1. Summary
Toll-like receptor (TLR) ligands are notable for their ability to induce APC maturation, which in turn facilitates optimal T cell mediated immune responses. Toll-like receptor ligands, such as CpG DNA, can also modulate immune responses by blocking the suppressive effects of CD4+CD25+ regulatory T cells (Tregs). Recently, we have demonstrated that CpG DNA, in addition to its actions on APCs and Tregs, can provide direct costimulatory signals to CD4+CD25- T cells. Here we show that this costimulatory effect is sufficient to abrogate suppression by Tregs. These data indicate a previously undefined role for TLR ligands in directly modulating CD4+ T cell responses.
Keywords: CpG DNA, Costimulation, CD4+CD25+ regulatory T cells
2. Introduction
Pathogen Associated Molecular Patterns (PAMPs) are microbial products known to indirectly promote optimal T cell mediated immune responses via stimulation of toll-like receptors (TLRs) on APCs [1]. In addition to these indirect effects on T cell responses a number of groups have demonstrated that distinct T cell subsets are also sensitive to direct stimulation with specific PAMPs [2-5]. Previously we have reported that the TLR ligand CpG DNA or PolyI:C could directly activate NF-κB signaling leading to enhanced activated CD4+ T cell survival [6]. More recently, we demonstrated that CpG DNA also provides a MyD88 dependent PI-3 kinase mediated costimulatory signal to CD4+ T cells that directly enhances IL-2 production and proliferation [7].
Although enhanced CD4+ T cell responses contribute to efficient T-cell mediated immunity, mechanisms also exist to restrict such responses and prevent the development of a potentially damaging autoimmune response. One such mechanism is through the activity of CD4+CD25+ regulatory T cells (Tregs), which play a central role in the establishment and maintenance of T cell tolerance. Tregs are known to suppress the development of autoimmunity in vivo and inhibit the responses of effector T cells in vitro, however their precise mode of action remains poorly understood [8]. The effects of specific PAMPs on the suppressive function of Tregs have been the subject of recent investigation. Notably it has been reported that pretreatment of human Treg cell lines with CpG DNA can partially abrogate their suppressive effects through direct stimulation of TLR8 [5]. However, the model system used during this study precluded determination of whether or not CpG DNA also exerted separate effects on the responder T cell population, thereby allowing them to escape suppression.
Given our recent finding that CpG DNA provides costimulatory signals to CD4+ T cells [7], here we examined the effects that CpG DNA stimulation of CD4+CD25-effector T cells would have on their susceptibility to suppression by CD4+CD25+ Tregs. Consistent with previous work, we found that CpG DNA can act directly on Tregs to block suppression. We show for the first time that the direct MyD88-dependent signal which CpG DNA provides to CD4+CD25- T cells is also sufficient to allow their escape from suppression. Together these results describe a previously undefined role for CpG DNA in modulating CD4+ T cell responses by allowing them to evade Treg suppression.
3. Materials and Methods
Mice
C57BL/6 mice were purchased from the Jackson Laboratory (Bar Harbor, ME). MyD88 deficient mice were a kind gift from S. Akira (Osaka University, Japan). All animals were maintained under specific pathogen-free conditions in the animal facilities of the University of Pennsylvania, Philadelphia, PA.
Media, reagents Abs and flow cytometry
Cells were cultured in RPMI 1640 supplemented with 10% FCS, 100U/ml penicillin, 100ug/ml streptomycin, 2mM L-glutamine, 10mM HEPES (Life Technologies, Rockland, MD) and 50μM 2-mercaptoethanol (Sigma-Aldrich, St. Louis, MO). Biotin-anti-CD25 (7D4), FITC-anti-CD4 (RM4-5) streptavidin-PE, purified anti-CD3 (2C11) were purchased from BD Pharmingen (San Diego, CA). Previously described control and stimulatory oligodeoxynucleotides [7], were synthesized on a phosphorothioate backbone and purified by HPLC (Life Technologies). Sequences are as follows, CpG DNA, TCCATGACGTTCCTGATGCT; the inverted GpC ODN, TCCATGA GCTTCCTGATGCT; and the nonstimulatory ODN, GCTTGATGACTCAGCCGG AA
CD4+CD25+ and CD4+CD25- T cell isolation
Spleen and lymph node cells were isolated from 6-8 week old mice. Cell preparations were stained with anti-CD4 FITC, anti-CD25 biotin, streptavidin-PE, CD11c APC and subsequently purified into CD4+CD25-CD11c- cells by flow cytometry on a FACSVantage Cell Sorter (BD Biosciences). Cell purity was routinely greater than 98%.
Suppression assays
2×105 FACS purified CD4+CD25- cells were stimulated for 72 hours, in the presence or absence of CD4+CD25+ Tregs, with latex beads (1 bead/1 T cell) coated with anti-CD3 (2C11) (1μg/ml) and anti-CD28 (2.5μg/ml). Cells were cultured in complete media and supplemented with either CpG DNA (3.3μM) or left untreated. Cells were pulsed with 0.5uCi of tritiated thymidine for the final 16 hours before being harvested. Alternatively, CD4+CD25- T cells with and without CD4+CD25+ Tregs (2:1), were stimulated with 8ng/ml anti-CD3 in the presence of MyD88 deficient APCs with the addition of CpG DNA or control oligodeoxynucleotides (0.3μM). Again cells were pulsed with 0.5uCi of tritiated thymidine for the final 16 hours before being harvested.
Foxp3 analysis
Purified CD4+CD25- T cells with or without CD4+CD25+ Tregs (1×106: 5×105) were stimulated with soluble anti-CD3 (8ng/ml) in the presence of irradiated MyD88 deficient APCs at a ratio of 1 T cell to 2 APCs. Cells were left untreated or cultured in the presence of either CpG DNA, control inverted GpC or non-stimulatory oligodeoxynucleotides (0.3μM) for 24hours. After 24 hours cells were stained for expression of Foxp3 using Foxp3 staining kit from e bioscience. Total live cells based on forward side scatter profiles were analysed for Foxp3 expression.
Fluoresence-linked immunosorbent assay
107 5μm latex beads (Interfacial Dynamics, Portland, Oregon) were coated with anti-IL-2 capture Ab (10μg/ml) (Pharmingen) in PBS for 90 mins at 37°C. 2×105 beads were added to 100μl of test supernatant or titrated amounts of rIL-2 in 100ul complete medium as standards. Bead bound IL-2 was detected using PE –labeled anti-IL-2 (Pharmingen) and subsequent FACS analysis.
4. Results and Discussion
CpG DNA can break CD4+CD25+ regulatory T cell mediated suppression in an APC free or APC unresponsive system
It has previously been reported that the addition of CpG DNA to a standard in vitro suppression assay significantly abrogates Treg mediated suppression. This effect can be mediated indirectly by the APC through TLR9 induced upregulation of costimulatory factors and cytokines, particularly IL-6 [9]. More recently, CpG DNA has been shown to act directly on human CD4+CD25+ regulatory T cell lines to inhibit their suppressive function [5].
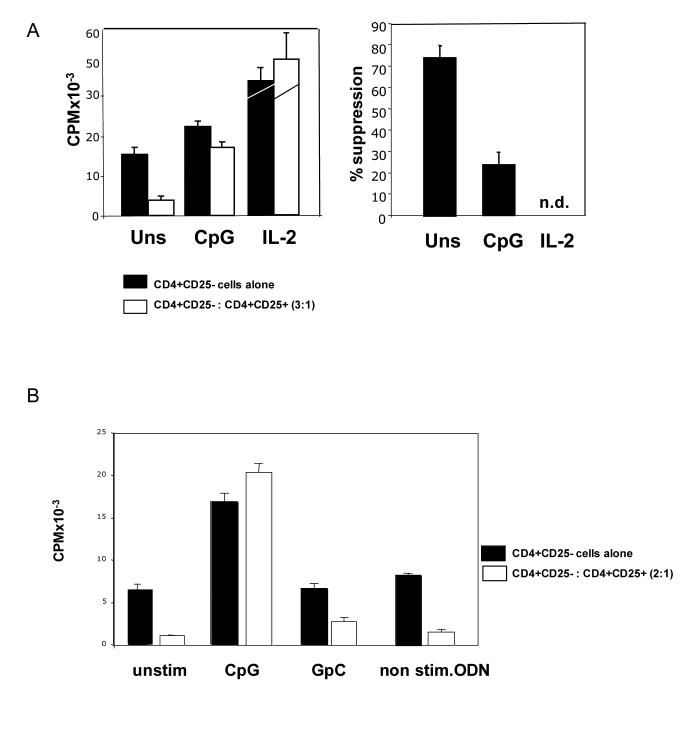
In order to examine further the effects of CpG DNA on Treg activity in our system we isolated highly pure (≥ 98%) Treg and effector CD4+ T cell subsets by FACS and examined the effects of CpG DNA on Treg mediated suppression in an APC free system. By replacing the APCs in a conventional suppression assay with latex beads coated with antibodies to CD3 and the co-stimulatory receptor CD28, we were able to determine if there were effects of CpG DNA stimulation directly on the T cell subsets. In agreement with previous reports, CpG DNA inhibited Treg mediated suppression as assayed by incorporation of tritiated thymidine to measure cell proliferation (Figure 1A). The addition of CD4+CD25+ Tregs at a ratio of 1:3 with CD4+CD25- effector cells resulted in significant inhibition of effector cell expansion (75%) and in the presence of CpG DNA this suppressive effect was reduced to only 22% (Figure 1A). The addition of exogenous rIL-2 (20U/ml) is reported to break Treg mediated suppression of effector cell proliferation and serves as a positive control [10].
Figure 1.
CpG DNA can inhibit CD4+CD25+ regulatory T cell suppression in an APC free system.
(A) Purified CD4+CD25- T cells were cultured in the presence or absence of CD4+CD25+ Tregs at a ratio of 3:1. Polystyrene beads coated with anti-CD3 (1μg/ml) and anti-CD28 (2.5μg/ml) were used as surrogate APCs, (1 beads for every T cell). Cells were left untreated or CpG DNA (3.3μM) or IL-2 (20U/mL) was added to cultures. Tritiated thymidine was added for the final 16 hours of culture before harvesting. Results are illustrated in terms of thymidine incorporation (CPM) and % suppression of effector T cell response. Graph shows mean +/− s.d. of triplicate cultures. Data are representative of four independent experiments. (B) Purified CD4+CD25- T cells with or without CD4+CD25+ Tregs (1×105: 5×104) were stimulated with soluble anti-CD3 (8ng/ml) in the presence of irradiated MyD88 deficient APCs at a ratio of 1 T cell to 2 APCs. Cells were left untreated or cultured in the presence of CpG DNA, control inverted GpC or non-stimulatory oligodeoxynucleotides (0.3μM) for 72 hours. Tritiated thymidine was added for final 16 hours of culture. Data presented as mean +/− s.d. of triplicate cultures and is representative of three independent experiments.
To determine whether the ability of CpG DNA to abrogate Treg mediated suppression was specific for oligodeoxynucleotides containing stimulatory CpG motifs, we carried out suppression assays using soluble anti-CD3 stimulation in the presence of MyD88-deficient APCs. We have previously used this approach to examine T-cell specific responses to costimulation with CpG DNA [7]. Studies of CpG-DNA-activated signaling pathways have revealed that the primary receptor through which CpG DNA mediates its effects is TLR9 [11]. While some evidence also exists indicating CpG oligodeoxynucleotides may exert some of its effects in a TLR9 independent fashion, possibly through DNA dependent protein kinases (DNA-PKcs) [12], most reports demonstrate a requirement for the TLR/IL-1 (TIR) domain containing adaptor molecule MyD88 in mediating costimulation by CpG DNA [13,14]. Indeed, Hemmi et al, using the same CpG oligonucleotide used in our studies, have previously demonstrated that while DNA-PKcs deficient APCs mature normally upon CpG DNA stimulation, MyD88 is required for both inflammatory cytokine synthesis and functional maturation [14]. Therefore we reasoned that this approach would allow us to examine the effects of CpG DNA stimulation specifically on T cell subsets, as the APCs present would not respond to CpG DNA in a manner which might indirectly affect T cell responses. As shown in figure 1B, while the presence of control oligodeoxynucleotides had no observable effects, CpG DNA completely abrogated Treg mediated suppression. These data confirm that phosphorothioate-modified oligodeoxynucleotides containing stimulatory CpG motifs can specifically inhibit the suppressive effects of CD4+CD25+ Tregs by acting directly on CD4+ T cell subsets.
MyD88 expression in effector CD4+CD25- T cells mediates CpG DNA induced abrogation of Treg mediated suppression
It has already been reported that CpG DNA can break Treg mediated suppression by acting directly on regulatory T cells themselves. Recently, we have described the costimulatory effects of CpG DNA on effector CD4+CD25- T cells. As costimulation of effector T cells through co-receptors such as CD28 and GITR can facilitate escape of these cells from the suppressive effects of Treg suppression [10,15], we hypothesized that CpG DNA may also break suppression by acting directly on the responding effector cells.
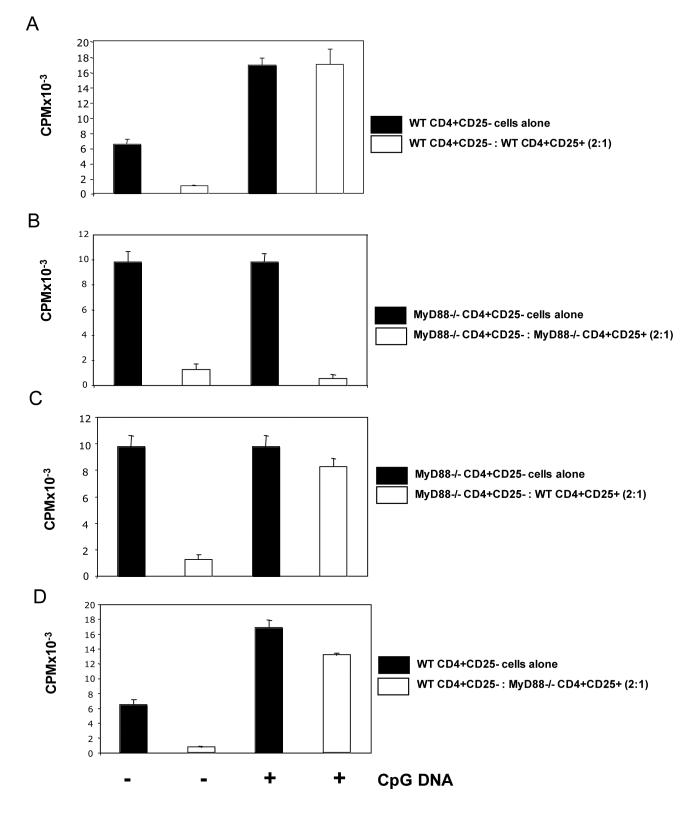
To address this question we isolated CD4+CD25- and CD4+ CD25+ cells from both wild-type and MyD88-deficient mice. Suppression assays were carried out with different combinations of either wild-type or MyD88-deficient T cell subsets all stimulated with soluble anti-CD3 in the presence of irradiated MyD88-deficient APCs. Importantly, when both CD4+CD25- and CD4+CD25+ T cells were isolated from MyD88 deficient animals, CpG DNA had no observable effects on suppression (Figure 2B). These data are consistent with our previous observations that CpG DNA can break suppression by acting directly on T cell subsets. Most significantly however, the effects of CpG DNA in abrogating Treg suppression were restored not only when Tregs were the only subset responsive to CpG DNA stimulation, but also when wild-type (CpG DNA responsive) CD4+CD25- effector cells were cocultured with MyD88-deficient (CpG DNA unresponsive) Tregs (Figure 2C&D). These data demonstrate that the direct costimulatory effects of CpG DNA can allow CD4+CD25- effector cells to escape from Treg mediated suppression.
Figure 2.
CpG DNA can inhibit Treg suppression by stimulating a MyD88 dependent signaling pathway in responder T cells.
Purified wild type (A&D) or MyD88 deficient (B&C) CD4+CD25- T cells (1×105) were stimulated with anti-CD3 (8ng/ml) plus irradiated MyD88 deficient APCs (2×105) for 72 hours in the presence or absence of MyD88 deficient (B&D) or wild type (A&C) CD4+CD25+Tregs (5×104). Cells were left untreated or CpG DNA (0.3μM) was added to cultures. Tritiated thymidine was added for the final 16 hours of culture before harvesting. Data presented as mean +/− s.d. of triplicate samples and are representative of three independent experiments.
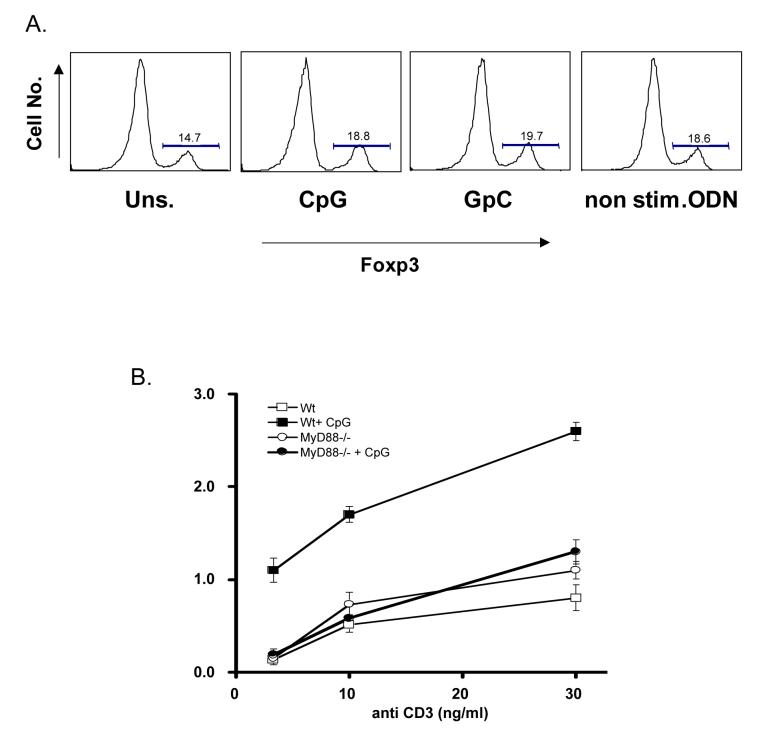
Interestingly, similar effects have recently been ascribed to another PAMP, the bacterial lipoprotein ligand for TLR2, which modulates Treg suppression through direct stimulation of both effector and regulatory T cell subsets [4]. TLR2 mediated downregulation of the transcription factor Foxp3, which is a critical mediator of the Treg phenotype, was proposed recently as a partial mechanism for these effects. However, we did not observe any such downmodulation in Foxp3 expression among Tregs, in the context of active suppression, in response to stimulation with CpG DNA (Fig. 3A). As such it will be of interest to further investigate the precise mechanisms by which direct CpG DNA stimulation Tregs abrogates suppression and how these events may differ from the costimulatory effects of CpG DNA on effector T cell subsets.
Figure 3.
CpG DNA induced expression of IL-2 requires MyD88.
(A) Purified CD4+CD25- T cells with or without CD4+CD25+ Tregs (1×106: 5×105) were stimulated with soluble anti-CD3 (8ng/ml) in the presence of irradiated MyD88 deficient APCs at a ratio of 1 T cell to 2 APCs. Cells were left untreated or cultured in the presence of either CpG DNA, control inverted GpC or non-stimulatory oligodeoxynucleotides (0.3μM) for 24hours. After 24 hours cells were stained for expression of Foxp3 and analyzed by flow cytometry. (B) Purified MyD88 deficient or wild type CD4+CD25- T cells were stimulated with graded doses of soluble anti-CD3 and MyD88 deficient APCs in the presence or absence of CpG DNA (0.3μM) for 24 hours. Supernatants were harvested and analysed for IL-2 secretion by ELISA. Results shown as mean +/− s.d. of triplicate samples. Data are representative of three independent experiments.
MyD88 expression is required for enhanced IL-2 expression by CD4+CD25- T cells in response to CpG DNA costimulation
Inhibition of IL-2 expression by responder CD4+ T cells has been implicated as a potential mechanism by which Tregs exert their immunosuppressive effects. As we have previously demonstrated that CpG DNA (3 μM) costimulation of effector T cells in conjunction with plate bound anti-CD3 leads to enhanced IL-2 secretion in a MyD88-dependent manner [7], we sought to confirm these observations under the stimulation conditions described above using soluble anti-CD3 and MyD88-deficient APCs in the presence of 0.3 μM CpG DNA. As shown in figure 3B, enhanced IL-2 expression by effector CD4+CD25- T cells in response to CpG DNA is almost completely diminished in the absence of MyD88. Enhanced IL-2 secretion provides an attractive mechanism through which effector cells escape suppression upon stimulation with CpG DNA. However, further analysis of this potential mechanism is complicated due to the pleiotropic nature of IL-2 responses reported for CD4+CD25+ Tregs. IL-2 has variously been reported to be essential for the development, homeostasis and function of Tregs. For example, it has been widely reported that IL-2 and IL-2R knock out mice are deficient in CD4+CD25+ Tregs while IL-2 neutralizing antibodies inhibit Treg function in vitro [16]. Also, as Tregs can act as a “sink” for IL-2 in vitro, measurement of relative levels of IL-2 protein secreted during suppression may not accurately reflect levels of suppression of effector CD4+ T cells. One potential way to circumvent this would be to examine the effects of CpG DNA on IL-2 deficient effector CD4+ T cells undergoing suppression by MyD88 deficient Tregs. Such an approach might indicate what role, if any, IL-2 plays in the escape of effector CD4+ T cells from Treg suppression induced by CpG DNA.
Together these data suggest a potential mechanism by which CpG DNA can inhibit Treg suppression by direct costimulation of the CD4+CD25- effector T cell subset. Since exposure of T cells to CpG DNA is likely to occur under conditions of microbial infection when optimal T-cell mediated immune responses are desirable, these data provide an attractive mechanism of effector T cell escape from the suppressive effects of naturally occurring Tregs.
Footnotes
This work was supported by the National Institutes of Health grant AI-43620 (to L.A.T.). and by an American Society of Transplantation Basic Science Fellowship grant (to P.T.W)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Akira S, Takeda K, Kaisho T. Nat Immunol. 2001;2:675–80. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 2.Crellin NK, Garcia RV, Hadisfar O, Allan SE, Steiner TS, Levings MK. J Immunol. 2005;175:8051–9. doi: 10.4049/jimmunol.175.12.8051. [DOI] [PubMed] [Google Scholar]
- 3.Komai-Koma M, Jones L, Ogg GS, Xu D, Liew FY. Proc Natl Acad Sci U S A. 2004;101:3029–34. doi: 10.1073/pnas.0400171101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu H, Komai-Koma M, Xu D, Liew FY. Proc Natl Acad Sci U S A. 2006;103:7048–53. doi: 10.1073/pnas.0601554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peng G, Guo Z, Kiniwa Y, Voo KS, Peng W, Fu T, Wang DY, Li Y, Wang HY, Wang RF. Science. 2005;309:1380–4. doi: 10.1126/science.1113401. [DOI] [PubMed] [Google Scholar]
- 6.Gelman AE, Zhang J, Choi Y, Turka LA. J Immunol. 2004;172:6065–73. doi: 10.4049/jimmunol.172.10.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gelman AE, Larosa DF, Zhang J, Walsh PT, Choi Y, Sunyer JO, Turka LA. Immunity. 2006 doi: 10.1016/j.immuni.2006.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakaguchi S. Nat Immunol. 2005;6:345–52. doi: 10.1038/ni1178. [DOI] [PubMed] [Google Scholar]
- 9.Pasare C, Medzhitov R. Science. 2003;299:1033–6. doi: 10.1126/science.1078231. [DOI] [PubMed] [Google Scholar]
- 10.Thornton AM, Shevach EM. J Exp Med. 1998;188:287–96. doi: 10.1084/jem.188.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. Nature. 2000;408:740–5. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 12.Dragoi AM, Fu X, Ivanov S, Zhang P, Sheng L, Wu D, Li GC, Chu WM. Embo J. 2005;24:779–89. doi: 10.1038/sj.emboj.7600539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaisho T, Akira S. Trends Immunol. 2001;22:78–83. doi: 10.1016/s1471-4906(00)01811-1. [DOI] [PubMed] [Google Scholar]
- 14.Hemmi H, Kaisho T, Takeda K, Akira S. J Immunol. 2003;170:3059–64. doi: 10.4049/jimmunol.170.6.3059. [DOI] [PubMed] [Google Scholar]
- 15.Shevach EM, Stephens GL. Nat Rev Immunol. 2006;6:613–8. doi: 10.1038/nri1867. [DOI] [PubMed] [Google Scholar]
- 16.Thornton AM, Donovan EE, Piccirillo CA, Shevach EM. J Immunol. 2004;172:6519–23. doi: 10.4049/jimmunol.172.11.6519. [DOI] [PubMed] [Google Scholar]