Abstract
The molybdenum cofactor (Moco), a highly conserved pterin compound coordinating molybdenum (Mo), is required for the enzymatic activities of molybdoenzymes. In all organisms studied so far Moco is synthesized by a unique and evolutionary old multistep pathway that requires the activities of at least six gene products. In eukaryotes, the last step of Moco synthesis, i.e., transfer and insertion of Mo into molybdopterin (MPT), is catalyzed by the two-domain proteins Cnx1 in plants and gephyrin in mammals. Both domains (E and G) of these proteins are able to bind MPT in vitro. Here, we show the identification and mutational dissection of functionally important regions within the Cnx1 G domain that are essential for MPT binding, the conversion of MPT to Moco, and Moco stabilization. By functional screening for mutants in the Cnx1 G domain that are no longer able to complement Escherichia coli mogA mutants, we found two classes of mutations in highly conserved amino acid residues. (i) The first class affects in vitro binding of MPT to the protein and the stabilization of Moco, the product of the G domain. (ii) The second class is represented by two independent mutations in the aspartate 515 position that is not affected in MPT binding and Moco stabilization; rather the conversion of MPT to Moco by using bound MPT and a yet unknown form of Mo is completely abolished. The results presented here provide biochemical evidence for a purified Cnx1 G domain catalyzing the insertion of Mo into MPT.
Molybdenum (Mo) plays an important role as the active center in molybdoenzymes (Mo-enzymes), catalyzing essential redox reactions in the global C, N, and S cycles (1). Mo-enzymes are important for such diverse metabolic processes as sulfur detoxification and purine catabolism in mammals (2), nitrate assimilation in autotrophs, and phytohormone synthesis in plants (3). With the exception of nitrogenase, in all Mo-enzymes studied so far Mo is activated and chelated by the so-called molybdenum cofactor (Moco). Moco consists of Mo covalently bound via a dithiolene group to the unique pterin compound molybdopterin (MPT) (4), a tetrahydro-pyranopterin (5, 6) that is highly conserved in eukaryotes, eubacteria, and archaebacteria. Biosynthesis of Moco requires the multistep synthesis of the MPT moiety followed by the subsequent transfer of Mo (3, 7, 8). A mutational block in Moco biosynthesis leads to the combined loss of function of all Mo-enzymes. Plant cells can no longer assimilate inorganic nitrogen and have changed levels of certain phytohormones (9), which is lethal for the organism. Human Moco deficiency as rare inborn error is characterized by loss of activity of sulfite oxidase, xanthine oxidase, and aldehyde oxidase. Affected patients die early postnatally because no therapy is yet available (10).
At least six gene products are involved in the biosynthesis of Moco as identified in humans (11–14), plants (15, 16), and bacteria (7). GTP, the proposed starting compound, is converted via precursor Z (17, 18) to the pyranopterin MPT. In eukaryotes, the final step of cofactor maturation, i.e., the transfer and incorporation of Mo into MPT, is catalyzed by the two-domain protein Cnx1 in plants (15) and by the neuroreceptor anchor protein (19) gephyrin in mammals (Fig. 1) (14). Both domains are highly homologous to the separately expressed Escherichia coli proteins MoeA and MogA. A mutation in proteins catalyzing the last step of Moco synthesis leads to a molybdate-repairable phenotype (20–23). The functional integrity and homology of the G domains in Cnx1 and gephyrin was demonstrated by heterologous complementation of the E. coli mogA mutant by using cnx1 (15) or gephyrin cDNAs (14). In contrast, we have not been able to demonstrate functional complementation of E. coli moeA mutants by the E domain of Cnx1 and gephyrin (G.S., unpublished data). Although in E. coli only a mutation in the mogA gene (or the molybdate transporter encoded by the mod locus) causes the molybdate repairability (24), recently a molybdate repairable phenotype was described for a mutation in the moeA gene of Rhodobacter capsulatus (25). This moeA mutation was only molybdate repairable for enzymes that contain the MPT-based cofactor (like that of eukaryotic enzymes), whereas activities of enzymes with a dinucleotide-based cofactor, as occurring in eubacteria and archaebacteria, were not restored by molybdate. These results suggest that in eukaryotes both the E and G domains are essential for the conversion of MPT to Moco. As biochemical support for this proposal we have shown that both eukaryotic proteins, Cnx1 and gephyrin, bind MPT with high affinity by their E and G domains (14, 26), indicating their functional role in the transfer of Mo to MPT. Because molybdate is sufficient to overcome a mutation in the last step of Moco synthesis, molybdate can be postulated as the source of Mo. However, binding of molybdate to Cnx1 has yet to be demonstrated. Results of Hasona et al. (27) suggest a role for MoeA in activation of molybdate. One can argue that the E domain in Cnx1 generates a Mo species that could be used by the adjacent G domain to convert MPT to Moco.
Figure 1.
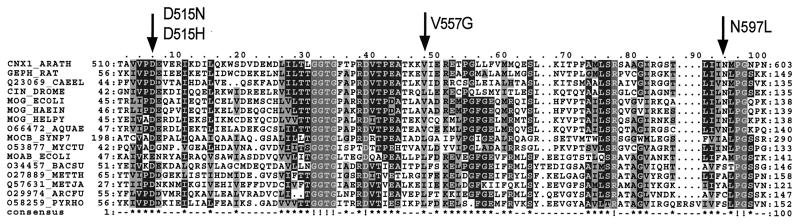
Multiple sequence alignment of Cnx1 G domain, homologous domains, and proteins from other organisms. The positions of the substituted amino acids in the four Cnx1 G domain mutants identified in this work are indicated. The aligned protein sequences are named by their Swiss-Prot accession numbers or identification and are marked with the starting and the ending amino acids in each row. The consensus sequences have been calculated with a threshold of 60%; completely conserved amino acids show a ! in the consensus sequence and are dark gray. Highly conserved amino acids are shown in black and low conserved amino acids in light gray. The protein sequences were derived from the following organisms in the order as listed in the figure: Arabidopsis thaliana, Rattus norvegicus, Caenorhabditis elegans, Drosophila melanogaster, E. coli, Haemophilus influenzae, Helicobacter pylori, Aquifex aeolicus, Synechococcus spec., Mycobacterium tuberculosis, E. coli, Bacillus subtilis, Methanobacterium thermoautotrophicum, Methauococcus jannaschii, Archeoglobus fulgidus, and Pyrrococcus horikoshii.
To identify amino acid residues essential for the function of Cnx1 G domain and in particular for high affinity MPT binding we took advantage of the described functional complementation of E. coli mogA mutants by Cnx1 (15). Here we report the random mutagenesis of Cnx1 G domain and subsequent functional screen for loss of mogA complementing activity. Different mutants were identified with mostly total loss of function that permitted us to dissect single steps within the catalyzed transfer of Mo to MPT. We isolated mutations in residues important for MPT binding and in residues that did not affect the pterin binding. However, in all cases the mutants were no longer able to introduce Mo into MPT to form active Moco. Finally, the mutants with reduced MPT binding have lost the third functional property of the G domain, namely the stabilization of the formed product by preventing the dissociation of Mo from Moco.
Experimental Procedures
Materials.
All chemicals used were from the highest grade available. Xanthine oxidase (EC 1.1.3.22) from buttermilk grade I (0.69 units/mg) was obtained from Sigma. Ni-NTA (nickel-nitrilotriacetic acid) superflow matrix was obtained from Qiagen (Hilden, Germany). Prepacked gel filtration PD10 and nick columns were used as recommended by the manufacturer (Amersham Pharmacia).
Plasmids, Bacterial Strains, and Growth Conditions.
E. coli strains were grown on LB medium, supplemented with 50 μg/ml kanamycin and/or 125 μg/ml ampicillin where appropriate. The mutator strain ES1578 [mutD5 ara-14 argE3(Oc) galK2 hisG4(Oc) kdgK51 lacY1 leuB6 mgl-51 mtl-1 rac rfbD1 rpsL31 supE44 thi-1 thr-1 tsx-33 xyl-S] (28) was routinely grown at 37°C, and the mogA mutant RK5206 (RK4353 chlG206∷Mu cts mogA) and the corresponding wild-type strain RK4353 [F− ara D139 Δ(argF-lac)U 169 deoC1 flb B5201 gyr A219 rel A1 rps150 non-9 pts F25] (20) were cultured at 30°C. For mutagenesis, functional complementation, and recombinant expression, the previously described expression construct of Cnx1 G domain (pQE60cnx1G) (26) was used. Aerobic growth was achieved by vigorous shaking (>200 rpm) on an orbital shaker. Anaerobic growth conditions were obtained under an atmosphere of nitrogen, hydrogen, and carbon dioxide in an anaerobic chamber, by using a palladium catalyst to remove residual oxygen (Gaspak, BBL). Screening of strains for chlorate resistance was carried out under anaerobic growth conditions as described (20), by using LB agar plates supplemented with 15 mM potassium chlorate and 0.2% glucose. For MPT copurification 50 ml of culture medium was inoculated with 2 ml of overnight culture followed by aerobic growth for 1 h at 30°C. Subsequently, expression of Cnx1 G domain was induced by addition of 1 mM isopropyl β-thiogalactoside (IPTG) and cells were grown for an additional 10–12 h before harvesting.
In Vivo Mutagenesis of Cnx1 G Domain.
Mutagenesis was achieved by passage of the Cnx1 G domain on plasmid pQE60cnx1G through the mutator strain ES1578. Briefly, single colonies of ES1578 that was freshly transformed with pQE60cnx1G were grown aerobically for 17–21 h in 5-ml cultures. Plasmid DNA subsequently was isolated and used to transform RK5206. For each series 1 μg mutagenized plasmid DNA was transformed into 200 μl competent mogA cells and was plated directly onto chlorate-selective media. Chlorate-resistant colonies also were screened for loss of nitrate reductase activity by colony overlay assay (29). Plasmid DNA subsequently was isolated, and cnx1G was sequenced on both strands to identify the mutation.
Determination of MPT Content.
MPT was detected and quantified by conversion to its stable oxidation product FormA-dephospho according to ref. 30. Oxidation, dephosphorylation, quarternary aminoethyl chromatography, and HPLC analysis were performed as described (26). FormA-dephospho was quantified by a standard isolated from xanthine oxidase by using the extinction coefficient of ɛ380 = 13,200 M-1⋅cm-1 (31).
Purification of Recombinant Proteins.
Large-scale purifications of wild-type and mutated Cnx1 G domains were performed after expression in M15 cells (Qiagen) according to ref. 26. For MPT copurification simultaneous protein purification was performed by using drop columns with 0.5 ml Ni-nitrilotriacetic acid matrix. Purification was subdivided into the following steps: loading of 1 ml crude extract, washing (2 × 3 ml), and elution (2 × 1 ml). The eluent immediately was desalted on a PD10 gel filtration column previously equilibrated with 100 mM Tris⋅HCl, pH 7.2 and either directly used for further experiments or shock-frozen in liquid nitrogen and stored at −70°C. Purity of the proteins was checked by using 12.5% SDS/PAGE (32). For CD spectroscopy further purification of the proteins was achieved by anion exchange chromatography on an Uno Q1 column (Bio-Rad). Pure fractions were pooled, concentrated (centricon; Amicon), and rebuffered on nick columns to 40 mM sodium phosphate buffer (pH 7.8).
CD Spectroscopy.
Purified proteins were used in a concentration of 0.1 mg/ml. Spectra were recorded on a J-600 CD-spectrometer (Jasco, Tokyo) at 10 nm/min scan speed with eight repetitions and a noise reduction of factor 10.
Enzyme Assays and MPT Binding Experiments.
Nitrate reductase activity in crude extracts was determined by a spectroscopic assay using benzyl viologen as described (33). MPT binding experiments were performed with protein-free MPT isolated from xanthine oxidase by ultra-filtration and gel filtration (26).
Nit-1 Reconstitution.
Neurospora crassa nit-1 extract was prepared as described (34) and stored in aliquots at −70°C. The reconstitution assay was performed in 60 μl reaction volume containing 40 μl of nongel-filtrated or 50 μl of gel-filtrated nit-1 extract in the presence of 4 mM reduced glutathione. Immediately before use, nit-1 extract was gel-filtrated on nick columns by loading 400 μl of extract and eluting 500 μl sample from the column previously equilibrated with 50 mM sodium phosphate, 200 mM NaCl, 5 mM EDTA, pH 7.2. Purified MPT source was added in different amounts (0.5–10 μl) according to the previously determined linear range of the reconstitution assay in the absence or presence of 10 mM sodium molybdate. Complementation was carried out anaerobically for 2 h at room temperature. After addition of 20 mM NADPH for 10 min, reconstituted NADPH-nitrate reductase activity was determined as described (34).
Results
Functional Screening for Mutants of Cnx1 G Domain.
To investigate the functional role of important amino acid residues in the G domain of Cnx1, mutants of the G domain were generated by random mutagenesis in the E. coli strain ES1578. For screening of functionally important mutations we have taken advantage of the finding that Cnx1 (15) and its G domain alone (G.S., unpublished data) are able to restore nitrate reductase activity in E. coli mogA mutants.
After exhaustive mutagenesis, we obtained 28 mutants that were unable to restore nitrate reductase activity in the E. coli mogA mutant RK5206. Nitrate reductase-negative clones subsequently were checked for expression of the Cnx1 G domain by immunoblotting. A significant proportion of the isolated mutants failed to express G domain protein, and these were not investigated further. However, we obtained four independent mutants that showed an unchanged expression level under noninducing (no IPTG) conditions when compared with the wild-type protein, and these were further analyzed. Sequence analysis of these mutants showed that in each case loss of function was caused by a single point mutation in the DNA sequence of the Cnx1 G domain. Each mutation altered a highly conserved amino acid residue (Fig. 1). The aspartate residue at codon 515 was mutated twice, resulting in nonconservative exchanges to asparagine and histidine (D515N, D515H). The two other mutants also show nonconservative exchanges where valine 557 is mutated to a glycine (V557G) and the asparagine 597 is changed to leucine (N597L).
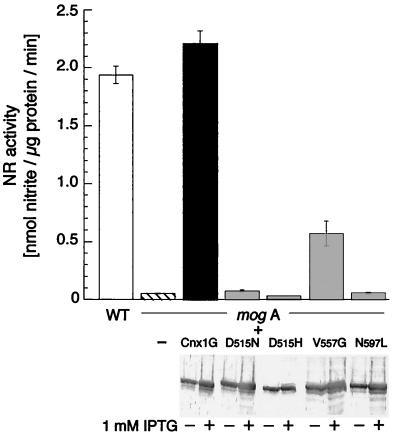
To quantify the effect of each mutation we measured the specific nitrate reductase activity of mogA mutants complemented with clones carrying wild-type and mutated proteins. The complementing activities of all four mutants in mogA crude extracts were strongly reduced (Fig. 2). Strains carrying Cnx1 G domains with the N597L and both aspartate 515 mutations had only basal levels of nitrate reductase activity, comparable to the mogA mutant alone. The V557G mutation gave a low, but measurable, rate of complementation, indicating a partial loss of function. The levels of expressed G domains were comparable in all strains (Fig. 2).
Figure 2.
Functional complementation of the E. coli mogA mutant RK5206 with wild-type and mutated Cnx1 G domains. The columns show nitrate reductase (NR) activity of the crude protein extract derived from E. coli wild-type (WT) strain RK4353 (white bar), E. coli mogA cells transformed with pQE60 control vector (hatched bar), E. coli mogA cells expressing Cnx1 G domain (Cnx1G, black bar), and the four G domain mutants (gray bars). Crude extract (1–20 μl) was used for each assay. Assays were performed in triplicate, and standard errors are shown by error bars. The expression level of wild-type and mutated G domains, detected by Western blot, in the presence (+) and absence (−) of IPTG is shown below the chart.
MPT Binding Properties of Purified Cnx1 G Domains.
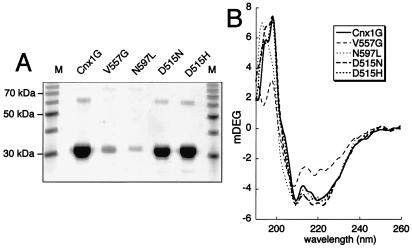
The mutated G domains and the wild-type protein were recombinantly overexpressed in E. coli and purified to homogeneity. The aspartate mutants showed the same level of expression as the wild-type protein under complementing (no IPTG induction) or overexpressing (IPTG induced) conditions that resulted in 1.2–1.45 mg protein/50 ml culture medium (Fig. 3A). The two other mutants, however, were expressed at a significantly lower level during overexpressing conditions with a yield of 0.12–0.13 mg protein/50 ml culture medium. Because the aspartate mutants and the wild type do not differ in their expression level, it is unlikely that these mutations significantly affect folding of the protein. This conclusion is strengthened by CD spectroscopy (Fig. 3B). The other two mutants (V557G and N597L) are expressed at lower levels, raising the possibility that the mutations could change the overall folding and structure of these protein domains. CD spectroscopic analysis (Fig. 3B) indicates that the asparagine mutant (N597L) shows signals comparable to the wild type (only a small shift at 195 nm) and the aspartate mutants, suggesting no significant change in the overall secondary structure of this mutant. In contrast, the V557G mutant exhibits marked changes in the secondary structure, suggesting that the protein may be partially unfolded because of its reduced CD signals at 195 and 210 nm.
Figure 3.
Purification and CD spectroscopy of Cnx1 G domains from wild type and mutants. (A) SDS/PAGE (12.5%) of the purified G domains (wild type and mutants) after overexpression and affinity purification. Twenty microliters of the 2 ml eluate from Ni-nitrilotriacetic acid column was analyzed. (B) CD spectra of 0.1 mg/ml protein.
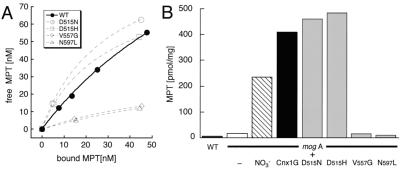
To examine differences in the biochemical properties of the four mutated proteins, MPT binding experiments were performed with 200 nM purified protein. Each of the mutant proteins was capable of binding MPT, but with different affinities (Fig. 4A). Thus, the aspartate mutants D515N and D515H bound MPT with the same affinity, or even more tightly than the wild-type G domain (Kd = 120 ± 15 nM, n = 195 ± 19 nM). In contrast, the V557G and N597L mutants showed a significant decrease in MPT binding capacity. The increase in Kd and decrease in binding sites of the V557G and N597L mutants could reflect a conformational change at the MPT binding pocket that may be correlated to a more structural character of the mutation that fits well with the lower level of IPTG-induced protein expression (Fig. 3A). One can suppose that the loss of function of these two mutants is caused, at least in part, by the decrease in MPT binding. On the other hand, because both D515 mutants show an MPT binding affinity similar to the wild-type protein, the mutant phenotypes must result from a loss of function of the G domain that is distinct from MPT binding. This second function could be the insertion of Mo into MPT, forming active Moco, or the stabilization of Moco itself to prevent the dissociation of Mo from the pterin.
Figure 4.
MPT binding properties of wild-type and mutant Cnx1 G domains. (A) MPT binding curves of Cnx1G and all four mutant proteins. For the binding assays, 200 nM of purified protein was used. The amount of free MPT was plotted against the amount of protein-bound MPT and fitted by using Michaelis–Menten binding curves. The two points of each mutant curve were fitted in the same way for better visualization. (B) MPT content of E. coli wild-type strain RK4353 (WT), E. coli mogA mutant RK5206 (white bar) grown under aerobic growth conditions, E. coli mogA mutant RK5206 grown under nitrate reductase-inducing conditions (NO3−, striped bar), and E. coli mogA cells overexpressing Cnx1 G domain (Cnx1G, black bar) and the four G domain mutants (gray bars). MPT content was determined by FormA HPLC analysis.
MPT Content of mogA Mutants Expressing Wild-Type and Mutant Cnx1 G Domains.
Because of the defect of E. coli mogA mutants in the last step of Moco synthesis, MPT as intermediate of the pathway is highly accumulated (G.S., unpublished data). In addition, the high-level expression of Cnx1 G domain resulted in an accumulation of MPT 10–20 times higher than the wild type (Fig. 4B; G.S., unpublished data) that could arise from its high affinity binding of MPT. Therefore, the ability of the G domain mutants to induce MPT synthesis was investigated (Fig. 4B). Crude cell extracts of the mogA strain expressing either of the aspartate 515 mutants (D515N and D515H) showed MPT contents comparable to strains carrying the wild-type G domain, confirming that the MPT binding behavior of these mutated proteins is unaffected in vivo. In contrast, the V557G and N597L mutants were not able to induce MPT synthesis in mogA mutants (Fig. 4B). This finding is in agreement with the results of the MPT binding experiments where a lower binding capacity of these two mutants was observed (Fig. 4A). Because expression levels achieved with 0.1 mM IPTG are sufficient to induce MPT synthesis (G.S., unpublished data), the lack of induction of MPT synthesis by expression of the V557G and N597L mutants reflects the reduced MPT binding capacity rather than the reduced expression level. These results suggest that the MPT binding and the ability to induce MPT synthesis are closely linked.
Purification of G Domains After Expression in mogA Mutants.
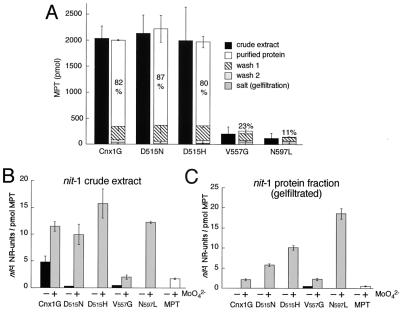
If there is a correlation between the MPT binding property of the G domain and induction of MPT synthesis, the major part of formed MPT (or Moco) should be bound to the G domain. To verify this, proteins were purified on small Ni-nitrilotriacetic acid drop columns as described in Experimental Procedures, and the amounts of cofactor present in crude cell extracts, wash fractions, and purified proteins were determined by FormA analysis. After purification, the main part (80–87%) of total MPT detectable in crude total cell extracts expressing either the wild-type G domain or the two aspartate 515 mutants (D515N and D515H) could be copurified with the recombinant proteins (Fig. 5A). The high recovery of MPT after purification is in line with the observed low dissociation of bound MPT from the G domain (26). These results confirm the in vivo binding of MPT to be similar among both aspartate mutants and the wild-type protein. Based on this finding it can be concluded that the induction of MPT synthesis by Cnx1 G domain is a result of the high affinity binding of MPT to the G domain. We already have demonstrated (Fig. 4B) that the MPT content in crude extracts of mogA cells expressing V557G or N597L is significantly lower. Fig. 5A shows that these proteins bind MPT with less capacity (23% MPT on V557G and 11% MPT on N597L).
Figure 5.
Copurification of MPT with wild-type and mutant Cnx1 G domains and its activity in the nit-1 reconstitution assay. (A) The columns show the MPT content of the fractions during affinity purification of Cnx1 G domain and the four mutant proteins. The percentage of protein-bound MPT is given in relation to the total MPT in crude cell extract. Average values for MPT crude extracts are derived from triplicate measurements. The error bars for each fraction of purification were calculated from the percentage of MPT values for each purification in relation to the corresponding crude extract. (B and C) Nit-1 reconstitution using either the nit-1 crude extract (B) or the protein fraction of gel-filtrated nit-1 (C) extract in the presence (+) or absence (−) of 10 mM molybdate with free MPT and MPT bound to Cnx1 G domains. The activity is given in nit-1–nitrate reductase (NR) units per pmol MPT. Standard errors were calculated from two different reconstitutions with at least two different MPT concentrations that were chosen from the linear range of the reconstitution assay.
Activity of Copurified MPT in the nit-1 Reconstitution Assay.
Based on the FormA analysis in the previous experiments it is unclear whether MPT or Moco was bound to purified G domains. To distinguish between the two possibilities, the purified protein/pterin complex was used for nit-1 reconstitution experiments. The protein was not denatured for release of cofactor because we assumed that the G domain has a function in transferring or generating Moco in vivo and should be able to donate the cofactor to a Mo-enzyme. The nit-1 reconstitution assay is a widely used and accepted method for the detection of Moco and MPT from biological sources by transfer to the nit-1 apo-nitrate reductase.
Reconstitutions were performed by using free MPT (isolated from xanthine oxidase) or MPT bound to the G domain with either the nit-1 crude extract or the protein fraction of gel-filtrated nit-1 extract (i) in the absence of external molybdate to determine active Moco or (ii) in the presence of 10 mM sodium molybdate to measure total MPT by chemical conversion to Moco (Fig. 5 B and C). The activities obtained were normalized to the MPT level bound on each protein. All proteins contained MPT that was detectable in nit-1 crude or gel-filtrated extracts in the presence of molybdate (Fig. 5 B and C, gray bars). These data indicate that the copurified MPT bound to wild-type or mutated G domains is biologically active. However, in the absence of molybdate only the wild-type G domain generated active Moco in nit-1 crude extract (Fig. 5B, molybdate, dark bars). This result is biochemical evidence that both D515N and D515H have lost this particular ability to donate active Moco. In addition, it is apparent that the V557G and N597L mutants also lack this function, which can be distinguished from MPT binding. The slight background activity of V557G is in line with its 30% residual mogA-complementing activity (Fig. 2), whereas N597L seems to be able to donate its bound MPT efficiently to the nit-1 apo-nitrate reductase. Keeping in mind that the residual amount of bound MPT on N597L was the lowest (11%), a high dissociation rate of MPT also would explain the high nit-1 reconstitution (Fig. 5C). Nevertheless, the absolute activity (per protein) of the V557G and N597L mutants was dramatically reduced compared to the wild type and the aspartate mutants (data not shown). Finally, free MPT isolated from xanthine oxidase gives an activity lower than any protein-bound MPT (Fig. 5 B and C, white bar).
In contrast, the observed activity of wild-type G domain to donate Moco to nit-1 apo-nitrate reductase is completely abolished when using gel-filtrated nit-1 protein extract (Fig. 5C). Because gel filtration of the nit-1 extract serves to remove low molecular weight compounds from the extract, including MPT precursors and metal ions, such a component seems to be essential for the wild-type G domain to generate active Moco. One can argue that the species bound by the purified G domain is MPT and not Moco itself, because active Moco can be transferred efficiently to gel-filtrated nit-1 protein extract (22, 23), which will be shown later (Fig. 6B). This result is suggestive evidence that the G domain catalyzes the formation of Moco from prebound MPT in the nit-1 crude extract by using a low molecular weight compound, whereas all of the mutants failed to convert their MPT to Moco.
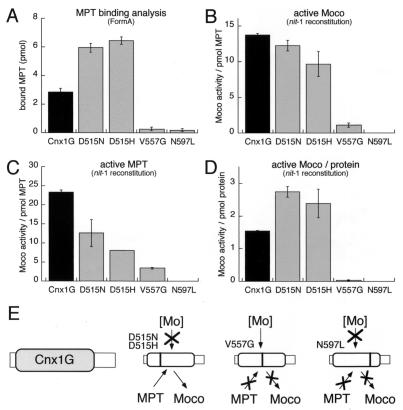
Figure 6.
Binding and stabilization of Moco by Cnx1 G domain. MPT (45 pmol) isolated from xanthine oxidase was incubated with 10 mM sodium molybdate to generate Moco that was supplied for binding to 30 pmol wild-type and mutated Cnx1 G domains or no protein as control. The binding mixture (400 μl) was separated by gel filtration and used for MPT determination (A) by FormA analysis (26) and nit-1 reconstitution in the absence (B and D) or presence (C) of 10 mM sodium molybdate. In all experiments the control (no protein) was subtracted from the measured values. The nit-1 activities determined kinetically by using different amounts of MPT source were plotted either against the MPT amount present in each sample (B and C) or against protein concentration (D) and are expressed in nit-1–nitrate reductase (NR) units per pmol protein-bound MPT (B and C) or nit-1–NR units per pmol protein (D). Standard errors were calculated from triplicates and the nit-1 activity was determined in the linear range of reconstitution by taking three kinetic points of measurement. (E) Model for the different effects of the mutations identified in this work, where both aspartate mutants are defective in Mo insertion and the V557G mutant is impaired in substrate and product binding whereas the N597L mutant has lost all three functional properties of the G domain.
Binding and Stabilization of Moco by the Cnx1 G Domain.
As the Cnx1 G domain is able to convert MPT to Moco we wanted to test whether or not the formed product can be bound and stabilized by the G domain. Moco was chemically generated from MPT previously isolated from xanthine oxidase in the presence of 10 mM sodium molybdate as described (35, 36) and was added to Cnx1 G domains. Subsequently, bound and free Moco as well as free molybdate were separated by gel filtration. In the protein fraction we determined (i) the total amount of bound MPT/Moco (FormA analysis) and (ii) the fraction of active Moco by reconstituting gel-filtrated nit-1 protein extract.
Comparing the amount of bound MPT/Moco between wild-type G domain and the mutants it can be seen again that the V557G and N597L mutants show a strong decrease in MPT binding (Fig. 6A). The higher affinity of both aspartate mutants for MPT is shown more clearly in the gel filtration experiment, indicating a decrease in MPT dissociation. Taking the detected MPT levels as a basis for normalizing the Moco content determined by nit-1 reconstitution, we can show that both aspartate mutants bind Moco in the same way as the wild-type G domain (Fig. 6B). However, the V557G and N597L mutants show a significant decrease in their Moco binding (Fig. 6B), whereas their biologically active MPT is almost similar to the other mutants (Fig. 6C). Taking together that the V557G and N597L mutants show a decrease in MPT binding as well as a loss of Moco stabilization, the combination of both impairments explains their dramatic loss of function when calculating Moco activities on protein level (Fig. 6D).
Discussion
The molybdate repairability of mutations in Cnx1 (15), together with ability of the isolated E and G domains to bind MPT (26), implies a role for the protein in the synthesis of active Moco. To test this function, we have taken advantage of the ability of the G domain to complement E. coli mogA (15) strains to isolate a number of mutations of the Cnx1 G domain that result in a loss of function.
We have clearly demonstrated that two of these mutations, V557G and N597L, show a strong reduction in the affinity for MPT, suggesting that the MPT binding site is affected in these mutants. Two additional mutations in the highly conserved aspartate at position 515 do not impair MPT binding to the protein. This finding indicates that the G domain has a second function in addition to the ability to bind MPT. To investigate this additional function, we overproduced and isolated the wild-type and mutant G domains. During these experiments we observed that expression of either the wild-type, D515H, or D515N G domains resulted in a dramatic induction in the cellular levels of MPT. The vast majority of MPT in these strains was found associated with the overexpressed proteins. These observations support our previous findings in that they suggest that MPT binding by the G domain in vivo is very tight (26).
The overexpressed and isolated wild-type and mutant G domains carrying bound MPT in the presence of molybdate were capable of activating the inactive nitrate reductase present in the nit-1 crude extract. However, in the absence of added molybdate, only the wild-type domain was capable of using a low molecular weight molybdenum compound present in the nit-1 crude extract to form active Moco. The nature of this Mo compound is not yet clear, but it is apparent that the D515H and D515N mutations are incapable of using this compound as a substrate for Moco synthesis.
The Cnx1 G domain is able to bind and stabilize active Moco as shown by molybdate-independent nit-1 reconstitution. However, binding of MPT and stabilizing of Moco in a donatable form are obviously different functional properties as can be seen in the behavior of the V557G and N597L mutants. Their ability to bind MPT is reduced (Fig. 6A), whereas their property to stabilize and donate active Moco is completely lost (Fig. 6D). Nevertheless, both properties are linked and must be regarded as two effects of one and the same structural region within the G domain.
The recently solved structure of the E. coli MogA protein (37) confirms our biochemical data on a structural level, assuming a similar folding of the Cnx1 G domain. In E. coli MogA, the residues corresponding to V557G and N597L both appear to be involved in mediating interactions between secondary structures. Further, V557 (V91 MogA) is located in β-strand 4 that is part of the trimer-forming interface in MogA. N597 is homolgous to N131 in E. coli MogA, which is the first residue of α-helix 5 that seems to be involved in forming the substrate binding pocket. In line with the MogA structure, the observed biochemical properties of the V557G and N597L mutants in Cnx1 G domain are suggested to have a structural influence and not to be directly involved in substrate binding and conversion to Moco.
Interestingly, the residue in MogA corresponding to the D515 was knocked out by Liu et al. (37) as well. On the basis of their observation it was assigned to a highly important region of the protein that is part of the putative active site. For this aspartate residue, direct substrate interactions are most likely because its position is near to an additional electron density that has been modeled as sulfate although it could be replaced principally by molybdate. These structural findings underline the functional importance of this residue, namely the conversion of MPT to Moco as indicated by our data.
Based on the mutations identified we assume that Cnx1 G domain shares different functional properties that are essential for catalyzing the conversion of MPT to active Moco: binding of MPT, inserting Mo into MPT, and stabilizing the so-formed Moco in its native state (Fig. 6E). It would be tempting for the future to investigate the nature of the Mo compound that is used for the generation of Moco by the Cnx1 G domain in nit-1 crude extracts. Finally, the functional role of the E domain in Cnx1 for transfer and/or activation of Mo remains to be elucidated.
Acknowledgments
We thank Dr. Gary Sayers for providing protocols and advice on the use of E. coli strain ES1578, Dr. Marissa Kalis for advice with the CD spectrometer, and Prof. Barry Smith for helpful comments on the CD spectra. The financial support of the Deutsche Forschungsgemeinschaft (R.R.M. and G.S.), the Fritz Thyssen Stiftung (J.K.), and the Royal Society (T.P.) is gratefully acknowledged.
Abbreviations
- Mo
molybdenum
- Moco
molybdenum cofactor
- MPT
molybdopterin
- Mo-enzymes
molybdoenzymes
- IPTG
isopropyl β-thiogalactoside
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.110568497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.110568497
References
- 1.Hille R. Chem Rev. 1996;96:2757–2816. doi: 10.1021/cr950061t. [DOI] [PubMed] [Google Scholar]
- 2.Kisker C, Schindelin H, Baas D, Retey J, Meckenstock R U, Kroneck P M. FEMS Microbiol Rev. 1998;22:503–521. doi: 10.1111/j.1574-6976.1998.tb00384.x. [DOI] [PubMed] [Google Scholar]
- 3.Mendel R R, Schwarz G. Crit Rev Plant Sci. 1999;18:33–69. [Google Scholar]
- 4.Kramer S P, Johnson J L, Ribeiro A A, Millington D S, Rajagopalan K V. J Biol Chem. 1987;262:16357–16363. [PubMed] [Google Scholar]
- 5.Romao M J, Archer M, Moura I, Moura J J, LeGall J, Engh R, Schneider M, Hof P, Huber R. Science. 1995;270:1170–1176. doi: 10.1126/science.270.5239.1170. [DOI] [PubMed] [Google Scholar]
- 6.Kisker C, Schindelin H, Pacheco A, Wehbi W A, Garrett R M, Rajagopalan K V, Enemark J H, Rees D C. Cell. 1997;91:973–983. doi: 10.1016/s0092-8674(00)80488-2. [DOI] [PubMed] [Google Scholar]
- 7.Rajagopalan K V, Johnson J L. J Biol Chem. 1992;267:10199–10202. [PubMed] [Google Scholar]
- 8.Mendel R R. Planta. 1997;203:399–405. doi: 10.1007/s004250050206. [DOI] [PubMed] [Google Scholar]
- 9.Seo M, Akaba S, Oritani T, Delarue M, Bellini C, Caboche M, Koshiba T. Plant Physiol. 1998;116:687–693. doi: 10.1104/pp.116.2.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson J L, Rajagopalan K V, Wadman S K. Adv Exp Med Biol. 1993;338:373–378. doi: 10.1007/978-1-4615-2960-6_77. [DOI] [PubMed] [Google Scholar]
- 11.Reiss J, Cohen N, Dorche C, Mandel H, Mendel R R, Stallmeyer B, Zabot M T, Dierks T. Nat Genet. 1998;20:51–53. doi: 10.1038/1706. [DOI] [PubMed] [Google Scholar]
- 12.Reiss J, Dorche C, Stallmeyer B, Mendel R R, Cohen N, Zabot M T. Am J Hum Genet. 1999;64:706–711. doi: 10.1086/302296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stallmeyer B, Schwarz G, Schulze J, Nerlich A, Reiss J, Kirsch J, Mendel R R. Proc Natl Acad Sci USA. 1999;96:1333–1338. doi: 10.1073/pnas.96.4.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stallmeyer B, Drugeon G, Reiss J, Haenni A L, Mendel R R. Am J Hum Genet. 1999;64:698–705. doi: 10.1086/302295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stallmeyer B, Nerlich A, Schiemann J, Brinkmann H, Mendel R R. Plant J. 1995;8:751–762. doi: 10.1046/j.1365-313x.1995.08050751.x. [DOI] [PubMed] [Google Scholar]
- 16.Hoff T, Schnorr K M, Meyer C, Caboche M. J Biol Chem. 1995;270:6100–6107. doi: 10.1074/jbc.270.11.6100. [DOI] [PubMed] [Google Scholar]
- 17.Wuebbens M M, Rajagopalan K V. J Biol Chem. 1995;270:1082–1087. doi: 10.1074/jbc.270.3.1082. [DOI] [PubMed] [Google Scholar]
- 18.Wuebbens M M, Rajagopalan K V. J Biol Chem. 1993;268:13493–13498. [PubMed] [Google Scholar]
- 19.Prior P, Schmitt B, Grenningloh G, Pribilla I, Multhaup G, Beyreuther K, Maulet Y, Werner P, Langosch D, Kirsch J, et al. Neuron. 1992;8:1161–1170. doi: 10.1016/0896-6273(92)90136-2. [DOI] [PubMed] [Google Scholar]
- 20.Stewart V, MacGregor C H. J Bacteriol. 1982;151:788–799. doi: 10.1128/jb.151.2.788-799.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomsett A B, Garrett R H. Genetics. 1980;95:649–660. doi: 10.1093/genetics/95.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendel R R, Alikulov Z A, Lvov N P, Müller A J. Mol Gen Genet. 1981;181:395–399. [Google Scholar]
- 23.Falciani F, Terao M, Goldwurm S, Ronchi A, Gatti A, Minoia C, Li Calzi M, Salmona M, Cazzaniga G, Garattini E. Biochem J. 1994;298:69–77. doi: 10.1042/bj2980069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joshi M S, Johnson J L, Rajagopalan K V. J Bacteriol. 1996;178:4310–4312. doi: 10.1128/jb.178.14.4310-4312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leimkühler S, Angermüller S, Schwarz G, Mendel R R, Klipp W. J Bacteriol. 1999;181:5930–5939. doi: 10.1128/jb.181.19.5930-5939.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwarz G, Boxer D H, Mendel R R. J Biol Chem. 1997;272:26811–26814. doi: 10.1074/jbc.272.43.26811. [DOI] [PubMed] [Google Scholar]
- 27.Hasona A, Ray R M, Shanmugam K T. J Bacteriol. 1998;180:1466–1472. doi: 10.1128/jb.180.6.1466-1472.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collins L A, Egan S M, Stewart V. J Bacteriol. 1992;174:3667–3675. doi: 10.1128/jb.174.11.3667-3675.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glaser J H, DeMoss J A. Mol Gen Genet. 1972;116:1–10. doi: 10.1007/BF00334254. [DOI] [PubMed] [Google Scholar]
- 30.Johnson J L, Rajagopalan K V. Proc Natl Acad Sci USA. 1982;79:6856–6860. doi: 10.1073/pnas.79.22.6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson J L, Hainline B E, Rajagopalan K V, Arison B H. J Biol Chem. 1984;259:5414–5422. [PubMed] [Google Scholar]
- 32.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 33.Jones R W, Garland P B. Biochem J. 1977;164:199–211. doi: 10.1042/bj1640199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nason A, Lee K Y, Pan S S, Ketchum P A, Lamberti A, DeVries J. Proc Natl Acad Sci USA. 1971;68:3242–3246. doi: 10.1073/pnas.68.12.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mendel R R. Phytochemistry. 1983;22:817–819. [Google Scholar]
- 36.Wahl R C, Hageman R V, Rajagopalan K V. Arch Biochem Biophys. 1984;230:264–273. doi: 10.1016/0003-9861(84)90107-3. [DOI] [PubMed] [Google Scholar]
- 37.Liu M T, Wuebbens M M, Rajagopalan K V, Schindelin H. J Biol Chem. 2000;275:1814–1822. doi: 10.1074/jbc.275.3.1814. [DOI] [PubMed] [Google Scholar]