Abstract
RNase E, an essential endoribonuclease in Escherichia coli, is involved in 9S rRNA processing, the degradation of many mRNAs, and the processing of the M1 RNA subunit of RNase P. However, the reason that RNase E is required for cell viability is still not fully understood. In fact, recent experiments have suggested that defects in 9S rRNA processing and mRNA decay are not responsible for the lack of cell growth in RNase E mutants. By using several new rne alleles, we have confirmed these observations and have also ruled out that M1 processing by RNase E is required for cell viability. Rather, our data suggest that the critical in vivo role of RNase E is the initiation of tRNA maturation. Specifically, RNase E catalytic activity starts the processing of both polycistronic operons, such as glyW cysT leuZ, argX hisR leuT proM, and lysT valT lysW valZ lysY lysZ lysQ, as well as monocistronic transcripts like pheU, pheV, asnT, asnU, asnV, and asnW. Cleavage by RNase E within a few nucleotides of the mature 3′ CCA terminus is required before RNase P and the various 3′ → 5′ exonucleases can complete tRNA maturation. All 59 tRNAs tested involved RNase E processing, although some were cleaved more efficiently than others.
Keywords: mRNA decay, rRNA processing, RNase P, RNase G, RNase T
Ribonuclease E (RNase E) of Escherichia coli was first characterized in a temperature-sensitive mutant (rne-3071) that accumulated 9S precursors of the 5S rRNA at 42°C (Ghora and Apirion 1978). Independently, the ams (altered mRNA stability) gene was identified because of its ability to affect the decay of total pulse-labeled RNA at elevated temperatures (Ono and Kuwano 1979). Subsequently, both loci were shown to encode RNase E (Mudd et al. 1990; Babitzke and Kushner 1991; Taraseviciene et al. 1991). The rne gene encodes a 1061-amino-acid protein (Casarégola et al. 1992, 1994) that has now been characterized as a 5′-end-dependent endoribonuclease (Mackie 1998). In addition, it has also been shown that RNase E is part of a multiprotein complex, called the degradosome, that includes polynucleotide phosphorylase (PNPase), the RhlB RNA helicase, and the glycolytic enzyme enolase (Carpousis et al. 1994; Py et al. 1994, 1996; Miczak 1996).
In vivo experiments with either the rne-1 or rne-3071 temperature-sensitive alleles have shown the accumulation of unprocessed 5S rRNA intermediates (Ghora and Apirion 1978; Babitzke et al. 1993) and a general slowing in the decay of specific mRNA transcripts (Arraiano et al. 1988; Mackie 1991; Régnier and Hajnsdorf 1991). As such it was assumed that the inviability associated with the inactivation of RNase E resulted from a defect in either 9S rRNA processing or mRNA decay. However, experiments by López et al. (1999) and Ow et al. (2000) suggest that this hypothesis is not correct. For example, using rne deletion mutations, both laboratories showed that 9S rRNA processing was almost normal under conditions in which mRNA decay was significantly impaired (López et al. 1999; Ow et al. 2000). Furthermore, Ow et al. (2000) characterized an extensive RNase E C terminus truncation mutation (rneΔ610) that was missing 609 amino acids, including the ARRBS (arginine-rich RNA-binding site) and the degradosome scaffolding region (Fig. 1). This protein was able to support cell viability at 37°C but not at 44°C. More importantly, decay of specific mRNAs was more defective at 37°C than in an rne-1 mutant shifted to the nonpermissive temperature. Therefore, it was concluded that inviability in the absence of RNase E was not associated with defects in either mRNA decay or 9S rRNA processing (Ow et al. 2000).
Figure 1.
Schematic representation of the rne+, rneΔ610, and rneΔ645 alleles. The rneΔ610 mutation encodes a truncated RNase E protein encompassing the first 427 amino acids of the N terminus plus the last 25 amino acids of the C terminus (Ow et al. 2000). The rneΔ645 revertant contains a C → A transversion on serine 418 (TCG → TAG), creating a premature stop codon to generate an RNase E polypeptide with only the first 417 amino acids of the N terminus. The various domains of the wild-type RNase E protein are as described by Vanzo et al. (1998). The figure is not drawn to scale.
Accordingly, we have sought to determine what other aspect of RNA metabolism requires the activity of this enzyme. One possibility was a defect in the processing of the M1 RNA subunit of RNase P (Gurevitz et al. 1983; Lundberg and Altman 1995). Because this is the only enzyme in E. coli that can generate the mature 5′ termini of tRNAs, the loss of its activity leads to cell inviability. However, because M1 RNA containing extra nucleotides at its 3′ terminus still retains catalytic activity (Liu and Altman 1995), this did not seem to be a likely explanation.
A more attractive candidate was tRNA processing. The E. coli genome contains 86 tRNAs, many of which exist in polycistronic operons (Berlyn 1998). Although these transcripts could be processed by a combination of endonucleolytic cleavage by RNase P at the 5′ end (Altman et al. 1995) and exonucleolytic degradation at the 3′ end by RNase II, RNase BN, RNase PH, RNase D, RNase T, and PNPase (Li and Deutscher 1996), it is also possible that RNase E is required to cleave within the intercistronic regions. Failure to process tRNAs properly would lead to a cessation of protein synthesis and concomitantly cell growth. In fact, Ono and Kuwano (1979) observed a drop-off in the rate of protein synthesis when an rne-1 strain (called ams-1 at that time) was shifted to the nonpermissive temperature. In addition, when Ray and Apirion (1981b) isolated small RNAs from an rne-3071 mutant shifted to 42°C, they found 9S rRNA precursors as well as molecules that contained both tRNALeu and tRNAHis. These two tRNAs are part of a four-gene tRNA transcription unit, argX hisR leuT proM (Berlyn 1998). Subsequently, they showed that if such tRNA precursors were treated in vitro with RNase E, then RNase P could cleave at the 5′ end (Ray and Apirion 1981a).
Using a series of RNase E mutants, we have examined the relationships among RNase E function, tRNA processing, and cell viability. Of particular importance was the fortuitous isolation of a temperature-resistant revertant of the rneΔ610 allele (Ow et al. 2000), called rneΔ645. The rneΔ645 mutation encodes an RNase E protein of only 417 amino acids, but unlike rneΔ610, permits cell growth at both 37°C and 44°C. Whereas the processing of both polycistronic and monocistronic tRNAs was impaired at 44°C in rne-1, rneΔ610, and rneΔ645 strains, transcripts matured 2.9- to 3.7-fold faster in the rneΔ645 mutant compared with the rne-1 and rneΔ610 strains. There was a direct correlation among tRNA processing rates, the steady-state levels of mature tRNAs, and cessation of cell growth. In addition, RNase P cleavage at the 5′ end of tRNA transcripts was dependent on prior RNase E processing at the 3′ terminus.
Results
Isolation and characterization of rneΔ645, a temperature-resistant revertant of the rneΔ610 allele
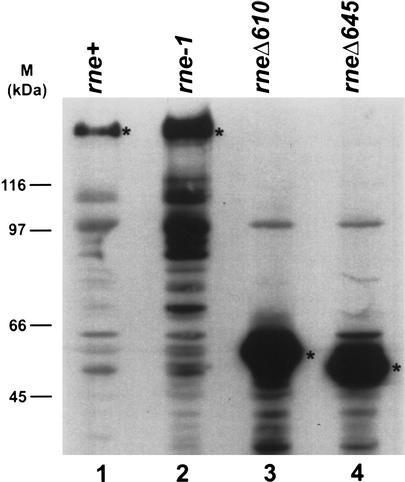
Because of our interest in mRNA decay, we constructed an rng::cat rneΔ610 double mutant (SK9982). When this strain was replica-plated to 44°C, a small number of temperature-resistant revertants were identified. Of eight that were tested, one (SK9987) arose from an alteration in the plasmid carrying the rneΔ610 allele. DNA sequencing of the complete rne gene in pMOK21 revealed a single nucleotide change, a C → A transversion within the codon for serine 418, resulting in a stop codon (Fig. 1). This new premature stop codon further shortened the protein from the RneΔ610 parent by 35 amino acids, generating a polypeptide, RneΔ645, of 417 amino acids that migrated as an ∼46-kD protein in an 8% SDS polyacrylamide gel (Figs. 1, 2). The autoregulation of RNase E synthesis, a phenomenon first described by Jain and Belasco (1995), was completely lost in the rneΔ645 strain (Fig. 2, lane 4). A similar loss of autoregulation has been observed previously with the rneΔ610 allele (Fig. 2, lane 3; Ow et al. 2000). To confirm that the C → A transversion was responsible for the suppression of lethality observed in SK9987, we used site-directed mutagenesis to reconstruct the rneΔ645 mutation in pMOK15 (rneΔ610 Cmr), creating pMOK29 (rneΔ645 Cmr). Transformation of pMOK29 into SK9705 and the subsequent displacement of the resident plasmid (pQLK26, rne+ Kmr) resulted in a strain, SK10103, whose properties were identical to SK9987.
Figure 2.
Western analysis of various RNase E proteins. Equivalent amounts of total protein lysates (50 μg) were electrophoresed in an 8% SDS/PAGE gel and electrotransferred onto an Immobilon PVDF membrane (Millipore). Membranes were probed for RNase E as described in Materials and Methods. Size standards are on the left. Asterisks denote the full-length RNase E proteins. (Lane 1) SK9714; (lane 2) SK9937; (lane 3) SK9957; (lane 4) SK9987.
Growth properties of the rneΔ645 mutant
On Luria agar medium at 44°C, SK9987 (rneΔ645) took 2 d to form large colonies compared with 1 d for the rne+ control (SK9714). In Luria broth, SK9987 continued to grow at 44°C (35 min generation time), albeit more slowly than the wild-type SK9714 control (26 min; Fig. 3A). In contrast, SK9957, the rneΔ610 progenitor, ceased growing at ∼4 h after the shift to the nonpermissive temperature (Fig. 3A). A control strain, SK9937, harboring the rne-1 temperature-sensitive allele ceased growing between 60 and 90 min after the shift to 44°C (Fig. 3A).
Figure 3.
Comparison of the growth properties, mRNA decay rates, and tRNA processing rates of strains carrying the rne+, rne-1, rneΔ610, and rneΔ645 alleles in either 6–8 copies/cell or single copy. Growth curves were carried out as described in Materials and Methods. (A) Growth curve with 6–8 copies/cell of each rne allele. (B) Growth curve with each rne allele in single copy. (C) Relative mRNA half-lives as a function of RNase E activity. (D) Relative tRNA precursor half-lives as a function of RNase E activity. The half-life of each mRNA (Table 1) and the half-life of each tRNA precursor obtained with the specific 3′-oligonucleotide probe (Table 3) in the rne-1 strain was set at 1.
Because these strains contained low-copy-number plasmids (6–8 copies/cell) carrying the rne+, rne-1, rneΔ610, and rneΔ645 alleles, we also determined the growth properties of cells carrying these mutations in a single copy. To accomplish this, the rne+, rne-1, rneΔ610, and rneΔ645 alleles were moved into pMOK40 (Ow et al. 2002), a single-copy vector carrying Smr/Spr. Under these conditions, both the rne-1 and rneΔ610 strains ceased growing at ∼120 min after the shift to 44°C, whereas the rneΔ645 and rne+ strains continued growing (Fig. 3B). Interestingly, the increased copy number of the rne-1 allele caused a more rapid cessation of cell growth (cf. SK9937 and SK10144 in Fig. 3A,B). In contrast, the generation times for the rneΔ645 strains at 44°C were 35 min (SK9987, 6–8 copies) versus 47 min (SK2685, single-copy). Based on Western blot analysis, there was a three-to fourfold decrease in the amount of the RneΔ610 and RneΔ645 proteins when the mutations were present in the single-copy plasmid compared with the 6–8-copy-number plasmid (data not shown).
mRNA decay properties of the rneΔ645 mutant
Previous analysis of the rneΔ610 allele had suggested that the conditional lethality associated with the rne-1 and rne-3071 mutations was not related to defects in mRNA decay (Ow et al. 2000). To determine if this hypothesis were correct, we examined the decay of two RNase E-dependent transcripts, rpsT and rpsO (Mackie 1991; Hajnsdorf et al. 1994) at both 37°C and 44°C in the rneΔ645 mutant. At 37°C, both the rpsO and rpsT mRNAs decayed between 1.5- and 1.7-fold more rapidly in SK9987 (rneΔ645) than in SK9957 (rneΔ610), but their decay rates were still much slower than those obtained in rne+ and rne-1 strains (Table 1A). In fact, the half-lives in the rneΔ645 strain at 37°C were still equal to or longer than those observed in an rne-1 mutant at the nonpermissive temperature (Ow et al. 2000).
Table 1.
mRNA half-lives at 37°C and 44°C
| Transcript A
|
Half-life (min)a
|
|||
|---|---|---|---|---|
|
rne+
|
rne-1b
|
rneΔ610
|
rneΔ645
|
|
| rpsOc | 1.8 ± 0.4 | 2.9 ± 0.3 | 16.3 ± 1.7 | 9.4 ± 2.1 |
| rpsTc | 1.5 ± 0.1 | 2.3 ± 0.4 | 4.4 ± 0.3 | 2.9 ± 0.0 |
| Transcript B | rne+ | rne-1b | rneΔ610 | rneΔ645 |
| rpsOc | ND | 8.4 ± 1.0 | >30 | 18.7 ± 4.0 |
| rpsTc | ND | 5.0 ± 1.4 | 6.1 ± 1.3 | 3.8 ± 0.3 |
| pnp | ND | 10.0 ± 1.2 | >20 | 8.4 ± 1.0 |
Half-lives for transcript A were determined at 37°C in strains carrying the various alleles in 6–8 copies/cell. Half-lives for transcript B were determined in strains carrying the various alleles in single copy after cultures had been shifted to 44°C for 120 min.
The data represent the average of two independent determinations.
rne-1 encodes a temperature sensitive RNase E protein that is unable to support cell viability at 44°C (Ono and Kuwano 1979; Babitzke and Kushner 1991). RNase E activity is partially inactivated at 37°C (Ow et al. 2000).
For rpsO, the half-life of the rpsO2 transcript (Ow et al. 2000) is presented. For rpsT, the half-life is the average of the two major transcripts (Mackie 1991).
ND, Not determined.
We also determined the half-lives of the rpsO, rpsT, and pnp transcripts at 44°C in strains carrying single copies of the rneΔ610 and rneΔ645 alleles (Table 1B). Under these conditions, mRNA decay in the rneΔ645 strain was either more defective than in the rne-1 mutant (rpsO) or not significantly better (rpsT and pnp; Table 1B). It should also be noted that with all three transcripts, the half-lives increased considerably in the rneΔ610 strain relative to rne-1, even though the rne-1 strain ceased growing sooner at 44°C (Fig. 3B). In contrast, the half-lives of all three transcripts in the rneΔ645 mutant were comparable to those observed in the rne-1 strain even though it continued growing at 44°C (Fig. 3B; Table 1).
Comparison of 9S rRNA processing in the rneΔ610 and rneΔ645 mutants
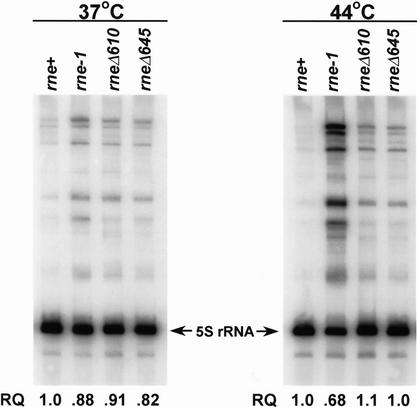
Because we had previously observed that 9S rRNA processing was relatively normal in the rneΔ610 mutant (Ow et al. 2000), we did not expect to see a significant difference between the rneΔ610 and rneΔ645 strains. To confirm this prediction, we probed a Northern blot containing total RNA, isolated from rne+, rne-1, rneΔ610, and rneΔ645 strains that had either been grown at 37°C or had been shifted to 44°C for 150 min, with an oligonucleotide complementary to 5S rRNA. As expected, in the rne-1 strain, larger rRNA precursors accumulated when the strain was shifted from 37°C to 44°C (Fig. 4). In contrast, there were significantly fewer precursors in both the rneΔ610 and rneΔ645 mutants at 44°C (Fig. 4). In fact, the steady-state levels of mature 5S rRNA were identical in the wild-type, rneΔ610, and rneΔ645 strains at 44°C compared with a 32% reduction in the rne-1 mutant (Fig. 4).
Figure 4.
Northern analysis of 9S rRNA processing. Five micrograms of total steady-state RNA isolated from either 37°C or 44°C as described in Materials and Methods was separated on a 6% PAGE/7 M urea gel, electrotransferred onto a nylon membrane (MSI MagnaCharge; Osmonics), and probed with a 32P-5′-end-labeled oligonucleotide complementary to the 5S rRNA (PB5S rRNA). The arrows indicate the 5S rRNA; the larger species are processing intermediates. RQ is the amount of mature 5S rRNA in each strain compared with the wild-type control.
Comparison of M1p processing in the rneΔ610 and rneΔ645 mutants
It has been shown that RNase E is involved in the maturation of the M1 RNA subunit of RNase P (Gurevitz et al. 1983; Lundberg and Altman 1995). Because this enzyme is required for the generation of the mature 5′ ends of tRNAs (Altman et al. 1995), we determined the half-life of the M1 precursor (M1p) in the rneΔ610 and rneΔ645 strains to ascertain whether a change in the processing of this transcript could account for the difference in cell growth at 44°C (Fig. 3A,B). In the rne+ control, processing occurred too rapidly to determine a half-life (Table 2). In contrast, when the rne-1, rneΔ610, and rneΔ645 alleles were present in 6–8 copies/cell, the half-life of the M1p was identical within experimental error (Table 2). When the rne alleles were present in single copy, the M1p half-life in the rne-1 and rneΔ645 strains was almost identical, whereas it was almost four times longer in the rneΔ610 mutant (Table 2).
Table 2.
Half-livesa of the M1 precursor of RNase P in various strains at 44°C
| Copy number
|
Genotype
|
|||
|---|---|---|---|---|
|
rne+
|
rne-1
|
rneΔ610
|
rneΔ645
|
|
| 6–8 | ND | 20.8 ± 6.0 | 20.1 ± 1.3 | 22.5 ± 10.9 |
| 1 | ND | 23.2b | 87.6 ± 12.7 | 19.1 ± 2.4 |
Data represent the average of two independent determinations. Cultures were grown at 37°C until they reached ∼1 × 108 cells/mL and were then shifted to 44°C. After 150 min of growth, half-life determinations (in minutes) were carried out as described in Materials and Methods.
This experiment was performed once only.
ND, The M1 precursor was not detected in the wild-type strain.
Processing of polycistronic transcripts containing tRNAs and tRNAs encoded within rRNA operons requires functional RNase E activity
Because the changes in mRNA decay rates as well as 9S rRNA and M1 processing did not seem sufficient to account for the ability of the rneΔ645 strain to grow at 44°C, we next analyzed the processing of tRNAs in rne+, rne-1, rneΔ610, and rneΔ645 strains at 44°C. If RNase E were required for tRNA processing, we assumed that its most likely role would be in the cleavage of polycistronic operons that contained several tRNAs. Initially, we chose to analyze the glyW cysT leuZ (Fig. 5A) and argX hisR leuT proM polycistronic transcripts (data not shown) because the cysT and hisR genes are unique.
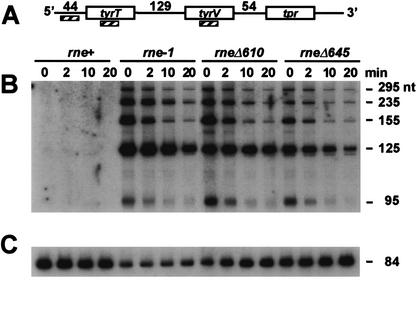
Figure 5.
Processing of the glyW cysT leuZ polycistronic transcript. Cultures were grown in LB medium at 44°C as described in Materials and Methods. Ten micrograms of total RNA isolated from SK9714 (rne+), SK9937 (rne-1), SK9957 (rneΔ610), and SK9987 (rneΔ645) at 0, 2, 5, 10, 20, and 30 min after rifampicin addition was loaded in each lane. The RNA samples were electrophoresed on a 6% PAGE/7 M urea gel and electrotransferred onto a nylon membrane (MSI MagnaCharge; Osmonics). The same membrane was used for the three Northern blots shown in this figure by following the recommendations of the membrane manufacturer for stripping the probe. (A) Schematic of the glyW cysT leuZ transcript. Numbers indicate the size (in nucleotides) of the spacer regions 5′ and 3′ of the tRNACys gene. Hatched rectangles indicate the oligonucleotide probes. All oligonucleotide probes were 5′-end-labeled with [γ-32P]ATP using T4 polynucleotide kinase. The diagram is not drawn to scale. (B) Processing of the 3′ end of tRNACys. Northern analysis was done using an oligonucleotide probe (cysT–leuZ) complementary to the 12-nt spacer region between tRNACys and tRNALeu. (C) Processing at the 5′ end of tRNACys. The Northern blot was stripped and reprobed with oligonucleotide glyW–cysT complementary to a section of the 54-nt 5′ spacer region of tRNACys. (D) Detection of the mature tRNACys using the cysT probe. Sizes of selected bands are indicated on the right.
Using an oligonucleotide probe (cysT–leuZ), we carried out a half-life experiment for the intercistronic region between the cysT and leuZ genes (Fig. 5A,B). Although no precursor transcripts were visible in the wild-type control, two large species of 460 (full length) and 290 nucleotides (nt) were observed in the rne-1, rneΔ610, and rneΔ645 mutants, along with a series of smaller intermediates ranging in size from 90 to 200 nt (Fig. 5B). Half-life determinations for the largest species showed that the rneΔ645 strain processed it twofold faster than the rneΔ610 mutant and 2.8-fold faster than the rne-1 strain (Table 3; Fig. 3D). As a control, the membrane was stripped and reprobed for the mature tRNACys (Fig. 5D).
Table 3.
Processing of polycistronic and monocistronic tRNA transcripts
| Transcript
|
Region
|
Half-lifea (min)
|
|||
|---|---|---|---|---|---|
|
rne+
|
rne-1
|
rneΔ610
|
rneΔ645
|
||
| glyW cysT leuZ | cysT-leuZ | ND | 9.9 ± 0.5 | 7.0 ± 0.4 | 3.5 ± 0.7 |
| glyW-cysTb | ND | 5.9 | NC | NC | |
| argX hisR leuT proM | hisR-leuT | ND | 14.5 ± 1.9 | 15.0 ± 5.4 | 4.1 ± 0.1 |
| argX-hisRb | ND | 12.5 | 10.4 | 3.4 | |
| pheU | pheU tRNA | ND | 31.8 ± 5.0 | 20.2 ± 3.0 | 8.6 ± 3.1 |
Half-lives were determined as the average of two independent experiments for the largest precursor detected (full-length) in each Northern blot. The various alleles were present in 6–8 copies/cell.
Experiments were performed only once.
ND, No intermediates were detected.
NC, Half-lives were not calculated because of the low level of intermediates that were observed.
To confirm that these observations were not unique to the glyW cysT leuZ operon, we next tested the argX hisR leuT proM polycistronic transcript. A probe (hisR–leuT) for the spacer region between hisR and leuT detected two large precursor transcripts of 530 (full length) and 335 nt in addition to smaller species between 85 and 225 nt in length in the rne mutants (data not shown). Again, no precursors were seen in the wild-type control. With this operon, the decay of the full-length precursor was almost identical in the rne-1 and rneΔ610 strains (Table 3; Fig. 3D) and much slower than that observed for the glyW cysT leuZ operon. However, the processing in the rneΔ645 strain was 3.7-fold faster than in either the rne-1 or rneΔ610 mutants (Table 3; Fig. 3D).
We also examined whether RNase E was involved in the processing of tRNA operons that contained multiple copies of specific tRNAs. For these experiments we chose the tyrT tyrV tpr and lysT valT lysW valZ lysY lysZ lysQ operons. In both cases, significant amounts of processing intermediates were observed in the three rne mutants compared with none in the wild-type control (data not shown). Similar differences in tRNA processing were observed when the rneΔ610 and rneΔ645 alleles were present in single copy (data not shown). As a final experiment, we tested the four glt tRNAs because they are all contained within rRNA operons (rrnB, rrnC, rrnE, and rrnB). As was seen with polycistronic tRNA transcripts, functional RNase E activity was required to obtain mature tRNAGlt (data not shown).
Processing of monocistronic tRNA transcripts also requires functional RNase E activity
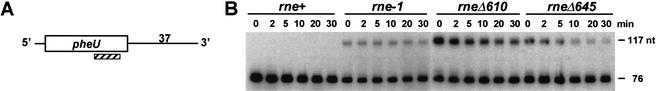
Because monocistronic transcripts have relatively short 3′ ends, we assumed that the multiple 3′ → 5′ exonucleases (RNase II, RNase BN, RNase D, RNase PH, RNase T, and PNPase) that have been shown to be involved in the generation of mature 3′ CCA termini (Li and Deutscher 1996) would be sufficient for maturation. To determine if this hypothesis were correct, we examined the processing of the pheU transcript because it contains 37 nt downstream of the mature CCA terminus (Fig. 6A) and it is almost identical in sequence organization to pheV (contains a 36-nt downstream sequence), the other phenylalanine tRNA in E. coli. Thus, analysis of the pheU transcript allowed us to simultaneously measure processing of both phenylalanine tRNAs.
Figure 6.
Processing of the pheU monocistronic transcript. The same conditions were used as described in Figure 5. (A) Schematic of the pheU monocistronic transcript. The hatched rectangle is the oligonucleotide probe (pheU2) complementary to the mature tRNAPhe. The diagram is not drawn to scale. (B) Northern analysis using the pheU2 oligonucleotide probe to detect both the mature tRNAPhe as well as the unprocessed tRNAPhe precursor. The sizes of both species are indicated on the right.
To our surprise, the 116–117-nt-long primary transcript of tRNAPhe was observed in all three rne mutant strains (Fig. 6B). Of even more interest were the longer half-lives for the pheU/pheV precursors compared with the polycistronic transcripts (e.g., Table 3, for rne-1 cf. the half-lives of 31.8 min for the pheU and pheV precursors with 9.9 min for cysT leuZ and 14.5 min for hisR leuT). However, as was seen for the polycistronic operons, processing was significantly faster in the rneΔ645 strain compared with either the rne-1 or rneΔ610 mutants (Table 3; Fig. 3D). No intermediates were detected in the rne+ strain (Fig. 6B). When these experiments were repeated with the rneΔ610 and rneΔ645 alleles in single copy, the 116–117-nt pheU/pheV precursors were processed fivefold more rapidly in the rneΔ645 strain compared with the rneΔ610 mutant (data not shown).
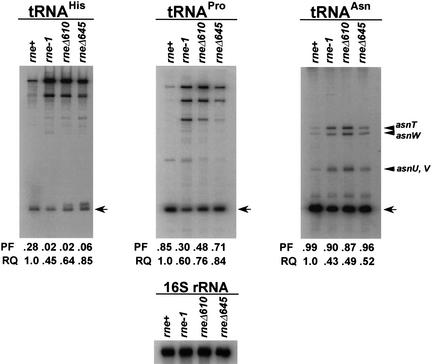
To show that pheU and pheV were not unique among the monocistronic tRNA transcripts, we also examined the maturation of tRNAAsn under steady-state conditions. There are four copies of this gene in the E. coli genome (asnT, asnU, asnV, and asnW), each transcribed as a monocistronic transcript of a different length. Inactivation of RNase E led to a large increase in the amount of the four precursor tRNAs (see Fig. 9, below).
Figure 9.
Analysis of steady-state levels of tRNAs in various RNase E mutants. Steady-state RNA was isolated from SK9714 (rne+) and SK9937 (rne-1) after cultures had been shifted to 44°C for 120 min as indicated in Materials and Methods. For SK9957 (rneΔ610) and SK9987 (rneΔ645), the cultures were grown for 240 min prior to isolating RNA. The Northern blots were run as described in Figure 5. The membranes were probed with oligonucleotides complementary to either tRNAHis (hisR), tRNAPro (proM), tRNAAsn (asn), or 16S rRNA (rrnB2448). PF denotes the processed fraction that is defined as the amount of a given mature tRNA relative to the total amount of that tRNA (processed and unprocessed). RQ indicates the amount of mature tRNA in each lane compared with that determined in the rne+ strain. As a normalization control a blot was probed for 16S rRNA.
When we compared the differences in tRNA processing rates (Fig. 3D) with the variations in the mRNA decay half-lives (Fig. 3C) and the growth profiles at 44°C associated with the rne-1, rneΔ610, and rneΔ645 strains (Fig. 3A,B), we observed a direct correlation between cell growth and tRNA-processing efficiency.
Maturation of the 5′ terminus of tRNA precursors requires prior RNase E cleavage
To determine if RNase E cleavage was the rate-limiting step in tRNA maturation, we performed two different types of experiments. Previous in vitro experiments by Ray and Apirion (1981a) suggested that the action of RNase E could influence RNase P cleavage at the 5′ terminus of an unprocessed tRNA transcript. Accordingly, to examine whether in vivo RNase P processing of the 5′ terminus was affected by RNase E cleavage at the 3′ end, we performed Northern analyses with probes designed to detect the presence of unprocessed 5′ termini. First, we tested for 5′-end processing of the tyrT tRNA, which is the first gene in the tyrT tyrV tpr transcript and contains a 44-nt 5′-terminal region (Fig. 7A). Although no processing intermediates were observed in the wild-type control, a variety of larger species were observed in the three rne mutants, indicating that RNase P cleavage required prior activity by RNase E at the 3′ end (Fig. 7B).
Figure 7.
Processing of the tyrT tyrV tpr polycistronic transcript. The same procedures were followed as described in Figure 5. Hatched rectangles indicate the oligonucleotide probes. (A) Diagrammatic representation of the operon (not to scale). (B) Processing of the 5′-end tyrT. Northern analysis was done using primer tyrT5′A, which was specific for the entire 44-nt 5′ unprocessed terminus of tyrT. (C) Detection of the mature tRNATyr using primer tyrTV3′. The sizes of the various tRNA transcripts are indicated on the right.
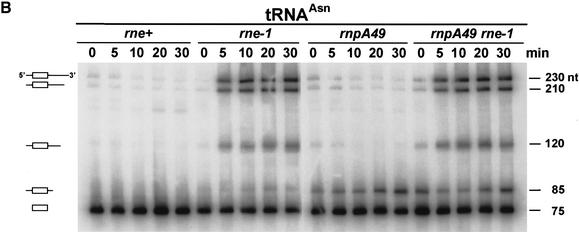
Although this result strongly suggested that RNase P did not process the 5′ end of the tyrT precursor prior to RNase E cleavage at the 3′ end, we wanted to confirm this observation using a simpler substrate. Accordingly, we examined the steady-state levels of six monocistronic transcripts (asnT, asnU, asnV, ansW, pheU, and pheV) in wild-type, rne-1, rnpA49, and rne-1 rnpA49 strains after a 150-min shift to 44°C. In the case of tRNAAsn, the four unprocessed monocistronic transcripts are 230 nt (asnT), 210 nt (asnW), and 120 nt (asnU and asnV) in length (Figs. 8A, 9). Each transcript contains ∼9–10 nt upstream of the mature 5′ end. In an rnpA49 mutant, which encodes a temperature-sensitive mutation in the protein subunit of RNase P (Apirion and Gitelman 1980), there was a large increase in an 85-nt species that represented all four tRNAAsn species that had been processed by RNase E at their 3′ ends but not by RNase P at their 5′ termini (Fig. 8A). In contrast, in the rne-1 strain, the full-length precursors (230, 210, and 120 nt) accumulated. If RNase P functioned prior to RNase E cleavage, one would have expected to see species of ∼220, ∼200, and ∼110 nt in the rne-1 mutant. The fact that the tRNAAsn band profile in the rnpA49 rne-1 double mutant was identical to those seen in the two single mutants further suggested that all potential processing intermediates were accounted for.
Figure 8.
Analysis of tRNA processing in RNase E and RNase P mutants. (A) Steady-state RNA was isolated from each strain following a 150-min shift to 44°C as described in Materials and Methods. (B) Total RNA was isolated from cultures shifted to 44°C for the specified times. Five micrograms of total RNA was loaded into each lane of a 6% polyacrylamide/7 M urea gel. Each blot was probed with an oligonucleotide complementary to the mature tRNA. Diagrams of the tRNA species are shown on the left. The sizes on the right were determined using RNA molecular size standards (Invitrogen).
A similar result was obtained with the pheU and pheV transcripts (Fig. 8A). In this case, the full-length transcripts are 116 nt (pheV) and 117 nt (pheU), respectively, and contain a very short 5′ leader (3–4 nt). In the rnpA49 mutant, two species (80–81 nt) differing in length by 1 nt accumulated, containing the immature 5′ end and 1–2 extra nucleotides at the 3′ end. In contrast, in the rne-1 mutant, only the full-length transcripts accumulated. Again, there was no evidence for an intermediate that contained a mature 5′ end and an immature 3′ end.
Although the steady-state analysis indicated that RNase P processing of tRNA precursors did not occur prior to RNase E cleavage, we also performed kinetic experiments with both tRNAAsn (Fig. 8B) and tRNAPhe (data not shown) following a shift to 44°C. At the time of the shift, the mature tRNAAsn was the major species in the rne+ and rne-1 strains (Fig. 8B). In the rnpA49 mutant there was also a low level of the 85-nt product, indicating that RNase P activity was slightly defective at 30°C. In the rnpA49 rne-1 double mutant, there were higher levels of all the precursors. More importantly, in the rne-1 strain the four full-length tRNAAsn transcripts accumulated over time, whereas the 85-nt species containing the unprocessed 5′ ends rapidly appeared in the rnpA49 strain. With the rnpA49 rne-1 double mutant, all of the precursors accumulated following the shift to 44°C (Fig. 8B). The kinetic analysis confirmed the results obtained with the steady-state experiment (Fig. 8A).
Analysis of tRNA processing efficiency at 44°C
In our Northern analysis of various tRNA transcripts, we noticed considerable differences in the half-lives among the various tRNA precursors (Table 3), suggesting a possible hierarchy of substrates for RNase E, whereby some were cleaved more efficiently than others. Accordingly, we examined the steady-state levels of 59 tRNA transcripts in the rne+, rne-1, rneΔ610, and rneΔ645 strains at 44°C under conditions such that each allele was present in 6–8 copies/cell. These experiments showed that there were three classes of tRNA substrates for RNase E. One group (Class 1) including tRNAAsn (Fig. 9), tRNAVal, tRNATyr, tRNAGly, tRNAPhe, and tRNAMet were very efficiently processed in the rne+ strain such that between ∼95% and ∼99% of the tRNA was present in the mature form (PF) under steady-state conditions. At the other extreme (Class 3), tRNAHis (Fig. 9), tRNACys, and tRNAArg were inefficiently processed such that only ∼28%–30% of the transcripts were mature in the wild-type strain under identical conditions. Class 2, represented by tRNAPro (Fig. 9), tRNAGlt, and tRNALeu, had PF values that ranged between 75% and 85%. Similar differences in PF values were observed when the rne alleles were present in single copy (data not shown).
Significant differences among the PF values for the various tRNAs were also observed in the rne-1, rneΔ610, and rneΔ645 mutants (Fig. 9; data not shown). For example, the PF in the rne-1 strain was always lower than that obtained in the rne+ control. In the case of tRNAHis, there was a 14-fold reduction, whereas with tRNAPro the decrease was 2.8-fold (Fig. 9). In addition, the PF was always greater in the rneΔ645 strain compared with the rne-1 and rneΔ610 mutants (Fig. 9; data not shown). Furthermore, irrespective of their relative processing efficiencies, the steady-state amounts (RQ) of each tRNA were always rne+ > rneΔ645 > rneΔ610 ≥ rne-1. This relationship correlated well with the growth profiles of these strains at 44°C (Fig. 3A,B). These data were also in complete agreement with the relative tRNA precursor half-lives described in Figure 3D.
Mapping RNase E cleavage sites within tRNA precursors
Because RNase E appeared to play an integral role in the processing of all the 59 tRNA transcripts we tested, we carried out a series of primer extension experiments to map some of the actual cleavage sites. When we tested the tyrT transcript, there was a single cleavage site located 1 nt downstream of the mature CCA terminus (CCAU↓AAUUCACC; Table 4; Fig. 10). This result was confirmed by performing a Northern blot for tyrT using steady-state RNA isolated from a Δrnt::kan rph-1 strain (SK10148), which should not be able to generate a mature 3′ end because of the loss of RNase T and RNase PH (Li and Deutscher 1994, 1996). No large precursors were observed, but an ∼86-nt species, 1 nt longer than the mature tRNATyr, was present in significant amounts (data not shown). In a second experiment, we determined the RNase E cleavage site between the lysY and lysZ genes. In this case, cleavage also occurred 1 nt downstream of the mature CCA terminus (CCAG↓UUUUA ACA; Table 4).
Table 4.
Analysis of 3′ termini of various tRNA transcripts
| tRNA gene
|
3′ terminal regiona
|
Processing efficiencyb
|
|---|---|---|
| tyrT | CCAU↓AAUUC | High |
| lysY | CCAG↓UUUUA | High |
| metZ | CCAAUUAAA | High |
| metW | CCAAUCAAA | High |
| metV | CCAAUUAUU | High |
| asnV | CCAAAUUCA | High |
| asnW | CCAAAUUUU | High |
| asnU | CCAAAUUCC | High |
| asnT | CCAAAUUCU | High |
| glyW | CCAGUUUAA | High |
| glyV | CCAAAAUUU | High |
| pheU | CCAAAUUCA | High |
| pheV | CCACUAAUU | High |
| valZ | CCAUCGG↑GU | High |
| hisR | CCAUUAUUA | Low |
| cysT | CCACUUUCU | Low |
| argX | CCAUUUAGU | Low |
| argV | CCAUAUUCU | Low |
| argY | CCAUCUCUU | Low |
| argZ and argQ | CCAUAUUCU | Low |
| argU | CCAUUACAA | Low |
| argW | CCAUUUAUC | Low |
| leuT | CCACGACUU | Intermediate |
| leuV | CCAAUUAUC | Intermediate |
| proM | CCAAUUUUG | Intermediate |
The mature CCA terminus is shown in bold. Known RNase E cleavage sites are indicated by the downward arrows. In the case of valZ, the RNase P cleavage site is given since the mature lysY 5′ end starts 4 nucleotides downstream of the valZ CCA sequence.
Processing efficiency is defined based on the observations described in Figure 9. High (Class 1) represents those tRNAs that are processed very efficiently (e.g., tRNAAsn) in wild-type E. coli. Low (Class 3) are those tRNAs that are processed inefficiently (e.g., tRNAHis) under identical conditions. Intermediate (Class 2) are those tRNAs (e.g., tRNAPro) that are processed at intermediate levels.
Figure 10.
Primer extension analysis of tyrT. Two micrograms of total steady-state RNA from SK9714 (rne+), SK9937 (rne-1), SK9957 (rneΔ610), or SK9987 (rneΔ645) was subjected to primer extension analysis using a 32P-end-labeled primer (tyrTV) complementary to a segment of the spacer region between tyrT and tyrV. RNA from SK9714 and SK9937 was isolated after a 120-min shift to 44°C, whereas RNA from SK9957 and SK9987 was isolated after a 240-min shift as described in Materials and Methods. The primer extension product, mapping 1 nt after the CCA terminus (Table 4), is indicated by the arrow. A sequencing ladder obtained by using the same primer is shown at the left.
Because both of the RNase E cleavage sites mapped very close to the mature CCA terminus and fell within an AU-rich region, we decided to compare the downstream sequences of the three classes of tRNAs identified by the experiments described in Figure 9. In particular, we wanted to see if there were any correlations between those tRNAs that were processed inefficiently (his, cys, and arg) and those that were processed efficiently (asn, val, tyr, gly, and met). As shown in Table 4, most of the 3′ regions are AU-rich. In general, in the efficiently processed tRNAs there is usually an A residue immediately downstream of the CCA, but in those inefficiently processed there is either a C or a U (Table 4). However, the significance of this observation is called into question by the fact that some of the intermediately processed tRNAs like leuV have a 3′-terminal region that is almost identical to that of metV and metZ (Table 4). In addition, there are a number of 3′ ends such as valZ that appear to be processed efficiently but are GC-rich (Table 4).
Discussion
Although previous work by Apirion and coworkers has suggested a possible role for RNase E in tRNA maturation (Ray and Apirion 1981a,b), use of several RNase E mutants has permitted us to show that in vivo, tRNA processing is the likely reason that RNase E is essential for cell viability. In addition, we have presented evidence that RNase E is required for the processing of every possible type of tRNA transcript (monocistronic, polycistronic, and tRNAs contained within rRNA operons). Our results also confirm that RNase E cleavage is the rate-limiting step in the maturation of tRNAs.
What is the evidence to support these conclusions? In the first place, even though RNase E is involved in the processing of 9S rRNA, the M1 subunit of RNase P, and the degradation of a large number of mRNAs, the data presented (Tables 1,2; Figs. 3C,4) show that these pathways are affected similarly in the rne-1, rneΔ610, and rneΔ645 mutants, irrespective of their growth properties at 44°C (Fig. 3A,B). In contrast, the defects in tRNA processing and the relative amounts of mature tRNAs present in the rne-1, rneΔ610, and rneΔ645 mutants directly correlated with their ability to grow at 44°C (Figs. 3A,B,D, 5–7, 9; Table 3).
With each allele present in 6–8 copies/cell, the rne-1 strain ceased growing between 1–2 h after the shift to 44°C, whereas it took the rneΔ610 mutant 3–4 h to stop growing. On the other hand, the rneΔ645 strain continued to grow with a slightly longer generation time than the wild-type control (Fig. 3A). If one now compares the half-lives of the precursor tRNA transcripts at 44°C (Table 3), the processing reactions catalyzed by RNase E were most impaired in the rne-1 strain, followed closely by the rneΔ610 mutant. The processing of the three tRNA substrates tested was between 2.0- and 3.7-fold faster in the rneΔ645 strain compared with the rneΔ610 parent (Table 3; Fig. 3D). These changes were far greater than any differences observed in mRNA decay (Table 1; Fig. 3C), M1 processing (Table 2), or 9S rRNA processing (Fig. 4). For example, the processing of the 9S rRNA precursor was identical, within experimental error, in both the rneΔ610 and rneΔ645 strains (Fig. 4). In addition, the half-life of the M1 precursor of RNase P was indistinguishable in the rne-1 and rneΔ645 strains (Table 2). In the case of mRNA decay, although only three specific transcripts were tested, in all cases the half-lives in the rneΔ645 temperature-resistant revertant were either comparable to or longer than those observed in the temperature-sensitive rne-1 strain at 44°C (Table 1B; Fig. 3C). When the rne mutations were present in single copy, a similar correlation between growth at 44°C and tRNA processing rates was also observed (data not shown).
Furthermore, the levels of mature tRNAs in the three mutants were in the order of rneΔ645 > rneΔ610 ≥ rne-1 (Fig. 9; data not shown). Thus, one would expect the rne-1 strain to experience a shortage of tRNAs at 44°C far more quickly than the rneΔ610 or rneΔ645 strains, leading to the more rapid cessation of cell growth that was observed (Fig. 3A). Additional support for this conclusion comes from the fact that when the copy number of rneΔ610 was reduced, the growth properties of the rneΔ610 and rne-1 strains were almost identical (Fig. 3B) and the half-lives of the pheU/pheV precursors were now longer than those observed in the rne-1 mutant (data not shown).
The analysis of the 5′ termini of tRNA precursors either by examining their degradation (Fig. 7) or the identification of tRNA processing intermediates in mutants defective in either RNase E or RNase P (Fig. 8), confirmed previous results (Ray and Apirion 1981a; Li and Deutscher 2002) that generation of the mature 5′ terminus by RNase P cleavage required prior processing by RNase E. For example, in the case of the asn tRNAs, there are four different monocistronic transcripts that all contain 9–10 nt at the 5′ end but vary in length because of differences at their 3′ termini. Inactivation of RNase P would be expected to result in the accumulation of a single species that contained the unprocessed 5′ end of each tRNAAsn and a few extra nucleotides at the 3′ terminus. In fact, a species of 85 nt was the most prominent species in the rnpA49 mutant (Fig. 8A). In the rne-1 strain, two types of precursor were possible: the full-length transcripts of 230, 210, and 120 nt, or processed intermediates that lacked their 5′ leaders but retained their unprocessed 3′ ends (220, 200, and 110 nt). Only the larger species were observed (Fig. 8). Similar results were seen for pheU, pheV (Fig. 8A), and met (data not shown). It thus appears that 5′ maturation is absolutely dependent on initial cleavages at the 3′ end by RNase E. As such, the inviability observed in RNase E mutants may be partially an indirect effect of preventing RNase P cleavage at the 5′ end of individual tRNAs.
Our results also suggest that RNase E is probably involved in the processing of all tRNAs, because we observed an effect on all 59 of the transcripts that we tested. These included the four glt tRNAs, which are contained in four different rRNA operons. What was of interest was that the tRNAs fell into three groups. The first group were those that were processed efficiently (tRNAAsn), such that ∼95%–99% of the steady-state tRNA existed in the mature form in the wild-type control (Fig. 9; Table 4). In comparison, there was a second group, exemplified by tRNAHis (Fig. 9; Table 4), that were inefficiently processed whereby ∼28%–30% of the tRNA existed in the mature form in the wild-type control. The third class of tRNAs (tRNAPro; Fig. 9; Table 4) were processed with an intermediate level of efficiency. However, in all cases, the relative amount of mature tRNA was greater in the temperature-resistant rneΔ645 strain compared with either the temperature-sensitive rne-1 or rneΔ610 mutants (Fig. 9; data not shown).
Taken together, these data indicate that tRNA processing in E. coli involves a series of steps that require initial cleavage events in the 3′ terminal regions by RNase E. Subsequently, RNase P and the various 3′ → 5′ exonucleases can complete the maturation process. Our results support the model for maturation of polycistronic tRNAs that was recently published (Li and Deutscher 2002) while this paper was in preparation. In addition, because RNase E also is required to process monocistronic tRNAs (Fig. 6), it is apparent that the multiple 3′ → 5′ exonucleases involved in generating the mature CCA terminus (Li and Deutscher 1994, 1996) normally do not remove more than a few nucleotides at the 3′ termini of tRNAs.
Several interesting questions are raised by the data reported here. One relates to the recognition of various substrates by RNase E. In all three rne mutants (rne-1, rneΔ610, and rneΔ645), tRNA processing was impaired compared with the wild-type control (Figs. 5–7, 9). The main differences among the three strains were the rate with which they processed the precursor transcripts (Table 3) and the fraction of mature tRNA in the total steady-state RNA population (Fig. 9). However, even though both the rne-1 and rneΔ610 strains stopped growing at 44°C (Fig. 3A,B), 9S rRNA processing was almost normal in the rneΔ610 mutant (Fig. 4). In contrast, the decay of mRNAs in the rneΔ610 and rneΔ645 strains was far more defective at 37°C than in the rne-1 mutant at 44°C (Table 1A,B; Ow et al. 2000). At 44°C, some of the differences were even more striking (Table 1B). Thus, RNase E appears to have different binding affinities for 9S rRNA versus mRNAs. In addition, based on the wide range of processing rates (Table 3) and the differences observed in steady-state levels (Fig. 9), some tRNAs are better substrates than others.
What can account for these differences? Ehretsmann et al. (1992) proposed a 5-nt recognition site for RNase E. Subsequent work has suggested that only a single-stranded AU-rich region is necessary (Cohen and McDowall 1997). By these criteria, however, it is not possible to distinguish why the tyrT processing site (CCAU↓AAUUC; Table 4; Fig. 10) is cleaved very efficiently but the hisR sequence (CCAUUAUUA) is not. In addition, examination of the 3′ termini of the three classes of tRNA substrates (Table 4) shows that the vast majority of them are AU-rich. It would, therefore, appear that secondary and tertiary structures must play an important role in site recognition.
Another point regards the nature of the rneΔ645 allele. Isolated as a revertant of the already extensive RNase E truncation mutant rneΔ610, rneΔ645 encodes an even shorter polypeptide than its progenitor (Figs. 1,2). Surprisingly, this revertant was able to grow at 44°C, albeit at a slower rate than the rne+ strain, in both 6–8 copies/cell and in single copy (Fig. 3A,B). Although the rneΔ645 mutation was initially isolated from a putative rng::cat rneΔ610 double mutant, we have subsequently obtained additional rneΔ645-like alleles in the rneΔ610 genetic background (data not shown).
Of most interest was that an even shorter version of the RneΔ610 protein could suppress the lethality observed at 44°C. A computer search using the amino acids missing in RneΔ645 (418–427; ser leu ser leu ser ile leu arg leu ile) did not yield any known motifs that would explain why the additional truncation generates a protein that is now able to maintain cell viability. The loss of the additional 35 amino acids could change the structure of the protein, resulting in an enzyme that more effectively processes tRNAs, hence rescuing the temperature sensitivity of the rneΔ610 parental strain. Interestingly, rneΔ645 is a recessive mutation (data not shown). Solving the crystal structure of RNase E would help explain this puzzle as well as aid investigators in defining how the various domains of RNase E interact with RNA substrates.
It is now clear that RNase E plays an important role in all aspects of RNA processing and decay in E. coli, including rRNAs, tRNAs, mRNAs, and catalytic RNAs. It is thus somewhat surprising that only the N-terminal catalytic domain of the protein has been evolutionarily conserved and that some organisms such as Bacillus subtilis do not even contain an RNase E homolog (Kaberdin et al. 1998).
Materials and methods
Bacterial strains and plasmid displacement
The E. coli strains used in this study are listed in Table 5. Strains SK2525, SK2534, SK2681, SK2685, SK9982, SK9987, SK10103, SK10143, SK10144, and SK10148 were constructed by a combination of P1 transduction and plasmid displacement (Ow et al. 2000, 2002). Details of these constructions are available upon request.
Table 5.
E. coli strains and plasmids
| Strain
|
Genotypea
|
Source or reference
|
|---|---|---|
| MG1693 | rph-1 | E. coli Genetic Stock Center |
| SK2525 | rnpA49 rph-1 rbsD296::Tn10 Tcr | This study |
| SK2534 | rnpA49 rne-1 rph-1 rbsD296::Tn10 Tcr | This study |
| SK2681 | rneΔ1018::bla rph-1 srlD300::Tn10 recA56 Tcr/pDHK2 (rneΔ610 Smr/Spr)/pWSK129 (Kmr) | This study |
| SK2685 | rneΔ1018::bla rph-1 srlD300::Tn10 recA56 Tcr/pDHK6 (rneΔ645 Smr/Spr)/;WSK129 (Kmr) | This study |
| SK9714 | rneΔ1018::bla rph-1 recA56 srlD300::Tn10 Tcr/pSBK1 (rne+ Cmr) | (Ow et al. 2000) |
| SK9937 | rneΔ1018::bla rph-1 recA56 srlD300::Tn10 Tcr/pMOK13 (rne-1 Cmr) | (Ow et al. 2000) |
| SK9957 | rneΔ1018::bla rph-1 recA56 srlD300::Tn10 Tcr/pMOK15 (rneΔ610 Cmr) | (Ow et al. 2000) |
| SK9982 | rneΔ1018::bla rng::cat rph-1 recA56 srlD300::Tn10 Tcr/pMOK19 (rneΔ610 Smr/Spr) | This study |
| SK9987 | rneΔ1018::bla rph-1 recA56 srlD300::Tn10 Tcr/pMOK21 (rneΔ645 Smr/Spr) | This study |
| SK10103 | rneΔ1018::bla rph-1 recA56 srlD300::Tn10 Tcr/pMOK29 (rneΔ645 Cmr) | This study |
| SK10143 | rneΔ1018::bla rph-1 srlD300::Tn10 recA56 Tcr/pMOK44 (rne+ Spr)/pWSK129 (Kmr) | (Ow et al. 2002) |
| SK10144 | rneΔ1018::bla rph-1 srlD300::Tn10 recA56 Tcr/pMOK45 (rne-1 Spr)/pWSK129 (Kmr) | This study |
| Sk10148 | rph-1 Δrnt::kan Kmr | This study |
| Plasmids | Genotype | Source or reference |
| PKRP13 | High-copy plasmid with Smr/Spr | (Reece and Phillips 1995) |
| pWSK29 | 6–8 copy plasmid with Apr | (Wang and Kushner 1991) |
| pWSK129 | 6–8 copy plasmid with Kmr | (Wang and Kushner 1991) |
| pMOK15 | 6–8 copy plasmid with rneΔ610 Cmr | (Ow et al. 2000) |
| pMOK18 | 6–8 copy plasmid with Smr/Spr | This study |
| pMOK19 | 6–8 copy plasmid with rneΔ610 Smr/Spr | This study |
| pMOK21 | 6–8 copy plasmid with rneΔ645 Smr/Spr | This study |
| pMOK29 | 6–8 copy plasmid with rneΔ645 Cmr | This study |
| pMOK40 | Single-copy plasmid with Smr/Spr | (Ow et al. 2002) |
| pMOK44 | Single-copy plasmid with rne+ Smr/Spr | (Ow et al. 2002) |
| pMOK45 | Single-copy plasmid with rne-1 Smr/Spr | This study |
| pDHK2 | Single-copy plasmid with rneΔ610 Smr/Spr | This study |
| pDHK6 | Single-copy plasmid with rneΔ645 Smr/Spr | This study |
All strains contain the thyA715 allele.
Plasmid construction
The plasmids used are listed in Table 5. Vector pMOK18 was made by inserting an SmaI/SmaI streptomycin/spectinomycin-resistance cassette from pKRP13 (Reece and Phillips 1995) into the ScaI site of pWSK29 (Wang and Kushner 1991). To make pMOK19 (rneΔ610 Smr), a KpnI/SacI DNA fragment from pMOK15 (rneΔ610 Cmr; Ow et al. 2000) containing the rneΔ610 allele was inserted into the KpnI/SacI sites of pMOK18. pMOK21 (rneΔ645 Smr) was isolated naturally from a 44°C replica plate of SK9982. pMOK29 (rneΔ645 Cmr) was made using site-directed mutagenesis using pMOK15 as the template. Plasmids pDHK2 and pMOK45 were constructed by digesting pMOK15 (rneΔ610) and pMOK13 (rne-1), respectively, with SacI and KpnI and ligating the DNA fragment containing the rne mutant allele into a SacI/KpnI-digested pMOK40 (Spr) vector (Ow et al. 2002). pDHK6 was made by inserting an EcoRI/NotI fragment carrying the rneΔ645 allele into the EcoRI/NotI sites in pMOK40.
Growth curves
Cultures were routinely grown in Luria broth with thymine (50 μg/mL) and chloramphenicol (20 μg/mL), kanamycin (50 μg/mL), spectinomycin (20 μg/mL), or streptomycin (20 μg/mL) at 37°C or 30°C with agitation until they reached 40–45 Klett units (1 × 108–1.1 × 108 cells/mL; No. 42 green filter). Once reaching this density, cultures were shifted to 44°C. Every 30 min thereafter, cells were diluted with fresh prewarmed medium. Klett values were adjusted with the appropriate dilution factor.
DNA sequencing
Sequencing of the rneΔ645 allele was done using an ABI Prism 310 Genetic Analyzer (PE Applied Biosystems) as instructed by the manufacturer using primers RNE+240, RNE+292, RNE+747, RNE+1006, and RNE+3628. These primers encompassed the rneΔ610 and rneΔ645 genes in their entirety.
Site-directed mutagenesis
The introduction of a stop codon into plasmid pMOK15 (rneΔ610) to generate the rneΔ645 allele (pMOK29) was done with the U.S.E. Mutagenesis Kit (Amersham Pharmacia) following the instructions of the manufacturer. The selection and the mutagenesis primers (which convert the codon for serine 418 from TCG to a TAG stop codon) were RNE-893 and RNE+1593T, respectively. DNA sequencing at the site of the C → A transversion and RNase E Western blot analysis on strain SK10103 were performed to verify the authenticity of the mutation.
Western analysis
Western analysis was performed as described previously (Ow et al. 2000, 2002).
Primer extension analysis
Primer extensions were carried out using Thermoscript RNase H− reverse transcriptase (Invitrogen) at 60°C according to the manufacturer's specifications. The sequences for the various primers used in the hybridization reactions are available upon request.
Northern analysis
For RNA isolation and Northern analysis, we followed the procedures described in O'Hara et al. (1995) and Burnett (1997). All of the RNAs used for the tRNA-processing Northern blots were obtained from 44°C cultures diluted as described for the growth curves so the cultures were maintained in exponential growth at all times. RNA was obtained from exponentially growing cultures that had been shifted to 44°C for 2 h (SK9937) and 4 h (SK9714, SK9957, and SK9987). These times were chosen based on the times after shift at which the rne-1 and rneΔ610 strains stopped growing (Fig. 3A). The 9S rRNA Northern was done using RNA extracted from cultures shifted to 44°C for 150 min. For strains carrying single copies of rne-1, rneΔ610, and rneΔ645 and the rnpA49 and rnpA49 rne-1 mutants, RNA was isolated after a 150-min shift to 44°C. RNAs from MG1693 (rph-1) and SK10148 (Δrnt::kan rph-1) were harvested from exponentially growing cultures at 37°C. Band densities were quantitated with a Phosphorimager series 400 (Molecular Dynamics). Decay rates for half-life determinations were calculated using linear regression analysis.
Acknowledgments
This work was supported in part by a grant from the NIGMS (GM57220) to S.R.K. M.C.O. was supported in part by a NIH predoctoral traineeship from NIGMS (GM07106). We are thankful to M. Wachi and S. Lovett for providing strains. We also thank B. Mohanty and T. Perwez for critical readings of the manuscript and R. Fabbri and D.-H. Chung for technical assistance.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL skushner@arches.uga.edu; FAX (706) 542-3910.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.983502.
References
- Altman S, Kirsebom L, Talbot S. Recent studies of RNase P. In: Soll D, Bhandary Raj, editors. tRNA: Structure and function. Washington, DC.: ASM Press; 1995. pp. 67–78. [Google Scholar]
- Apirion D, Gitelman D. Decay of RNA in RNA processing mutants of Escherichia coli. Mol Gen Genet. 1980;177:139–154. doi: 10.1007/BF00267448. [DOI] [PubMed] [Google Scholar]
- Arraiano CM, Yancey SD, Kushner SR. Stabilization of discrete mRNA breakdown products in ams pnp rnb multiple mutants of Escherichia coli K-12. J Bacteriol. 1988;170:4625–4633. doi: 10.1128/jb.170.10.4625-4633.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babitzke P, Kushner SR. The Ams (altered mRNA stability) protein and ribonuclease E are encoded by the same structural gene of Escherichia coli. Proc Natl Acad Sci. 1991;88:1–5. doi: 10.1073/pnas.88.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babitzke P, Granger L, Kushner SR. Analysis of mRNA decay and rRNA processing in Escherichia coli multiple mutants carrying a deletion in RNase III. J Bacteriol. 1993;175:229–239. doi: 10.1128/jb.175.1.229-239.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlyn MKB. Linkage map of Escherichia coli K-12, Edition 10: The traditional map. Microbiol Mol Biol Rev. 1998;62:814–984. doi: 10.1128/mmbr.62.3.814-984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett WV. Northern blotting of RNA denatured in glyoxal without buffer recirculation. Bio Techn. 1997;22:668–671. doi: 10.2144/97224st01. [DOI] [PubMed] [Google Scholar]
- Carpousis AJ, Van Houwe G, Ehretsmann C, Krisch HM. Copurification of E. coli RNAase E and PNPase: Evidence for a specific association between two enzymes important in RNA processing and degradation. Cell. 1994;76:889–900. doi: 10.1016/0092-8674(94)90363-8. [DOI] [PubMed] [Google Scholar]
- Casarégola S, Jacq A, Laoudj D, McGurk G, Margarson S, Tempête M, Norris V, Holland IB. Cloning and analysis of the entire Escherichia coli ams gene. ams is identical to hmp-1 and encodes a 114 kDa protein that migrates as a 180 kDa protein. J Mol Biol. 1992;228:30–40. doi: 10.1016/0022-2836(92)90489-7. [DOI] [PubMed] [Google Scholar]
- ————— Cloning and analysis of the entire Escherichia coli ams gene. ams is identical to hmp1 and encodes a 114 kDa protein that migrates as a 180 kDa protein. J Mol Biol. 1994;238:867. doi: 10.1006/jmbi.1994.1344. [DOI] [PubMed] [Google Scholar]
- Cohen SN, McDowall KJ. RNase E: Still a wonderfully mysterious enzyme. Mol Microbiol. 1997;23:1099–1106. doi: 10.1111/j.1365-2958.1997.tb02593.x. [DOI] [PubMed] [Google Scholar]
- Ehretsmann CP, Carpousis AJ, Krisch HM. Specificity of Escherichia coli endoribonuclease RNase E: In vivo and in vitro analysis of mutants in a bacteriophage T4 mRNA processing site. Genes & Dev. 1992;6:149–159. doi: 10.1101/gad.6.1.149. [DOI] [PubMed] [Google Scholar]
- Ghora BK, Apirion D. Structural analysis and in vitro processing to p5 (5S) rRNA of a 9S RNA molecule isolated from an rne mutant of E. coli. Cell. 1978;15:1055–1066. doi: 10.1016/0092-8674(78)90289-1. [DOI] [PubMed] [Google Scholar]
- Gurevitz M, Jain SK, Apirion D. Identification of a precursor molecule for the RNA moiety of the processing enzyme RNase P. Proc Natl Acad Sci. 1983;80:4450–4454. doi: 10.1073/pnas.80.14.4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnsdorf E, Steier O, Coscoy L, Teysset L, Régnier P. Roles of RNase E, RNase II and PNPase in the degradation of the rpsO transcripts of Escherichia coli: Stabilizing function of RNase II and evidence for efficient degradation in an ams pnp rnb mutant. EMBO J. 1994;13:3368–3377. doi: 10.1002/j.1460-2075.1994.tb06639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain C, Belasco JG. RNase E autoregulates its synthesis by controlling the degradation rate of its own mRNA in Escherichia coli: Unusual sensitivity of the rne transcript to RNase E activity. Genes & Dev. 1995;9:84–96. doi: 10.1101/gad.9.1.84. [DOI] [PubMed] [Google Scholar]
- Kaberdin VR, Miczak A, Jakobsen JS, Lin-Chao S, McDowall KJ, von Gabain A. The endoribonucleolytic N-terminal half of Escherichia coli RNase E is evolutionarily conserved in Synechocystis sp. and other bacteria but not the C-terminal half, which is sufficient for degradosome assembly. Proc Natl Acad Sci. 1998;95:11637–11642. doi: 10.1073/pnas.95.20.11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Deutscher MP. The role of individual exoribonucleases in processing at the 3′ end of Escherichia coli tRNA precursors. J Biol Chem. 1994;269:6064–6071. [PubMed] [Google Scholar]
- ————— Maturation pathways for E. coli tRNA precursors: A random multienzyme process in vivo. Cell. 1996;86:503–512. doi: 10.1016/s0092-8674(00)80123-3. [DOI] [PubMed] [Google Scholar]
- ————— RNase E plays an essential role in the maturation of Escherichia coli tRNA precursors. RNA. 2002;8:97–109. doi: 10.1017/s1355838202014929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Altman S. Inhibition of viral gene expression by the catalytic RNA subunit of RNase P from Escherichia coli. Genes & Dev. 1995;9:471–480. doi: 10.1101/gad.9.4.471. [DOI] [PubMed] [Google Scholar]
- López PJ, Marchand I, Joyce SA, Dreyfus M. The C-terminal half of RNase E, which organizes the Escherichia coli degradosome, participates in mRNA degradation but not rRNA processing in vivo. Mol Microbiol. 1999;33:188–199. doi: 10.1046/j.1365-2958.1999.01465.x. [DOI] [PubMed] [Google Scholar]
- Lundberg U, Altman S. Processing of the precursor to the catalytic RNA subunit of RNase P from Escherichia coli. RNA. 1995;1:327–334. [PMC free article] [PubMed] [Google Scholar]
- Mackie GA. Specific endonucleolytic cleavage of the mRNA for ribosomal protein S20 of Escherichia coli requires the products of the ams gene in vivo and in vitro. J Bacteriol. 1991;173:2488–2497. doi: 10.1128/jb.173.8.2488-2497.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Ribonuclease E is a 5′-end-dependent endonuclease. Nature. 1998;395:720–723. doi: 10.1038/27246. [DOI] [PubMed] [Google Scholar]
- Miczak A, Kaberdin VR, Wei C-L, Lin-Chao S. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc Natl Acad Sci. 1996;93:3865–3869. doi: 10.1073/pnas.93.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd EA, Krisch HM, Higgins CF. RNase E, an endoribonuclease, has a general role in the chemical decay of Escherichia coli mRNA: Evidence that rne and ams are the same genetic locus. Mol Microbiol. 1990;4:2127–2135. doi: 10.1111/j.1365-2958.1990.tb00574.x. [DOI] [PubMed] [Google Scholar]
- O'Hara EB, Chekanova JA, Ingle CA, Kushner ZR, Peters E, Kushner SR. Polyadenylation helps regulate mRNA decay in Escherichia coli. Proc Natl Acad Sci. 1995;92:1807–1811. doi: 10.1073/pnas.92.6.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M, Kuwano M. A conditional lethal mutation in an Escherichia coli strain with a longer chemical lifetime of mRNA. J Mol Biol. 1979;129:343–357. doi: 10.1016/0022-2836(79)90500-x. [DOI] [PubMed] [Google Scholar]
- Ow MC, Liu Q, Kushner SR. Analysis of mRNA decay and rRNA processing in Escherichia coli in the absence of RNase E-based degradosome assembly. Mol Microbiol. 2000;38:854–866. doi: 10.1046/j.1365-2958.2000.02186.x. [DOI] [PubMed] [Google Scholar]
- Ow MC, Liu Q, Mohanty BK, Andrew ME, Maples VF, Kushner SR. RNase E levels in Escherichia coli are controlled by a complex regulatory system that involves transcription of the rne gene from three promoters. Mol Microbiol. 2002;43:159–171. doi: 10.1046/j.1365-2958.2002.02726.x. [DOI] [PubMed] [Google Scholar]
- Py B, Causton H, Mudd EA, Higgins CF. A protein complex mediating mRNA degradation in Escherichia coli. Mol Microbiol. 1994;14:717–729. doi: 10.1111/j.1365-2958.1994.tb01309.x. [DOI] [PubMed] [Google Scholar]
- Py B, Higgins CF, Krisch HM, Carpousis AJ. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996;381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- Ray BK, Apirion D. RNAase P is dependent on RNAase E action in processing monomeric RNA precursors that accumulate in an RNAase E− mutant of Escherichia coli. J Mol Biol. 1981a;149:599–617. doi: 10.1016/0022-2836(81)90349-1. [DOI] [PubMed] [Google Scholar]
- ————— Transfer RNA precursors are accumulated in Escherichia coli in the absence of RNase E. Eur J Biochem. 1981b;114:517–524. doi: 10.1111/j.1432-1033.1981.tb05175.x. [DOI] [PubMed] [Google Scholar]
- Reece KS, Phillips GJ. New plasmids carrying antibiotic-resistance cassettes. Gene. 1995;165:141–142. doi: 10.1016/0378-1119(95)00529-f. [DOI] [PubMed] [Google Scholar]
- Régnier P, Hajnsdorf E. Decay of mRNA encoding ribosomal protein S15 of Escherichia coli is initiated by an RNase E-dependent endonucleolytic cleavage that removes the 3′ stabilizing stem and loop structure. J Mol Biol. 1991;187:23–32. doi: 10.1016/0022-2836(91)90542-e. [DOI] [PubMed] [Google Scholar]
- Taraseviciene L, Miczak A, Apirion D. The gene specifying RNase E (rne) and a gene affecting mRNA stability (ams) are the same gene. Mol Microbiol. 1991;5:851–855. doi: 10.1111/j.1365-2958.1991.tb00758.x. [DOI] [PubMed] [Google Scholar]
- Vanzo NF, Li YS, Py B, Blum E, Higgins CF, Raynal LC, Krisch HM, Carpousis AJ. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes & Dev. 1998;12:2770–2781. doi: 10.1101/gad.12.17.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RF, Kushner SR. Construction of versatile low-copy-number vectors for cloning, sequencing and expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]