) Cell nucleus.
GUS protein was clearly detected in the chloroplasts of AG8 mt+ gametes by immunofluorescent staining. Immediately after mating, GUS was localized only in mt+ chloroplasts, and both mt+ and mt− cp nucleoids were still visible. Active digestion of mt− cpDNA occurred at 60–90 min after mating, whereas GUS remained exclusively in the mt+ chloroplasts. GUS diffusion began 120 min after mating, and 30 min later both mt+ and mt− chloroplasts were stained in most of the zygotes. These results indicate that mt+ and mt− chloroplasts are separated during the active digestion of mt− cpDNA.
Analysis of nuclease activities in chloroplasts by the native-PAGE/in gelo assay
The mt− cpDNA of young zygotes is extensively digested within 60–90 min after mating. The most simple and generally accepted explanation for this digestion is the activation of nucleases in the chloroplasts during zygote maturation (Kuroiwa 1985; Gillham 1994). To test this hypothesis, nuclease activities in the chloroplasts of young zygotes were surveyed using the native-PAGE/in gelo assay.
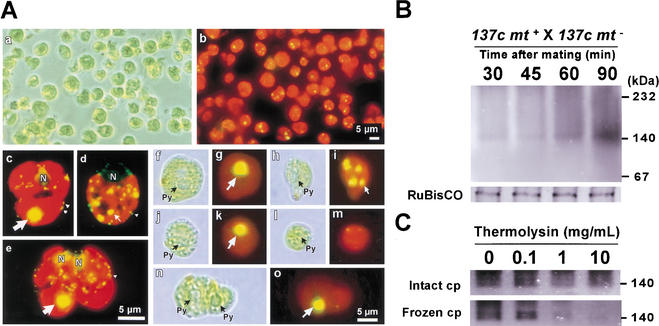
To date, various techniques to isolate chloroplasts from C. reinhardtii have been reported (Belknap 1983; Klein et al. 1983; Goldschmidt-Clermont et al. 1989; Mason et al. 1991). The mt+ and mt− chloroplasts of young zygotes fuse, which makes them extremely fragile and complicates their isolation by conventional methods. To circumvent this problem, we employed a procedure in which the cells were sprayed with an airbrush. This procedure breaks the zygotes gently and homogeneously without producing heat. Phase contrast microscopy was used to verify that the chloroplasts were intact (Fig. 3A). The mt+ and mt− chloroplasts were distinguished using moc mutants as the mt+ parents. Intact chloroplasts at various stages of development were isolated using the airbrush method. mt+ chloroplasts with moc-type cp nucleoids, mt− chloroplasts with wild-type cp nucleoids, and chloroplasts without cp nucleoids were isolated, as were fused chloroplasts. This result shows that the airbrush method is highly suitable for the isolation of chloroplasts from young zygotes, and the fact that chloroplasts without cp nucleoids were observed confirms that the active digestion of mt− cpDNA occurs well before the fusion of the mt+ and mt− chloroplasts (Fig. 3A).
Figure 3.
(A) Phase-contrast (a) and fluorescent (b) images of intact chloroplasts isolated from zygotes using an airbrush. To distinguish mt+ and mt− chloroplasts, mocA84 mt+ and 137c mt− gametes were crossed to form zygotes. Fluorescent images of SYBR Green I-stained gametes (mocA84 mt+ [c]; 137c mt− [d]) and a zygote (mocA84 mt+ × 137c mt− [e]). moc-type, wild-type cp nucleoids, and mt nucleoids are indicated by large arrows, small arrows, and white arrowheads, respectively. Phase contrast (f,h,j,l,n) and fluorescent images (g,i,k,m,o) of isolated chloroplasts stained by SYBR Green I. Intact mt+ (f,g,j,k) and mt− (h,i,l,m) chloroplasts before (f–i) and after (j–m) active digestion, and fused chloroplasts (n,o) were observed. (Py) Pyrenoid. (B) Ca2+-dependent nuclease activity in chloroplasts detected by native-PAGE/in gelo assay. Chloroplasts were isolated from zygotes (137c mt+ × 137c mt−) at 30, 45, 60, and 90 min after mating. As a control, CBB-stained RuBisCO bands are shown under each lane. (C) Intact chloroplasts and frozen chloroplasts were treated with 0, 0.1, 1, and 10 mg/mL thermolysin. Their Ca2+ nuclease activities were investigated by native-PAGE/in gelo assay.
Intact chloroplasts were isolated from zygotes at 30, 45, 60, and 90 min after mating, and changes in the levels of nuclease activity were investigated using the native-PAGE/in gelo assay (Fig. 3B). Initially, very little nuclease activity was detected in the isolated chloroplasts. A Ca2+-dependent nuclease activity with apparent molecular mass of 140 kD appeared and rapidly increased during the 60–90 min period after mating, which corresponded precisely to the timing of mt− cpDNA digestion. This was the only chloroplast nuclease activity detected by our native-PAGE/in gelo assay system, and no nuclease activity was detected in the presence of other metal ions (Mg2+, Zn2+, and Mn2+). The chloroplasts were treated with a thermolysin to examine whether the Ca2+-dependent nuclease activity was contained within the chloroplasts (Fig. 3c). Chloroplasts were isolated from zygotes 90 min after mating. The isolated fraction was divided in two: one portion was frozen and thawed to disrupt the chloroplast membranes, and the other was kept intact. Both fractions were treated with 0, 0.1, 1, and 10 μg/mL themolysin at 4°C for 20 min, and the nuclease activities were examined by the native-PAGE/in gelo assay. When frozen and thawed chloroplasts were treated with thermolysin, the Ca2+-dependent nuclease activity (the band of ∼140 kD) diminished dramatically. In contrast, when intact chloroplasts were treated with thermolysin, the Ca2+-dependent nuclease activity was unaffected, even in the presence of 10 μg/mL thermolysin. These results confirm that the Ca2+-dependent nuclease is taken up by, or activated in, chloroplasts during the 60–90 min period after mating.
Comparison of Ca2+-dependent nuclease activities between mt+ and mt− chloroplasts in zygotes
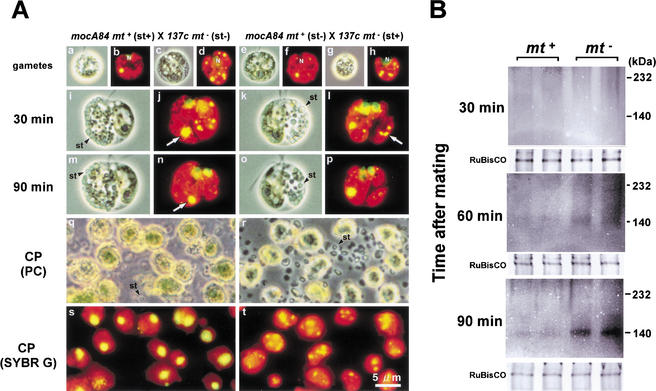
As shown in Figure 2, the active digestion of mt− cpDNA occurs well before the fusion of mt+ and mt− chloroplasts. This suggests that the separation of mt+ and mt− chloroplasts is important in the preferential digestion of mt− cpDNA. To test this possibility, the nuclease activities were compared in mt+ and mt− chloroplasts during the maturation of young zygotes. However, the chloroplast fraction from zygotes is typically a mixture of mt+ and mt− chloroplasts. We took advantage of the starch granules that accumulate in chloroplasts to separate these two components. The quantity of starch granules in Chlamydomonas chloroplasts can be increased by prolonged incubation in Snell's medium. When gametes with extensive starch granulation are crossed with normal gametes, the resulting zygotes show large amounts of starch granules in either mt+ or mt− chloroplasts, but not in both (Fig. 4A).
Figure 4.
(A) Phase-contrast (a,c,e,g,i,k,m,o,q,r) and fluorescent (b,d,f,h,j,l,n,p,s,t) images of SYBR Green I-stained gametes (mocA84 mt+ [a,b,e,f]; 137c mt− [c,d,g,h]), zygotes (30 min [i–l]; 90 min [m–p] after mating), and mt+ (q,s) and mt− (r,t) chloroplasts isolated from zygotes at 30 min after mating. Gametes that are incubated for 10 d accumulate large quantities of starch granules in their chloroplasts (st+; a,b,g,h), whereas gametes that are incubated for 2 d accumulate few starch granules (st−; c,d,e,f). By crossing the two kinds of gametes, zygotes can be formed that accumulate large numbers of starch granules only in mt+ chloroplasts (i,j,m,n) or mt− chloroplasts (k,l,o,p). The presence of a large amount of starch did not affect the active digestion of mt− cpDNA (m–p). Chloroplasts containing large amounts of starch granules were isolated and selectively separated by Percoll step-gradient centrifugation. The purity of separated chloroplasts was verified by fluorescence microscopy (q–t). (St) Starch granules. (B) Comparison of Ca2+-dependent nuclease activity in mt+ and mt− chloroplasts using the native-PAGE/in gelo assay. The mt+ and mt− chloroplasts that were isolated from zygotes at 30, 60, and 90 min after mating were analyzed. Experiments were repeated twice to ensure reproducibility. As a control, CBB-stained RuBisCO bands are shown under each lane.
Chloroplasts were isolated from zygotes and separated on a Percoll step gradient. Chloroplasts with starch accumulations (st+) were selectively pelleted to the bottom of centrifugation tubes along with the starch granules, whereas the chloroplasts without starch (st−) were collected at the interface of the 45%–80% Percoll step gradient. The purity of the mt+ or mt− chloroplasts was verified by fluorescent microscopy using the moc-type cp nucleoid as a marker for mt+ chloroplasts (Fig. 4A). The st+ chloroplast fractions showed negligible contamination with either st− chloroplasts or undisrupted cells. Therefore, the st+ fractions from crosses involving A84 mt+ (st+) × 137c mt− (st-) and A84 mt+ (st−) × 137c mt− (st+) were collected and used in the native-PAGE/in gelo assays. Thus, we compared the nuclease activities of mt+ or mt− chloroplasts from zygotes taken at 30, 45, 60, and 90 min after mating (Fig. 4B). Once again, our native-PAGE/in gelo assay detected a notable increase in the level of the Ca2+-dependent nuclease at 60–90 min after mating. Importantly, this increase was observed only in mt− chloroplasts, that is, where cpDNA digestion occurred. These observations strongly support the possibility that a sequence-nonspecific Ca2+-dependent nuclease (of ∼140 kD) is responsible for the preferential digestion of mt− cpDNA that leads to maternal inheritance.
Developmental control of MDN activity during gametogenesis, mating, and zygote maturation
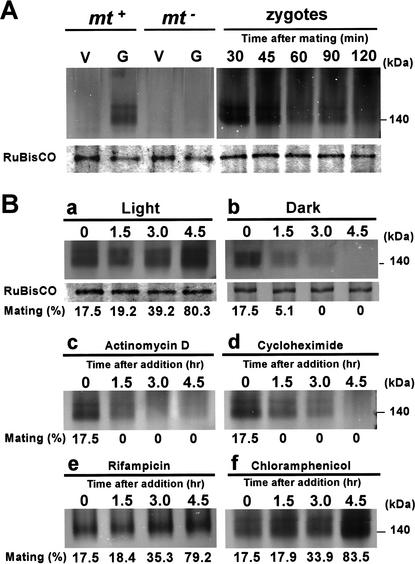
The Ca2+-dependent nuclease activity was compared between gametes and vegetative cells (Fig. 5). Interestingly, the nuclease activity was detected in mt+ gametes, but not in mt− gametes or vegetative cells of either mating type, suggesting that the nuclease might be activated during the maturation of mt+ gametes. From this observation, the Ca2+-dependent nuclease was designated as MDN (Mt+-specific DNase).
Figure 5.
(A) Comparisons of MDN activities between 137c vegetative cells (V; mt+, mt−), gametes (G; mt+, mt−), and zygotes (30, 45, 60, 90, and 120 min after mating). (B) MDN activity in 137c mt+ cells that were incubated in the light (a) or the dark (b). The CBB-stained RuBisCO bands detected from the same gels are shown below each lane, as a control. The MDN activity level in 137c mt+ gametes that were treated with actinomycin D (c; 20 μg/mL), rifampicin (e; 80 μg/mL), cycloheximide (d; 10 μg/mL), or chloramphenicol (f; 80 μg/mL) in the light, is indicated. Cells were harvested and analyzed at 0, 1.5, 3.0, and 4.5 h after the addition of the drugs. The number indicated under each lane represents the mating efficiency (in percent) of the mt+ cells with matured mt− gametes. The white arrowheads indicate a minor nuclease activity signal with apparent molecular mass of ∼190 kD.
We speculated that coregulation of gametogenesis and nuclease activation represented a strategy that ensured the efficient digestion of mt− cpDNA and guaranteed that only mt+ gametes that were ready to digest mt− cpDNA could mate with mt− gametes. Based on previous reports that gametes could be reversibly activated or inactivated under light or dark conditions, respectively (Beck and Haring 1996), we compared the MDN activities of mt+ cells that were incubated in either light (∼6000 lux) or darkness (Fig. 5B). The mt+ cells were harvested from Snell's plates, suspended in Tsubo mating buffer, and incubated for 2 h under light (6600 lux) to let the cells protrude their flagella and start swimming. The cell suspension was then incubated in the dark, and changes in MDN activity were compared with those of cells that had been incubated continuously in light. Initially, the cells could not mate efficiently with mature mt− gametes (17.5%). However, an additional 3-h exposure to light irradiation resulted in the almost full maturation of mt+ cells (mating efficiency 80%). On the other hand, the cells that were incubated in the dark lost the ability to mate. MDN activity was measured after 0, 1.5, 3.0, and 4.5 h of incubation in the dark. The MDN activity of light-incubated cells increased gradually, accompanied by an increase in the number of mating-competent gametes. In contrast, the MDN activity of the dark-incubated cells decreased rapidly. These results further support the hypothesis that MDN activity is reversibly regulated simultaneously with the maturation of mt+ gametes (Fig. 5).
It should be noted that when whole-cell extracts of C. reinhardtii were analyzed, MDN appeared as a doublet; the additional minor band had an apparent molecular mass of 190 kD (Fig. 5). The intensity of the minor band changed under the same conditions as the major band, suggesting that the two bands have common components. Currently, we have no information regarding the significance of this minor band.
To further identify factors that regulate MDN activity, mt+ gametes were treated with the cytoplasmic transcription inhibitor actinomycin D, the cytoplasmic translation inhibitor cycloheximide, the organelle transcription inhibitor rifampicin, or the organelle translation inhibitor chloramphenicol. In separate experiments, mating efficiency was also monitored (Fig. 5). The levels of MDN activity increased in untreated gametes as the period of gametogenesis was prolonged. The addition of rifampicin or chloramphenicol had no apparent effect on MDN activity. However, when mt+ gametes were treated with actinomycin D or cycloheximide, MDN activity decreased rapidly and almost completely disappeared within 4.5 h. These results suggest that MDN activity is developmentally controlled and maintained by de novo RNA and protein synthesis from nuclear-encoded genes, whose expression may be triggered by gametogenesis in mt+ cells.
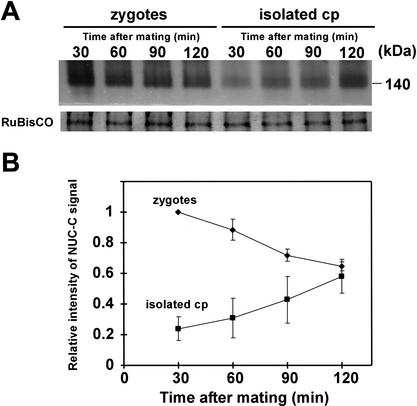
We then compared the MDN activities of young zygotes at 30, 45, 60, 90, and 120 min after mating (Figs. 5A, right, and 6A). MDN activity gradually decreased as the zygotes matured. The activation pattern of MDN, which increased during gametogenesis and decreased during zygote maturation, was highly similar to the mRNA-accumulation pattern of certain gamete-specific genes (von Gromoff and Beck 1993). Paradoxically, MDN activity in whole-cell extracts of zygotes decreased during zygote maturation, whereas the MDN levels in chloroplasts increased in the 60–90-min period after mating (Figs. 3B and 6A). To address this apparent contradiction, MDN activity levels were compared between whole zygotes and isolated chloroplasts (Fig. 6B). The loadings of samples were adjusted to give the same density of CBB-stained RuBisCO band, and the MDN activities in whole zygotes and isolated chloroplasts were compared on the same gels. The duration of electrophoresis was shortened to 25 min in this experiment, to obtain sharp bands that were suitable for precise quantification. The experiments were repeated three times to ensure reproducibility (Fig. 6B). The level of MDN activity was generally higher in whole zygotes than in isolated chloroplasts, even at 120 min after mating. Our results suggest the following mode of action for MDN (Fig. 7): MDN is activated during the maturation of mt+ gametes and accumulates in the cytoplasm; upon zygote formation, MDN enters the unprotected mt− chloroplast during the 60–90 min after mating, which precisely corresponds to the timing of mt− cpDNA digestion.
Figure 6.
Quantitative comparison of MDN activities in whole zygotes and isolated chloroplasts at 30, 60, 90, and 120 min after mating. (A) MDN activities detected by the native-PAGE/in gelo assay for whole zygotes and isolated chloroplasts at 30, 60, 90 and 120 min after mating. Samples that gave the identical densities of RuBisCO bands were used, and their MDN activities were compared on the same gel. The experiment was repeated three times to ensure reproducibility. (B) Relative intensity of MDN activity normalized to the MDN activity of the whole zygote at 30 min. Whole zygotes (♦) and isolated chloroplasts (▪).
Figure 7.
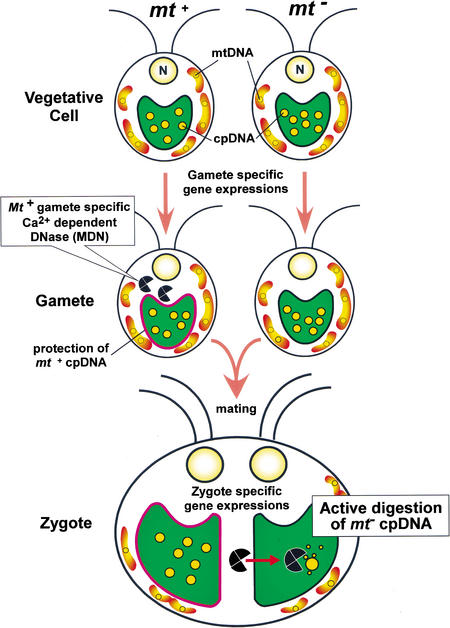
Model for the molecular mechanism of uniparental inheritance in C. reinhardtii based on the active digestion of mt− cpDNA by MDN. In vegetative cells, MDN is absent or inactivated in both mating types. During gametogenesis, MDN is synthesized or activated only in mt+ cells. At the same time, mt+ cpDNA becomes resistant to the action of MDN. During gamete fusion, MDN obtains access to unprotected mt− chloroplasts and digests mt− cpDNA, leading to the uniparental inheritance of cpDNA. Several factors may mediate the successful digestion of mt− cpDNA after zygote formation: (1) entry of MDN into mt− chloroplasts; (2) efficient access of MDN to cpDNA molecules; and (3) an increase in Ca2+ inside mt− chloroplasts. Zygote-specific gene expression may be crucial to these processes.
Discussion
In the unicellular green algae C. reinhardtii, mt− cpDNA molecules are digested in young zygotes, and this process is considered to be important for the maternal inheritance of cpDNA (Sager and Lane 1972; Kuroiwa 1982; Nishimura et al. 1999; Umen and Goodenough 2001). The nuclease that participates in the active digestion of mt− cpDNA has long been sought (Burton et al. 1977; Ogawa and Kuroiwa 1985a,b). We have identified a Ca2+-dependent mt+-specific nuclease (MDN) that is potentially involved in the active digestion of mt− cpDNA.
A nuclease that is specifically activated in the mt− chloroplasts of young zygotes
In this report, we examined the process of mt− cpDNA digestion in young zygotes using ultrasensitive microscopic photometry, VIMPCS. This analysis revealed that the mt− cpDNA was retained for ∼30 min and subsequently (60–90 min after mating) was extensively digested. In contrast, the mt+ cpDNA content was unchanged (Fig. 1). Microscopic analysis using the A84 mt+ transformant showed that digestion was completed before the fusion of mt+ and mt− chloroplasts (Fig. 2). Based on these observations, we surveyed nuclease activities in chloroplasts that were isolated from young zygotes, and identified one sequence-nonspecific, Ca2+-dependent nuclease, MDN. The timing of MDN activation (60–90 min after mating) and its localization (in mt− chloroplasts) precisely corresponded to the preferential digestion of mt− cpDNA. This finding lends strong support to the possibility that MDN is the driving force behind the process of mt− cpDNA digestion, which leads to the maternal transmission of cpDNA in C. reinhardtii.
The observation that MDN activity is detected only in mt− chloroplasts provides an important clue as to the mechanism of protection of mt+ cpDNA (Fig. 4). Our preliminary results suggest three alternatives: (1) the nuclease is not incorporated into mt+ chloroplasts; (2) it is inactivated; or (3) it is digested within mt+ chloroplasts. Because chloroplast fusion occurs after the digestion of mt− cpDNA, explanations 2 and 3 appear to be more likely. Proteins that bind and inactivate specific nucleases include: the immunity protein (Im9), which inactivates the bacterial toxin nuclease; E9 colicin in Escherichia coli (Wallis et al. 1994); and ICAD, which inhibits CAD (caspase-activated deoxyribonuclease) in mammals (Enari et al. 1998). A survey of chloroplast proteases and chloroplast proteins that inactivate nucleases might elucidate the molecular mechanisms that protect mt+ cpDNA from digestion.
The apparent molecular mass of MDN (∼140 kD; Fig. 3) is high compared with other nucleases, such as DNase I (31 kD), micrococcal nuclease (16.8 kD), and BAL31 nuclease (73–83 kD); high-molecular-mass nucleases do exist, such as the RecBCD nuclease of E. coli, which is a heterotrimer (330 kD) of RecB (134 kD), RecC (129 kD), and RecD (67 kD; Kuzminov 1999). Because the protein mobility in native gels depends on native electric charge, we could not exclude the possibility that the value for MDN might be an overestimate. The SDS-PAGE/in gelo assay failed to detect MDN activity, and SDS treatment of the native gel diminished MDN activity, indicating that MDN is sensitive to denaturation (data not shown). Therefore, the precise molecular mass and constitution of MDN remains unclear. Purification and characterization of MDN would reveal the molecular components of MDN, and this information could be used to determine the regulatory mechanism of MDN activity.
MDN activation during mt+ gametogenesis
Several lines of evidence support the possibility that the MDN activity is coregulated with the activation of mt+ gametogenesis: (1) MDN activity is detected only in mature mt+ gametes, and not in mt− gametes or vegetative cells of either mating type; (2) MDN activity is inactivated under dark growth conditions, which also inactivate mt+ gametes; and (3) MDN activity is inactivated by inhibitors of nuclear gene transcription and translation, and this occurs simultaneously with the loss of mating competency by mt+ cells. Taken together, these results suggest that MDN activity is controlled by mt+ gamete-specific nuclear gene expression. The expression of several gamete-specific genes has been reported; von Gromoff and Beck (1993) found that gas genes were expressed in gametes, but that expression was absent in vegetative cells and mature zygotes. Furthermore, the kinetics of the mRNA expression of some gamete-specific genes (i.e., gas3 and gas18) are highly similar to the activation patterns of MDN; mRNA levels of these genes increase during gametogenesis and decrease along with the maturation of zygotes just like MDN activity. In addition, the gsp1 gene, which encodes the homeodomain protein, is expressed only in mt+ gametes (Kurvari et al. 1998; Wilson et al. 1999; Pan and Snell 2000; Zhao et al. 2001). Some of these genes may be responsible for the regulation of MDN activity. Ogawa and Kuroiwa showed that the cp nucleoids of mt+ cells became resistant to Ca2+-dependent nucleases (Ogawa and Kuroiwa 1985b), indicating that the mechanisms used to protect mt+ cpDNA might also operate in mt+ cells during gametogenesis. Further investigations of mt+ gamete-specific genes and insertional mutagenesis of mt+ cells are needed to elucidate the mechanisms underlying the developmental regulation of MDN activity and the protection of mt+ cpDNA against nucleases.
A model for maternal inheritance that is based on MDN
Based on our observations, we propose a tentative model to explain the molecular mechanism behind the uniparental inheritance of mt− cpDNA (Fig. 7). During gametogenesis, synthesis or activation of MDN is triggered only in mt+ cells, presumably by mt+ gamete-specific gene expression. Simultaneously, mt+ cpDNA becomes resistant to MDN, just as to Nuclease C (Ogawa and Kuroiwa 1985b). When the mt+ and mt− gametes fuse, MDN is actively transported into the unprotected mt− chloroplast. MDN then eliminates the mt− cpDNA, which ensures uniparental inheritance of cpDNA. Coupling of gametogenesis and nuclease synthesis or activation appears to be an effective strategy for the digestion of mt− cpDNA, because it ensures that mt+ gametes ready to digest mt− cpDNA can mate with mt− gametes.
The active digestion of mt− cpDNA is a multistep process that includes mt+ cpDNA protection, MDN activation, MDN transportation into chloroplasts, and MDN recruitment to cpDNA strands. Because MDN requires Ca2+ for full activation, regulation of Ca2+ levels in chloroplasts may also be important for this process. There are many reports regarding zygote-specific gene expression (Wegener and Beck 1991; Armbrust et al. 1993; Uchida et al. 1993; Kuriyama et al. 1999; Uchida et al. 1999; Suzuki et al. 2000). Some of these genes may participate in the active digestion of mt− cpDNA. Interestingly, the product of the ezy1 gene has been shown to localize to cp nucleoids of both mating types, just after mating (Armbrust et al. 1993). Therefore, EZY1 may play a role in the digestion of mt− cpDNA by recruiting MDN to cpDNA strands.
On the other hand, Umen and Goodenough (2001) found that treatment of mt+ cells with 200 μM methylation inhibitor 5-aza Cyd, for periods that were longer than those used in previous experiments (Feng and Chiang 1984), perturbed uniparental inheritance of cpDNA. They proposed that the higher replication rate of methylated mt+ cpDNA in mature and germinating zygotes would ensure the uniparental inheritance of cpDNA (Umen and Goodenough 2001). Two distinct mechanisms may operate cooperatively to accomplish uniparental transmission of cpDNA in C. reinhardtii.
Uniparental inheritance of organellar DNA is a feature that is common to numerous taxa of sexual eukaryotes, including higher plants, mosses, ferns, algae, fungi, and animals, including humans. There would be various mechanisms that guarantee maternal inheritance (Birky 1995). In higher plants, the mechanism of maternal inheritance of cpDNA involves exclusion of plastids from generative cells during first pollen mitosis, degeneration of plastids in generative cells during maturation, and the stripping off of paternal plastids during fertilization (Hagemann and Schröder 1989). However, the preferential disappearance of plastid nucleoids has been observed in various lineages of higher plants (Miyamura et al. 1987; Corriveau and Coleman 1988; Nagata et al. 1999), ferns (Kuroiwa 1988), and algae (Kuroiwa and Hori 1986; Kuroiwa et al. 1991). Most notably, it was reported that the paternal mitochondrial DNA (mtDNA) of mammals was actively digested in eggs just after fertilization (Kaneda et al. 1995), although the behavior of the mitochondrial nucleoids was not determined in that particular study. These observations suggest that active digestion of paternal organellar genomes may be one of the universal molecular mechanisms that control maternal inheritance. Further elucidation of the molecular mechanisms that control uniparental inheritance of cpDNA in Chlamydomonas will help to clarify maternal inheritance in the numerous lineages of sexual eukaryotes.
Materials and methods
Strains, culture conditions, and the mating reaction
The wild-type, mating-type plus (mt+), and mating-type minus (mt−) strains were derived from strain 137c of C. reinhardtii. Cells were grown separately on agar plates (1.2% agar in Snell's medium; Snell 1982; Nakamura et al. 1986) at 22°C for 12 h in light followed by 12 h in darkness, to synchronize cell division. The light intensity was ∼6600 lux at the surface of the flat culture container. The moc mutant strain (A84; isolated by Misumi et al. 1999), which has only one condensed chloroplast nucleoid, was maintained under the same conditions. The mating reaction was induced by suspending the cells in nitrogen-free Tsubo mating buffer (1.2 mM HEPES at pH 6.8, 1 mM MgSO4) at a concentration of ∼5 × 106 cells/mL. After a 5–8-h incubation under cool white light (∼6600 lux), equal numbers of mt+ and mt− gametes were mixed and allowed to mate. The chloroplast transformant AG8 mt+ was obtained using the particle bombardment technique (Ishikura et al. 1999). This strain harbors an atpA–uidA chimeric gene in its chloroplast genome, and expresses β-glucuronidase (GUS) in its chloroplast.
DNA staining and microscopic quantification of cpDNA using VIMPCS
Cells were fixed with 1% (w/v) glutaraldehyde in TAN buffer that contained 0.25 M sucrose, 20 mM Tris-HCl (pH 7.6), 0.5 mM EDTA, 1.2 mM spermidine, 7 mM 2-mercaptoethanol, and 1.4 mM phenylmethylsulfonylfluoride (PMSF). The cell suspension was stained with 0.5 μg/mL DAPI in TAN buffer. Cells were observed under ultraviolet (UV) excitation with a fluorescence microscope (Olympus BHS-RFK). Vital staining of DNA in living C. reinhardtii cells was performed with SYBR Green I (Molecular Probes) using a method described previously (Nishimura et al. 1998). The fluorescent intensity of chloroplast nucleoids was quantified using VIMPCS (Kuroiwa et al. 1986). The photons emitted from the fluorescent chloroplast nucleoids were counted every 2 sec and expressed in terms of arbitrary units, T. The number of photons emitted by a DAPI-stained T4 phage (170 kb) was defined as T = 1.
Indirect immunofluorescence microscopy
GUS protein was detected in AG8 mt+ chloroplasts by immunofluorescent staining according to a previously described method (Kuriyama et al. 1999). Cells were harvested and fixed in 4% formaldehyde at room temperature for 10 min. After washing with PBS, the cells were gently permeabilized with 0.1% Triton X-100 at room temperature for 20 min. They were then resuspended in TBS buffer (5% bovine serum albumin [BSA], 0.25% Tween 20 in PBS) and kept at room temperature for 30 min. The cells were then incubated at 37°C for 1 h with the primary antibody (rabbit anti-β-glucuronidase IgG; Sigma) at a 1:2000 dilution. After washing, the cells were resuspended in TBS buffer at room temperature for 30 min. The secondary antibody (FITC-conjugated goat anti-rabbit IgG) was added at a 1:1000 dilution and incubated at 37°C for 1 h. The cells were washed, stained with 1% DAPI, and observed by fluorescent microscopy under UV (for DAPI) or blue (for FITC) light.
Isolation of intact chloroplasts using an airbrush
Chloroplasts were isolated from zygotes using an airbrush. Zygotes were harvested and resuspended in 30 mM sucrose solution at room temperature. The zygotes were then broken by a single passage through an airbrush at a pressure of 0.7 kg/cm2 (0.2-mm aperture airbrush [HP-62B, OLYMPOS, Japan] connected to an air regulator and air compressor [TC-65-21, OLYMPOS]). Cell lysates were collected in a precooled, 300-mL beaker. After centrifugation at 1000g at 4°C for 3 min, the precipitated chloroplasts were resuspended in suspension buffer (30 mM sucrose, 5% polyethylene glycol 6000, 1.2 mM HEPES-KOH at pH 6.8, 1 mM MgSO4, and 0.1% BSA) and maintained at 4°C. For further purification, cell lysates were layered on top of a discontinuous Percoll gradient (45%–80% Percoll containing 30 mM sucrose, 1.2 mM HEPES-KOH at pH 6.8, 1 mM MgSO4). Percoll gradient centrifugation was performed using 2-mL centrifugation tubes. After centrifugation at 10,000g at 4°C for 30 min, the green band that appeared at the interface between the 45% and 80% layers of Percoll was collected. The chloroplast fraction was diluted fourfold using the suspension buffer and then centrifuged at 1000g at 4°C for 3 min. The precipitated chloroplasts were used for the native-PAGE analysis.
Preparation of soluble protein extracts
Cells were suspended in extraction buffer (10 mM Tris-HCl at pH 8.5, 0.1 mM EDTA, 0.2 mM PMSF) and ground into a fine powder using liquid N2 in a prechilled mortar and pestle. The powder was collected in sampling tubes and incubated at 37°C for 10 min. After centrifugation at 18,000g at 4°C for 15 min, the supernatants were used as crude soluble protein extracts in enzyme assays. Isolated chloroplasts were treated similarly. The soluble protein concentrations of extracts were determined with a protein assay kit (Bio-Rad) using BSA as a standard.
Native-PAGE/in gelo assay for nucleases
The analysis of nucleases was performed in native-PAGE gels using the following modifications to the SDS-PAGE/in gelo assay method (Ogawa and Kuroiwa 1985a). Soluble protein extracts were mixed with an equal volume of native sampling buffer (63 mM Tris-HCl at pH 6.8, 30% glycerol, 0.01% bromophenol blue). Proteins were separated using a stacking gel buffer (3.3% [w/v] acrylamide, 63 mM Tris-HCl at pH 6.8, 2 mM EDTA) and a separating gel buffer (5.0% [w/v] acrylamide, 375 mM Tris-HCl at pH 8.8, 2 mM EDTA, 10 μg/mL salmon testes DNA). After electrophoresis (150 V at 4°C for 33 min) in nondenaturing electrophoresis buffer (25 mM Tris, 192 mM glycine at pH 8.3), the gels were washed in an incubation buffer (0.2 mM PMSF, 0.1 mM DTT) for 30 min and then incubated overnight in the reaction buffer (10 mM Tris-HCl at pH 8.5, 0.2 mM PMSF, 0.1 mM DTT, 10 mM CaCl2). Nuclease activity was visualized by staining the gels with ethidium bromide and viewing under UV light. In some experiments, the intensities of the activity signals were quantified using an ATTO Lane Analyzer System (ATTO). A high-molecular-weight calibration kit for native electrophoresis (Amersham Pharmacia Biotech) was used as the molecular weight marker in the native-PAGE/in gelo assay. This kit contains a mixture of thyroglobulin (669 kD), ferritin (440 kD), catalase (232 kD), lactase dehydrogenase (140 kD), and bovine serum albumin (67 kD). In these experiments, duplicates of each sample were electrophoresed on the same gel. One of the duplicate samples was used for in gelo detection of nuclease activity, and the other was stained with CBB to detect RuBisCO bands. The RuBisCO band was a prominent band on CBB-stained native-PAGE gels, and was positively detected by Western blotting using a rabbit antibody raised against the large subunit of RuBisCO (data not shown).
Acknowledgments
We gratefully acknowledge the support of the Japan Society for the Promotion of Science (Research Fellowship number 13-5096 to Y.N.), the Ministry of Education, Culture, Sports, Science, and Technology of Japan (Grants 12440222 and 13206011 to T.K.), and the Program for the Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN grant to T.K.). We also thank Ikuo Nishida, Kazumi Nakabayashi, Narie Sasaki, and Yutaka Miyazawa for helpful technical advice.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL yoshiki@biol.s.u-tokyo.ac.jp; FAX 81-3-3814-1408.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.979902.
References
- Armbrust EV, Ferris PJ, Goodenough UW. A mating type-linked gene cluster expressed in Chlamydomonas zygotes participates in the uniparental inheritance of the chloroplast genome. Cell. 1993;74:801–811. doi: 10.1016/0092-8674(93)90460-8. [DOI] [PubMed] [Google Scholar]
- Beck CF, Haring MA. Gametic differentiation of Chlamydomonas. Int Rev Cytl. 1996;168:259–302. [Google Scholar]
- Belknap WR. Partial purification of intact chloroplasts from Chlamydomonas reinhardtii. Plant Physiol. 1983;72:1130–1132. doi: 10.1104/pp.72.4.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birky CW., Jr Uniparental inheritance of mitochondrial and chloroplast genes: Mechanism and evolution. Proc Natl Acad Sci. 1995;92:11331–11338. doi: 10.1073/pnas.92.25.11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolen BL, Grant DM, Swinton D, Boynton JE, Gillham NW. Extensive methylation of chloroplast DNA by a nuclear gene mutation does not affect chloroplast gene transmission in Chlamydomonas. Cell. 1982;28:335–343. doi: 10.1016/0092-8674(82)90351-8. [DOI] [PubMed] [Google Scholar]
- Burton WG, Roberts RJ, Myers PA, Sager R. A site-specific single-strand endonuclease from the eukaryote Chlamydomonas. Proc Natl Acad Sci. 1977;7:2687–2691. doi: 10.1073/pnas.74.7.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton WG, Gravowy CT, Sager R. Role of methylation in the modification and restriction of chloroplast DNA in Chlamydomonas. Proc Natl Acad Sci. 1979;76:1390–1394. doi: 10.1073/pnas.76.3.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Electron microscopy of zygospore formation in Chlamydomonas reinhardtii. Protoplasma. 1976;87:297–315. doi: 10.1007/BF01624002. [DOI] [PubMed] [Google Scholar]
- Correns C. Vererbungsversuche mit blaβ-(gelb-)-grünen und buntblättrigen Sippen bei Mirabilis jalapa, Urtica pilulifera und Lunaria annua. Z Abst u Vererbl. 1909;1:291–329. [Google Scholar]
- Corriveau JS, Coleman AW. Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperms species. Am J Bot. 1988;75:1443–1458. [Google Scholar]
- Ebersold W, Levine RP, Levine EE, Olmsted MA. Linkage maps in Chlamydomonas reinhardtii. Genetics. 1962;47:531–543. doi: 10.1093/genetics/47.5.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- Feng T-Y, Chiang K-S. The persistence of maternal inheritance in Chlamydomonas despite hypomethylation of chloroplast DNA induced by inhibitors. Proc Natl Acad Sci. 1984;81:3438–3442. doi: 10.1073/pnas.81.11.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles RE, Blanc H, Cann HM, Wallace DC. Maternal inheritance of human mitochondrial DNA. Proc Natl Acad Sci. 1980;77:6715–6719. doi: 10.1073/pnas.77.11.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillham NW. Organelle heredity. New York, NY: Raven Press; 1978. [Google Scholar]
- ————— . Organelle genes and genomes. New York, NY: Oxford University Press; 1994. [Google Scholar]
- Goldschmidt-Clermont M, Malnoë P, Rochaix JD. Preparation of Chlamydomonas chloroplasts for the in vitro import of polypeptide precursors. Plant Physiol. 1989;89:15–18. doi: 10.1104/pp.89.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann R, Schröder MB. The cytological basis of the plastid inheritance in angiosperm. Protoplasma. 1989;152:57–64. [Google Scholar]
- Harris EH. Chlamydomonas sourcebook. San Diego, CA: Academic Press; 1989. [Google Scholar]
- Hutchison CA, III, Newbold JE, Potter SS, Edgell MH. Maternal inheritance of mammalian mitochondrial DNA. Nature. 1974;251:536–538. doi: 10.1038/251536a0. [DOI] [PubMed] [Google Scholar]
- Ishikura K, Takaoka Y, Sekine M, Yoshida K, Shinmyo A. Expression of a foreign gene in Chlamydomonas reinhardtii chloroplast. J Biosci Bioeng. 1999;87:307–314. doi: 10.1016/s1389-1723(99)80037-1. [DOI] [PubMed] [Google Scholar]
- Kaneda H, Hayashi J, Takahama S, Taya C, Lindahl KF, Yonekawa H. Elimination of paternal mitochondrial DNA in intraspecific crosses during early mouse embryogenesis. Proc Natl Acad Sci. 1995;92:4542–4546. doi: 10.1073/pnas.92.10.4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano S, Anderson RW, Nanba T, Kuroiwa T. Polymorphism and uniparental inheritance of mitochondrial DNA in Physarum polycephalum. J Gen Microbiol. 1987;133:3175–3182. doi: 10.1099/00221287-133-11-3175. [DOI] [PubMed] [Google Scholar]
- Kirk JTO, Tilney-Basset RAE. The plastids: Their chemistry, structure, growth, and inheritance. 2nd ed. Oxford, UK: Elsevier/North-Holland Biomedical Press; 1978. [Google Scholar]
- Klein U, Chen C, Gibbs M, Platt-Aloia KA. Cellular fractionation of Chlamydomonas reinhardtii with emphasis on the isolation of the chloroplast. Plant Physiol. 1983;72:481–487. doi: 10.1104/pp.72.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyama H, Takano H, Suzuki L, Uchida H, Kawano S, Kuroiwa H, Kuroiwa T. Characterization of Chlamydomonas reinhardtii zygote-specific cDNAs that encode novel proteins containing ankyrin repeats and WW domains. Plant Physiol. 1999;119:873–884. doi: 10.1104/pp.119.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa H, Sugai M, Kuroiwa T. Behavior of chloroplasts and chloroplast nuclei during spermatogenesis in the fern, Pteris vittata L. Protoplasma. 1988;146:89–100. [Google Scholar]
- Kuroiwa T. Mechanisms of maternal inheritance of chloroplast DNA: An active digestion hypothesis. Microbiol Sci. 1985;2:267–272. [PubMed] [Google Scholar]
- ————— The replication, differentiation, and inheritance of plastids with emphasis on the concept of organelle nuclei. Int Rev Cytl. 1991;128:1–60. [Google Scholar]
- Kuroiwa T, Hori T. Preferential digestion of male chloroplast nuclei and mitochondrial nuclei during gametogenesis of Bryopsis maxima Okamura. Protoplasma. 1986;133:85–87. [Google Scholar]
- Kuroiwa T, Suzuki T, Ogawa K, Kawano S. The chloroplast nucleus: Distribution, number, size, and shape, and a model for the multiplication of the chloroplast genome during chloroplast development. Plant Cell Physiol. 1981;22:381–396. [Google Scholar]
- Kuroiwa T, Kawano S, Nishibayashi S, Sato C. Epifluorescent microscopic evidence for maternal inheritance of chloroplast DNA. Nature. 1982;298:481–483. doi: 10.1038/298481a0. [DOI] [PubMed] [Google Scholar]
- Kuroiwa T, Miyakawa S, Kawano S, Hizume M, Toh-e A, Miyakawa I, Sando N. Cytological characterization of NOR in the bivalent of Saccharomyces cerevisiae. Exp Cell Res. 1986;165:199–206. doi: 10.1016/0014-4827(86)90544-6. [DOI] [PubMed] [Google Scholar]
- Kuroiwa T, Kawano S, Watanabe M, Hori T. Preferential digestion of chloroplast DNA in male gametangia during the late stage of gametogenesis in the anisogamous alga Briopsis maxima. Protoplasma. 1991;163:102–113. [Google Scholar]
- Kurvari V, Grishin NV, Snell WJ. A gamete-specific, sex-limited homeodomain protein in Chlamydomonas. J Cell Biol. 1998;143:1971–1980. doi: 10.1083/jcb.143.7.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage λ. Microbiol Mol Biol Rev. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason CB, Matthews S, Bricker TM, Moroney JV. Simplified procedure for the isolation of intact chloroplasts from Chlamydomonas reinhardtii. Plant Physiol. 1991;97:1576–1580. doi: 10.1104/pp.97.4.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misumi O, Suzuki L, Nishimura Y, Sakai A, Kawano S, Kuroiwa H, Kuroiwa T. Isolation and phenotypic characterization of Chlamydomonas reinhardtii mutants defective in chloroplast DNA segregation. Protoplasma. 1999;209:273–282. [Google Scholar]
- Mitchell MB, Mitchell HK. A case of “maternal” inheritance in Neurospora crassa. Proc Natl Acad Sci. 1952;38:442–449. doi: 10.1073/pnas.38.5.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Lam E. Identification, characterization, and purification of a tobacco endonuclease activity induced upon hypersensitive response cell death. Plant Cell. 1995;7:1951–1962. doi: 10.1105/tpc.7.11.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamura S, Kuroiwa T, Nagata T. Difference of plastid and mitochondrial nucleoids during the formation of generative cells of higher plants revealed by fluorescence microscopy. Protoplasma. 1987;141:149–159. [Google Scholar]
- Nagata N, Saito C, Sakai A, Kuroiwa H, Kuroiwa T. The selective increase or decrease of organellar DNA in generative cells just after pollen mitosis one controls cytoplasmic inheritance. Planta. 1999;209:53–65. doi: 10.1007/s004250050606. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Itoh S, Kuroiwa T. Behavior of chloroplast nucleus during chloroplast development and degradation in Chlamydomonas reinhardtii. Plant Cell Physiol. 1986;27:775–784. [Google Scholar]
- Nishimura Y, Higashiyama T, Suzuki L, Misumi O, Kuroiwa T. The biparental transmission of the mitochondrial genome in Chlamydomonas reinhardtii visualized in living cells. Eur J Cell Biol. 1998;77:124–133. doi: 10.1016/S0171-9335(98)80080-0. [DOI] [PubMed] [Google Scholar]
- Nishimura Y, Misumi O, Matsunaga S, Higashiyama T, Yokota A, Kuroiwa T. The active digestion of uniparental chloroplast DNA in a single zygote of Chlamydomonas reinhardtii is revealed by using the optical tweezers. Proc Natl Acad Sci. 1999;96:12577–12582. doi: 10.1073/pnas.96.22.12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa K, Kuroiwa T. Nuclease C: Polymorphism of calcium-dependent nucleases in Chlamydomonas reinhardtii. Plant Cell Physiol. 1985a;26:481–491. [Google Scholar]
- ————— Destruction of chloroplast nuclei of the male gamete by calcium-dependent nucleases in Chlamydomonas reinhardtii. Plant Cell Physiol. 1985b;26:493–503. [Google Scholar]
- Pan J, Snell WJ. Signal transduction during fertilization in the unicellular green alga, Chlamydomonas. Curr Opin Microbiol. 2000;3:596–602. doi: 10.1016/s1369-5274(00)00146-6. [DOI] [PubMed] [Google Scholar]
- Royer HD, Sager R. Methylation of chloroplast DNAs in the life cycle of Chlamydomonas. Proc Natl Acad Sci. 1979;76:5794–5798. doi: 10.1073/pnas.76.11.5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R. Mendelian and non-Mendelian inheritance of streptomycin resistance in Chlamydomonas reinhardtii. Proc Natl Acad Sci. 1954;40:356–363. doi: 10.1073/pnas.40.5.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R, Lane D. Molecular basis of maternal inheritance. Proc Natl Acad Sci. 1972;69:2410–2413. doi: 10.1073/pnas.69.9.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H, Gravowy CT, Sager R. Differential activity of DNA methyltransferase in the life cycle of Chlamydomonas reinhardtii. Proc Natl Acad Sci. 1981;78:3118–3122. doi: 10.1073/pnas.78.5.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz DC, Samad A. Optical mapping approaches to molecular genomics. Curr Opin Biotechnol. 1997;8:70–74. doi: 10.1016/s0958-1669(97)80160-7. [DOI] [PubMed] [Google Scholar]
- Sears BB. Elimination of plastids during spermatogenesis in the plant kingdom. Plasmid. 1980;4:233–255. doi: 10.1016/0147-619x(80)90063-3. [DOI] [PubMed] [Google Scholar]
- Snell WJ. Study of the release of cell wall degrading enzymes during adhesion of Chlamydomonas gametes. Exp Cell Res. 1982;138:109–119. doi: 10.1016/0014-4827(82)90096-9. [DOI] [PubMed] [Google Scholar]
- Sutovsky P, Schatten G. Paternal contribution to the mammalian zygote: Fertilization after sperm–egg fusion. Int Rev Cytl. 2000;195:1–65. doi: 10.1016/s0074-7696(08)62703-5. [DOI] [PubMed] [Google Scholar]
- Suzuki L, Woessner JP, Uchida H, Kuroiwa H, Yuasa Y, Waffenschmidt S, Goodenough UW, Kuroiwa T. A zygote-specific protein with hydroxyproline-rich glycoprotein domains and lectin-like domains involved in the assembly of the cell wall of Chlamydomonas reinhardtii (Chlorophyta) J Phycol. 2000;36:571–583. doi: 10.1046/j.1529-8817.2000.99112.x. [DOI] [PubMed] [Google Scholar]
- Thelen MP, Northcote DH. Identification and purification of a nuclease from Zinnia elegans L.: A potential molecular marker for xylogenesis. Planta. 1989;179:181–195. doi: 10.1007/BF00393688. [DOI] [PubMed] [Google Scholar]
- Uchida H, Kawano S, Sato N, Kuroiwa T. Isolation and characterization of novel genes which are expressed during the very early stage of zygote formation in Chlamydomonas reinhardtii. Curr Genet. 1993;24:296–300. doi: 10.1007/BF00336779. [DOI] [PubMed] [Google Scholar]
- Uchida H, Suzuki L, Anai T, Doi K, Takano H, Yamashita H, Oka T, Kawano S, Tomizawa K, Kawazu T, et al. A pair of invertedly repeated genes in Chlamydomonas reinhardtii encodes a zygote-specific protein whose expression is UV-sensitive. Curr Genet. 1999;36:232–240. doi: 10.1007/s002940050495. [DOI] [PubMed] [Google Scholar]
- Umen JG, Goodenough UW. Chloroplast DNA methylation and inheritance in Chlamydomonas. Genes & Dev. 2001;15:2585–2597. doi: 10.1101/gad.906701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbano A, McCaffrey R, Foss F. Isolation and characterization of NUC70, a cytoplasmic hematopoietic apoptotic endonuclease. J Biol Chem. 1998;273:34820–34827. doi: 10.1074/jbc.273.52.34820. [DOI] [PubMed] [Google Scholar]
- von Gromoff ED, Beck CF. Gene expressed during sexual differentiation of Chlamydomonas reinhardtii. Mol Gen Genet. 1993;241:415–421. doi: 10.1007/BF00284695. [DOI] [PubMed] [Google Scholar]
- Wallis R, Reilly A, Barnes K, Abell C, Campbell DG, Moore GR, James R, Kleanthous C. Tandem overproduction and characterization of the nuclease domain of colicin E9 and its cognate inhibitor protein Im9. Eur J Biochem. 1994;220:447–454. doi: 10.1111/j.1432-1033.1994.tb18642.x. [DOI] [PubMed] [Google Scholar]
- Wang C-C, Lu S-C, Chen H-L, Liao T-H. Porcine spleen deoxyribonuclease II: Covalent structure, cDNA sequence, molecular cloning, and gene expression. J Biol Chem. 1998;273:17192–17198. doi: 10.1074/jbc.273.27.17192. [DOI] [PubMed] [Google Scholar]
- Wegener D, Beck CF. Identification of novel genes specifically expressed in Chlamydomonas reinhardtii zygotes. Plant Mol Biol. 1991;16:937–946. doi: 10.1007/BF00016066. [DOI] [PubMed] [Google Scholar]
- Whatley JM. Ultrastructure of plastid inheritance: Green algae to angiosperms. Biol Rev. 1982;57:527–569. [Google Scholar]
- Wilson NF, O'Connell JS, Lu M, Snell WJ. Flagellar adhesion between mt(+) and mt(−) Chlamydomonas gametes regulates phosphorylation of the mt+ specific homeodomain protein GSP1. J Biol Chem. 1999;274:34383–34388. doi: 10.1074/jbc.274.48.34383. [DOI] [PubMed] [Google Scholar]
- Xu Y, Hanson MR. Programmed cell death during pollination-induced petal senescence in petunia. Plant Physiol. 2000;122:1323–1333. doi: 10.1104/pp.122.4.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Lu M, Singh R, Snell WJ. Ectopic expression of a Chlamydomonas mt+-specific homeodomain protein in mt− gamete initiates zygote development without gamete fusion. Genes & Dev. 2001;15:2767–2777. doi: 10.1101/gad.919501. [DOI] [PMC free article] [PubMed] [Google Scholar]