Abstract
Axin, APC, and the kinase GSK3β are part of a destruction complex that regulates the stability of the Wnt pathway effector β-catenin. In C. elegans, several Wnt-controlled developmental processes have been described, but an Axin ortholog has not been found in the genome sequence and SGG-1/GSK3β, and the APC-related protein APR-1 have been shown to act in a positive, rather than negative fashion in Wnt signaling. We have shown previously that the EGL-20/Wnt-dependent expression of the homeobox gene mab-5 in the Q neuroblast lineage requires BAR-1/β-catenin and POP-1/Tcf. Here, we have investigated how BAR-1 is regulated by the EGL-20 pathway. First, we have characterized a negative regulator of the EGL-20 pathway, pry-1. We show that pry-1 encodes an RGS and DIX domain-containing protein that is distantly related to Axin/Conductin. Our results demonstrate that despite its sequence divergence, PRY-1 is a functional Axin homolog. We show that PRY-1 interacts with BAR-1, SGG-1, and APR-1 and that overexpression of PRY-1 inhibits mab-5 expression. Furthermore, pry-1 rescues the zebrafish axin1 mutation masterblind, showing that it can functionally interact with vertebrate destruction complex components. Finally, we show that SGG-1, in addition to its positive regulatory role in early embryonic Wnt signaling, may function as a negative regulator of the EGL-20 pathway. We conclude that a highly divergent destruction complex consisting of PRY-1, SGG-1, and APR-1 regulates BAR-1/β-catenin signaling in C. elegans.
Keywords: Axin, GSK3β, Wnt signaling, C. elegans
The Wnt family of secreted glycoproteins controls a wide range of developmental processes, ranging from cell-fate specification to determination of cellular polarity and migration (for review, see Cadigan and Nusse 1997; Bienz and Clevers 2000). A key effector of the pathway that transduces the Wnt signal into the expression of specific Wnt target genes is β-catenin. In the absence of Wnt signaling, β-catenin is targeted for destruction by a complex consisting of the scaffold protein Axin, the kinase GSK3β, and the tumor suppressor gene product APC. GSK3β phosphorylates β-catenin, which in turn becomes a substrate for the ubiquitin ligase β-TrCP/Slimb and is degraded by the proteasome. In the presence of Wnt signaling, the Axin/APC/GSK3β complex is inhibited through the action of Dishevelled and the binding of Axin to the Wnt coreceptor LRP/Arrow (Mao et al. 2001). The unphosphorylated β-catenin is now free to move to the nucleus, in which it interacts with transcription factors of the Tcf/LEF-1 family to activate Wnt target gene expression.
In C. elegans, the expression of the homeobox gene mab-5 in the Q neuroblast lineage is controlled by the Wnt EGL-20 (Harris et al. 1996; Maloof et al. 1999; Whangbo and Kenyon 1999). The Q cells are migratory neuroblasts that are born at similar positions on the left and right sides of the animal. Although both Q cells generate an identical set of descendants, their migration is different. The QL cell and its descendants migrate toward the posterior, whereas the QR cell and its daughter cells move toward the anterior. This difference in migration is induced by the asymmetric expression of mab-5 (Kenyon 1986; Salser and Kenyon 1992). Only the QL cell is sensitive to EGL-20/Wnt and expresses mab-5 (Whangbo and Kenyon 1999). mab-5 in turn directs the migration of the QL descendants (collectively denoted as QL.d) into the posterior. The QR cell, on the other hand, does not respond to EGL-20 and its daughter cells migrate in the default anterior direction. Detailed analysis of mutations that disrupt the expression of mab-5 in the QL lineage has suggested that a canonical Wnt pathway transduces the EGL-20 signal. Thus, mutation of lin-17/Fz (Harris et al. 1996), mig-5/Dsh (Guo 1995), bar-1/β-catenin (Maloof et al. 1999), and RNA mediated interference (RNAi) of pop-1/Tcf function (Herman 2001) all result in loss-of mab-5 expression and anterior migration of the QL.d. Furthermore, it was shown that an interaction between BAR-1 and POP-1 is required for mab-5 expression, suggesting that BAR-1 and POP-1 (like β-catenin and Tcf) form an active bipartite transcription factor (Korswagen et al. 2000).
There are still major gaps in our understanding of this pathway. For example, it is unknown whether BAR-1/β-catenin signaling is regulated by a conserved Axin/APC/GSK3β complex. First, no Axin homologs were identified in the C. elegans genome sequence (Ruvkun and Hobert 1998). Second, the APC-like protein APR-1 is only distantly related to Drosophila and vertebrate APC, and may lack the structural features required for β-catenin binding (Rocheleau et al. 1997; Korswagen et al. 2000). Third, both APR-1 and the GSK3β-like protein SGG-1 have been shown to function as positive, rather than negative regulators of a Wnt pathway in the early embryo (Rocheleau et al. 1997, 1999; Schlesinger et al. 1999). At the four-cell stage, a MOM-2/Wnt signal from the blastomere P2 polarizes its neighbor, EMS, to generate daughter cells with different fates. The anterior daughter cell MS will form muscle cells, whereas the posterior daughter cell E will form the intestine. The main determinant of the difference in fate between MS and E is POP-1/Tcf (Lin et al. 1995, 1998). POP-1 is present at high levels in MS and interacts with UNC-37/Groucho and the histone deacetylase HDA-1 to repress the endoderm-specific transcription factor gene end-1 (Calvo et al. 2001). In response to the MOM-2/Wnt pathway and a parallel mitogen-activated protein kinase (MAPK) pathway consisting of MOM-4/TAK1 and LIT-1/NLK, POP-1 levels are down-regulated in E (Meneghini et al. 1999; Rocheleau et al. 1999). The lower POP-1 level results in derepression of end-1 and the induction of endoderm development. Genetic and biochemical evidence suggests that the down-regulation of POP-1 is mediated by a complex consisting of the β-catenin WRM-1 and LIT-1/NLK. Upon activation by MOM-4/TAK1, this complex phosphorylates POP-1, which may in turn result in a loss-of-DNA binding and nuclear localization of POP-1 (Ishitani et al. 1999; Rocheleau et al. 1999; Shin et al. 1999). Like mutation of mom-2/Wnt, disruption of apr-1 or sgg-1 function by RNAi results in a failure to induce endoderm (Rocheleau et al. 1997, 1999; Schlesinger et al. 1999). The mechanism of this positive requirement of APR-1 and SGG-1 in endoderm induction is unknown.
To gain insight into the regulation of the EGL-20/Wnt pathway, we have studied the putative negative regulator pry-1 (Maloof et al. 1999). Mutation of pry-1 results in ectopic expression of mab-5 in the QR lineage and posterior migration of the QR descendants. This activity is dependent on bar-1, indicating that pry-1 is a negative regulator of BAR-1/β-catenin signaling. Loss of pry-1 function also induces widespread ectopic expression of the Hox genes lin-39 and egl-5, suggesting that pry-1 is a general repressor of Hox gene expression (Maloof et al. 1999). Here we show that pry-1 encodes a divergent, but functional Axin homolog. Furthermore, our results indicate that SGG-1 may have an additional, negative regulatory role in the EGL-20/Wnt pathway. We propose that in C. elegans, an Axin/APC/GSK3β-related complex negatively regulates BAR-1/β-catenin signaling.
Results
pry-1 encodes an Axin-related protein
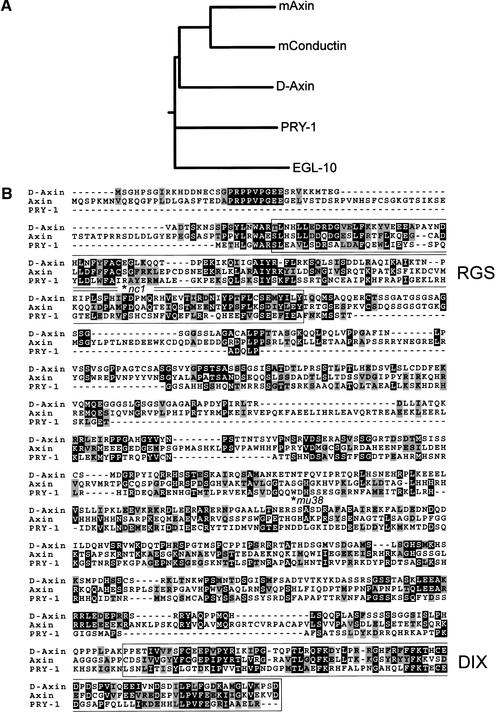
We identified PRY-1 as a protein that specifically interacted with BAR-1 in a yeast two-hybrid screen. A mixed stage C. elegans cDNA library (Walhout et al. 2000) was screened using the Armadillo repeat region of BAR-1 as bait. One of the interacting proteins was encoded by the gene C37A5.9. A full-length cDNA of 1894 bp was isolated, and was shown to include the trans-splice leader SL1, a 6-bp 5′ untranslated region (UTR) and a 106-bp 3′ UTR. C37A5.9 encodes a predicted protein of 587 amino acids. We noted that the extrapolated genetic map position of C37A5.9 is within 1 map unit of the reported map position of pry-1 (Maloof et al. 1999). We sequenced the C37A5.9-coding region in the two existing pry-1 mutant alleles mu38 and nc1 (Maloof et al. 1999; Shioi et al. 2001), and found that both introduce nonsense mutations; nc1 changes CGA/Arg 48 to TGA/Stop and mu38 changes TGG/Trp 268 to TGA/Stop (Fig. 1B). In addition, we found that a transgene containing the C37A5.9 genomic sequence fused to a Green Fluorescent Protein (GFP) reporter gene rescued the pry-1(mu38 and nc1) mutant phenotype (see below). We conclude that pry-1 is the gene C37A5.9.
Figure 1.
PRY-1 is related to Axin/Conductin. (A) Multiple sequence alignment dendrogram of murine mAxin (GenBank accession no. AAC53285), mConductin (AAC26047), Drosophila D-Axin (Q9V407), EGL-10 (P49809), and PRY-1 (AF468834), generated with CLUSTALW by use of default program parameters. (B) Alignment of PRY-1 with D-Axin and mAxin. The RGS and DIX domains are boxed. The positions of the pry-1(nc1) and mu38 mutations are indicated by an asterisk.
The predicted PRY-1 protein shows limited similarity to Axin and the related protein Conductin, with 18%–21% overall amino acid sequence identity to vertebrate and Drosophila Axin/Conductin family members. The similarity of PRY-1 to Axin/Conductin is mainly within two domains, a putative regulator of G-protein signaling (RGS) domain and a DIX domain, a conserved region that is found in both Axin and Dishevelled (Zeng et al. 1997; Fig. 1B). Thus, the PRY-1 RGS domain is 27% identical (48% similar) to the RGS domain of Drosophila D-Axin and the PRY-1 DIX domain is 31% identical (49% similar) to the respective domain of D-Axin (Hamada et al. 1999; Willert et al. 1999). Axin family members also contain defined binding domains for GSK3β and β-catenin (Behrens et al. 1998; Ikeda et al. 1998; Sakanaka et al. 1998; Hedgepeth et al. 1999). On the basis of sequence similarity searches, we could not identify these binding regions in the PRY-1 sequence. Furthermore, PRY-1 does not cluster with Axin/Conductin family members in a multiple sequence alignment that also includes the unrelated RGS domain containing protein EGL-10 (Koelle and Horvitz 1996; Fig. 1A). This shows that PRY-1 is only distantly related to the Axin/Conductin family at the sequence level.
PRY-1 physically interacts with BAR-1/β-catenin, SGG-1/GSK3β, the APC-like protein APR-1, and MIG-5/Dsh
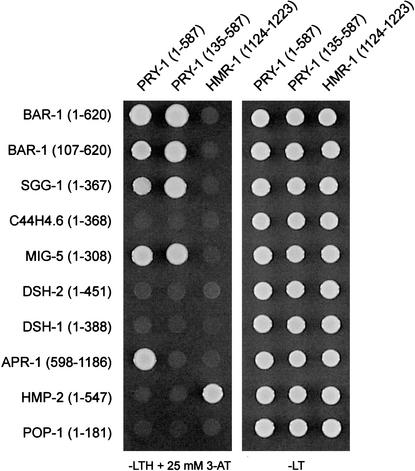
Axin functions as a scaffold protein that assembles the β-catenin destruction complex, and allows efficient phosphorylation of β-catenin by GSK3β. The RGS domain of Axin binds APC (Behrens et al. 1998; Kishida et al. 1998), defined domains of Axin are required for binding GSK3β and β-catenin (Behrens et al. 1998; Ikeda et al. 1998; Sakanaka et al. 1998; Hedgepeth et al. 1999) and the carboxy-terminal DIX domain and more internal sequences are required for Axin homodimerization and binding to Dsh and LRP/Arrow (Hsu et al. 1999; Kishida et al. 1999; Li et al. 1999; Smalley et al. 1999; Mao et al. 2001). Like Axin/Conductin, PRY-1 contains an RGS and DIX domain. Therefore, we investigated whether PRY-1 can physically interact with components of a putative C. elegans β-catenin destruction complex. Using a yeast-two-hybrid assay, we found that (as expected) PRY-1 binds the Armadillo repeat region of BAR-1/β-catenin (Fig. 2). PRY-1 also binds the GSK3β homolog SGG-1, but not a second GSK3β-related protein encoded by C44H4.6 (Ruvkun and Hobert 1998). In addition, we found that PRY-1 interacts with the APC-related protein APR-1. This interaction requires the RGS domain of PRY-1 (deleted in PRY-1[135–587]) and the carboxy-terminal half of APR-1, which contains two repeats that resemble the conserved SAMP repeats found in Drosophila and vertebrate APC (Rocheleau et al. 1997). The SAMP repeats of human APC have been shown to interact directly with conserved residues in the RGS domain of Axin (Behrens et al. 1998; Spink et al. 2000). This suggests that the structural basis of the interaction between PRY-1 and APR-1 is similar. Indeed, modeling of the interaction of the PRY-1 RGS domain with the APR-1 SAMP repeats has shown that despite considerable sequence differences, the interaction is comparable with the human Axin/APC interaction (W. Weiss, pers. comm.). PRY-1 also interacts with the Dishevelled homolog MIG-5, but not with two additional Dishevelled homolog, DSH-1 and DSH-2.
Figure 2.
Yeast two-hybrid assay showing specific interactions between PRY-1 and BAR-1/β-catenin, SGG-1/GSK3β, MIG-5/Dsh, and the APC-like protein APR-1. The intracellular domain of the cadherin HMR-1 was used as a negative control. HMR-1 specifically interacts with the adhesion-specific β-catenin HMP-2 (Korswagen et al. 2000).
These results suggest that PRY-1 assembles a complex of APR-1, SGG-1, and BAR-1. This complex may in turn be regulated by the EGL-20 pathway through the interaction of PRY-1 with MIG-5. We found that PRY-1 does not bind the adhesion-specific β-catenin HMP-2 (Costa et al. 1998; Korswagen et al. 2000; Natarajan et al. 2001; Fig. 2). This is in agreement with a function of PRY-1 in Wnt signaling rather than cellular adhesion.
pry-1 functions downstream of egl-20 and mig-5, but upstream of bar-1, pop-1, and mab-5
The interaction of Axin with APC, GSK3β, and β-catenin allows the efficient phosphorylation and destruction of β-catenin in the absence of Wnt signaling. Consequently, Axin functions as an important negative regulator of the Wnt pathway, as observed in Drosophila, in which D-Axin mutants resemble animals in which Wg is overexpressed (Hamada et al. 1999; Willert et al. 1999). The phenotype of pry-1 suggests that it may have a similar negative regulatory function. In the Q neuroblast lineage, only the Q cell on the left side of the animal (QL) responds to EGL-20/Wnt and activates the expression of the Hox gene mab-5. As a consequence, the QL daughter cells (QL.d) migrate in a posterior direction. The QR cell does not respond to physiological levels of EGL-20, and its daughter cells migrate into the default, anterior direction. When EGL-20 is overexpressed, however, the QR cell also activates mab-5. As a result, its daughter cells migrate toward the posterior (Whangbo and Kenyon 1999). A similar Wnt overactivity phenotype is observed in pry-1 mutants (Maloof et al. 1999). Furthermore, double-mutant analysis has shown that pry-1 functions downstream of egl-20, but upstream of bar-1/β-catenin and mab-5, showing that pry-1 functions as a negative regulator of the EGL-20 pathway (Maloof et al. 1999).
To determine additional epistatic relationships with components of the EGL-20 pathway, we examined the pry-1 phenotype in different mutant backgrounds. We could phenocopy the pry-1 QR.d migration defect by RNAi and assayed the final positions of the QL and QR daughter cells AVM and PVM using a mec-7∷gfp reporter transgene (Table 1). As was reported previously, we observed that in egl-20(n585) mutants, pry-1(RNAi) still induced a QR.d migration defect. We found that the pry-1 phenotype was also unaffected by disruption of mig-5/Dsh, indicating that pry-1 functions downstream of mig-5 as well. The pry-1(RNAi) phenotype was, however, completely suppressed in bar-1 and mab-5 null mutants. We have shown previously that BAR-1 functions together with POP-1/Tcf to activate mab-5 expression in the Q cell lineage (Korswagen et al. 2000). Therefore, we also determined the pry-1(RNAi) phenotype in a pop-1(hu9) mutant background. hu9 was isolated as a suppressor of a constitutively active bar-1 mutant, and contains a mutation (E47K) in the BAR-1-binding domain of POP-1 that may disrupt the interaction between the two proteins. hu9 is so far the only viable pop-1 allele and shows a low penetrance QL.d migration defect (Table 1; H. Korswagen, unpubl.). We found that pop-1(hu9) completely suppressed the pry-1(RNAi) QR.d migration defect. Taken together, these results suggest that PRY-1 is a negative regulator of the EGL-20-dependent expression of mab-5 in the Q neuroblast lineage and that PRY-1 functions downstream of EGL-20 and MIG-5, but upstream of BAR-1 and POP-1. This places PRY-1 at a position in the EGL-20/Wnt pathway that is in agreement with a function of PRY-1 as an Axin-like protein.
Table 1.
Genetic interactions between pry-l and components of theegl-20/Wnt pathway
| Genotype
|
%
QL descendants in posterior
|
% QR descendants in
posterior
|
|---|---|---|
| Wild type | 100 | 0 |
| pry-l(RNAi) | 100 | 48 |
| egl-20(n585) | 0 | 0 |
| egl-20(n585); pry-l (RNAi) | 82 | 34 |
| bar-1(ga80) | 1 | 0 |
| bar-1(ga80); pry-l(RNAi) | 0 | 0 |
| pop-1(hu9) | 91 | 0 |
| pop-1(hu9); pry-l(RNAi) | 100 | 2 |
| mab-5(el239) | 0 | 0 |
| mab-5(el239); pry-l (RNAi) | 6 | 0 |
| mig-5(RNAi) | 21 | 0 |
| pry-l(mu38) | 100 | 55 |
| pry-l(mu38); mig-5(RNAi) | 100 | 46 |
The RNA interference experiment was performed by feeding animals on E. coli expressing double-stranded RNA of pry-l, mig-5, or the empty vector L4440 as described (Kamath et al. 2001; Timmons et al. 2001). In each case, n = 100. The final positions of QL.paa (PVM) and QR.paa (AVM) were scored in L4 larvae using a mec-7::gfp reporter transgene. Final position was scored as anterior or posterior to the vulva.
Overexpression of PRY-1 and SGG-1 induces a Wnt loss-of-function phenotype
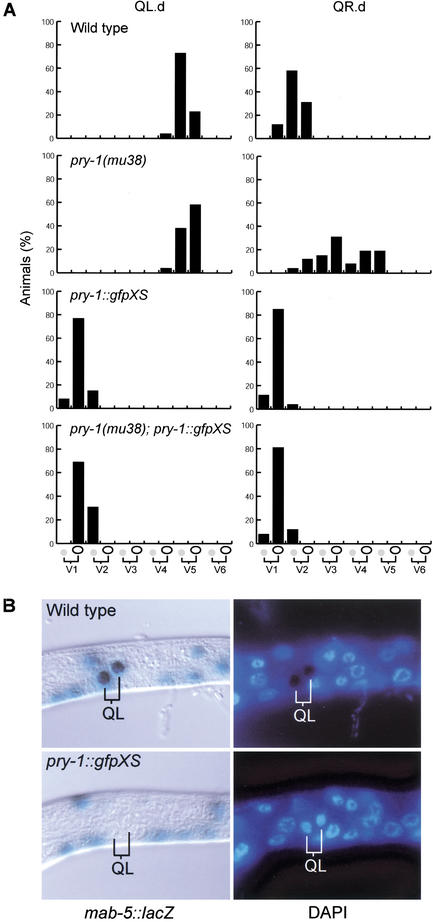
Overexpression of Axin in the dorsal blastomeres of the early Xenopus embryo inhibits signaling through the Wnt pathway and induces a strong ventralization phenotype (Zeng et al. 1997). Axin overexpression may exert this effect by allowing more efficient GSK3β-mediated phosphorylation of β-catenin (Behrens et al. 1998; Ikeda et al. 1998; Kishida et al. 1998; Sakanaka et al. 1998). To investigate whether overexpression of PRY-1 induces a Wnt loss-of-function phenotype as well, we generated multicopy transgenes containing the pry-1 genomic sequence. Overexpression of PRY-1 or a PRY-1∷GFP fusion protein resulted in a highly penetrant QL.d migration defect (Fig. 3A; data not shown). To accurately determine the effect of PRY-1 overexpression on QL.d migration, we scored the final positions of the QL.pa daughter cells, which can be easily recognized, relative to the positions of the six seam cells, V1-V6 (Harris et al. 1996). In wild-type and pry-1(mu38) animals, the QL.pa daughter cells were found at their normal position (near V5.a), but in animals that overexpress PRY-1, the QL.pa daughter cells localized far into the anterior. This phenotype is similar to egl-20/Wnt and mab-5 null mutants (Harris et al. 1996) and suggests that PRY-1 overexpression inhibits the EGL-20-dependent expression of mab-5. To verify this, we used a mab-5∷lacZ reporter gene that has been shown to closely mimic the expression of the endogenous mab-5 gene (Salser and Kenyon 1992). We found that 40/40 wild-type animals expressed the mab-5∷lacZ reporter in the QL daughter cells (Fig. 3B). In animals that overexpress PRY-1, no mab-5∷lacZ expression was observed in the QL.d (n = 40), indicating that PRY-1 overexpression inhibits mab-5 expression. We also observed that the QL and QR daughter cells migrated further into the anterior than the QR.d in wild-type animals (Fig. 3A), a phenotype that is also observed in lin-17/Fz and bar-1 mutants (Whangbo et al. 2000). The localization of other migratory neurons, such as the HSN and CAN neurons, was not affected (n = 26) by PRY-1 overexpression.
Figure 3.
Overexpression of PRY-1 inhibits mab-5 expression. (A) Overexpression of a PRY-1∷GFP fusion protein caused anterior migration of the QL.d. Black bars indicate the proportion of cells at each position relative to the positions of the six seam cells V1-V6 (in each case, n = 26). (B) Overexpression (XS) of the PRY-1∷GFP fusion protein inhibited expression of a mab-5∷lacZ reporter gene (Salser and Kenyon 1992) in the QL.d. Whole-mount larvae, synchronized at 4–5 h after hatching, were fixed and stained for β-galactosidase expression and the DNA stain DAPI.
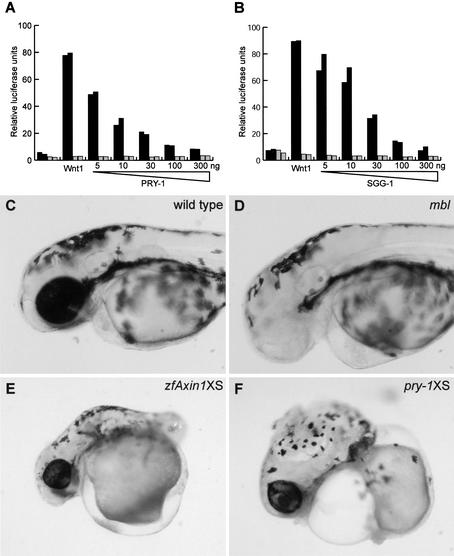
Does PRY-1 function as a negative regulator of mab-5 expression by enabling the phosphorylation of BAR-1 by the GSK3β homolog SGG-1? The amino terminus of BAR-1 contains several consensus GSK3β phosphorylation sites (Eisenmann et al. 1998; Korswagen et al. 2000). Furthermore, overexpression of a truncated BAR-1 protein, in which these phosphorylation sites have been removed, results in a Wnt pathway activation phenotype that is similar to the pry-1 mutant phenotype (Gleason et al. 2002; data not shown). This suggests that BAR-1 is a substrate of GSK3β-mediated down-regulation. However, instead of acting as a negative regulator of Wnt signaling, SGG-1 has been shown to act positively in the MOM-2/Wnt pathway (Schlesinger et al. 1999). Because SGG-1 interacts physically with PRY-1, and because SGG-1 inhibits the Wnt-1-induced transcriptional activation of a Tcf reporter gene in mammalian cells (Fig. 4B), we investigated whether SGG-1 has an additional function as a negative regulator of the EGL-20/Wnt pathway. We could not directly investigate the role of sgg-1 in Q neuroblast migration by disrupting sgg-1 function, as sgg-1(RNAi) is embryonic lethal (Schlesinger et al. 1999). Therefore, we assayed the effect of SGG-1 overexpression. As is shown in Table 2, overexpression of SGG-1 (like overexpression of PRY-1) induced anterior migration of the QL daughter cells. Taken together, these results suggest that both PRY-1 and SGG-1 are negative regulators of the EGL-20/Wnt pathway and that an Axin/GSK3β/APC-related complex regulates the stability of BAR-1.
Figure 4.
PRY-1 is a functional Axin homolog. (A) PRY-1 inhibits the Wnt-1-dependent activation of a TCF reporter gene in 293-T cells. (Black bars) Activation of the Tcf reporter TOP; (gray bars) activation of the negative control FOP. (B) SGG-1 also inhibits the Wnt-1-dependent activation of the Tcf reporter. Expression of PRY-1 resulted in a phenotypic rescue of the optic and anterior brain structure defects of (D) zebrafish mbl/axin1 mutants that was indistinguishable from (C) wild-type embryos (see Table 3). Overexpression (XS) in wild-type zebrafish embryos of (E) Axin1 or (F) PRY-1 induced identical phenotypes, such as deletion of the trunk posterior to the otic vesicle. Overexpression of PRY-1 and Axin1 was carried out by microinjection of 25 or 150 pg of the respective mRNAs into one-cell stage zebrafish embryos.
Table 2.
Overexpression of SGG-1 induces anterior migration of the QL descendants
| Genotype
|
Heat shock
|
% QL
descendants in anterior
|
|---|---|---|
| Wild type | no | 0.7 ± 0.5 |
| yes | 1 ± 0.8 | |
| hs::sgg-1(huIs24) | no | 0.5 ± 0.5 |
| yes | 70 ± 11 |
Synchronized L1 larvae were heat shocked for 2 h at 33°C. The final positions of QL.paa (PVM) and QR.paa (AVM) were scored in L4 larvae using a mec-7::gfp reporter transgene. Final position was scored as anterior or posterior to the vulva. In each, n = 100.
PRY-1 is a functional Axin homolog
As discussed above, PRY-1 is only distantly related to Axin/Conductin. Our results indicate, however, that despite this sequence divergence, PRY-1 has an Axin-like function in C. elegans. We investigated whether this functional homology extends to the vertebrate system. We first tested whether overexpression of PRY-1 inhibits the Wnt-1-induced activation of a Tcf-reporter gene in 293T cells (van de Wetering et al. 1997; Korinek et al. 1998; Sakanaka et al. 1998). As is shown in Figure 4A, coexpression of Wnt-1 with PRY-1 efficiently inhibited Tcf-reporter gene activation in a dose-dependent manner, showing that overexpression of PRY-1, like Axin/Conductin, inhibits Wnt signaling in vertebrate cells. We next investigated whether PRY-1 can complement the zebrafish axin1 mutation masterblind (mbl) (Heisenberg et al. 2001; van de Water et al. 2001). The mbl mutation is characterized by the absence or reduction in size of the telencephalon, the optic vesicles, and the olfactory placodes (Fig. 4D). These defects are fully rescued by Axin1 overexpression. We tested whether overexpression of PRY-1 has a similar activity. One-cell stage progeny of an incross between heterozygous mbl parents was injected with pry-1 mRNA and the resulting phenotypes were scored (Table 3). Whereas in the noninjected embryos, all genotypically mbl homozygotes showed the mutant phenotype, two-thirds (4/6) of the pry-1-injected mbl embryos were fully rescued. We also noted that about three-quarters (15/21) of the sibling embryos that were injected with pry-1 mRNA showed an overexpression phenotype (Table 3). To confirm this observation, we injected pry-1 or zebrafish axin1 mRNA in wild-type embryos. Overexpression of PRY-1 resulted in defects in the generation of posterior trunk structures, a phenotype that was also observed in embryos that overexpress zfAxin1 (Fig. 4E,F). Truncation of posterior structures ranged from deletion of all structures posterior to the otic vesicles to deletion of the tail only. These defects are likely caused by interference with the role of Wnt signaling in the establishment of anterior-posterior polarity and posterior fates (van de Water et al. 2001). Taken together, these results show that PRY-1 can interact with the vertebrate destruction complex components, establishing the functional homology of PRY-1 with vertebrate Axin.
Table 3.
Expression of pry-l rescues the eye phenotype of zebrafish masterblind/axin1
| Phenotypeb
|
Noninjectedan
= 17
|
25 pg pry-l
mRNAan = 27
|
||
|---|---|---|---|---|
| No. of
sib
|
No. of Mut
|
No. of Sib
|
No. of Mut
|
|
| Wt | 10 (59%) | 6 (22%) | 4 (15%) | |
| Se | 1 (6%) | 2 (7%) | ||
| Ne | 6 (35%) | |||
| Oe | 15 (56%) | |||
A random sample of eggs, derived from a single clutch of a heterozygous mbl incross, was injected with pry-l mRNA. Another sample was used as a noninjected control.
Embryos were genotyped as homozygous mbl (Mut) or sibling (Sib) as described (van de Water et al. 2001).
The observed phenotypes were as follows (Wt) wild type; (Se) small eyes; (Ne) eyeless; (Oe) overexpression phenotype. No rescue of the mbl eyeless phenotype and no overexpression phenotypes were observed after injection of 275 pg of a frame-shifted pry-l mRNA (n = 33).
PRY-1 is widely expressed throughout development
To determine the expression pattern of pry-1, we constructed a translational fusion of the pry-1 genomic sequence and a GFP reporter gene. The expression pattern of this construct was similar in independent transgenic lines, but the subcellular localization of the PRY-1∷GFP fusion protein differed, ranging from localization at the plasma membrane and in cytoplasmic dots to diffuse cytoplasmic and nuclear staining. This difference in subcellular localization may be a consequence of variations in expression levels of the fusion protein in different transgenic lines. For ease of cell identification, a transgenic line showing diffuse cytoplasmic and nuclear staining was selected. This transgene fully rescued the lethality, the multivulva phenotype, and the QR.d migration defect of pry-1(mu38 and nc1). This suggests that the PRY-1∷GFP fusion protein is functional and is correctly expressed in cells in which PRY-1 is essential.
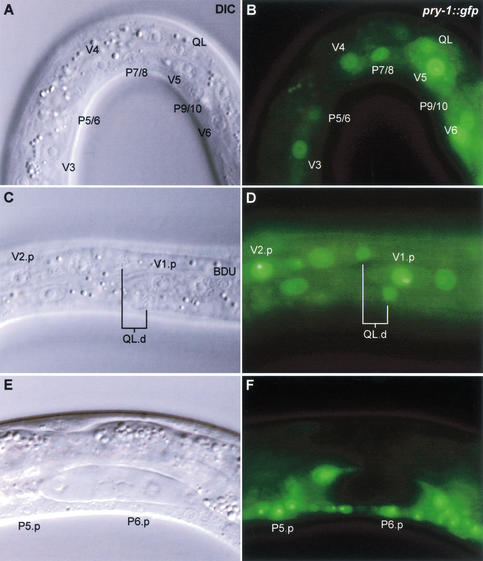
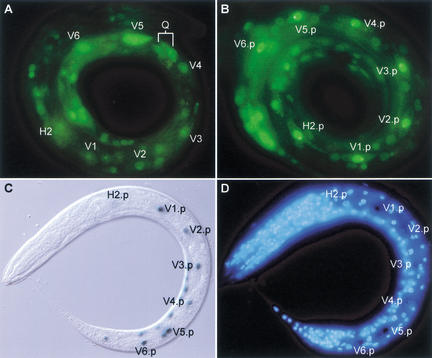
The pry-1 reporter gene is widely expressed throughout development. Expression starts halfway through embryogenesis and is mainly localized to the ventral and lateral hypodermal cells. At the early L1 stage, pry-1 is expressed at high levels in the lateral hypodermal cells (or seam cells) V5 and V6 and in the Q neuroblasts QL and QR (Figs. 5A,B, 6A). pry-1 is also expressed in the ventral hypodermal (P) cells P7/8 to P11/12, body wall muscle cells, and neurons in the head, the tail, and the ventral nerve cord. We did not observe differences in pry-1 expression levels between QL and QR (data not shown), but this may be a result of PRY-1∷GFP overexpression. At the end of the L1 stage, pry-1 is expressed at high levels in all seam cells (Fig. 6B; see below). Expression was also observed in the QL and QR daughter cells (Fig. 5C,D). At later larval stages, pry-1 is expressed at high levels throughout the animal, including hypodermal cells, body wall muscle cells, and many neurons in the ventral nerve cord and head and tail ganglia. In addition, pry-1 is expressed in the vulva precursor (Pn.p) cells (Fig. 5E,F) and in the developing vulva and male tail (data not shown).
Figure 5.
Expression of a pry-1∷gfp reporter construct in the seam cells, the Q cell lineage, and the vulva precursor cells. (A,B) Early L1 larva, 1–2 h after hatching. pry-1 is expressed at high levels in the seam cells V5 and V6, in the Q neuroblast QL, and in the lateral hypodermal cells P7/8 to P11/12. Faint expression was observed in more anterior seam cells. (C,D) Late L1 larva, ∼10 h after hatching. pry-1 is expressed in the QL.pa daughter cells. Note that overexpression of PRY-1∷GFP fusion protein causes anterior localization (near V1.p) of the QL.d. (E,F) Expression of pry-1 in the vulva precursor (P4–8.p) cells. P5.p and P6.p are shown.
Figure 6.
Up-regulation of pry-1 expression levels in the Vn.p cells correlates with ectopic mab-5 expression in pry-1 mutants. (A) Early L1 stage, 1–2 h after hatching. pry-1 is expressed at high levels in V5 and V6, but at low levels in more anterior seam cells. (B) Late L1 stage, ∼10 h after hatching. pry-1 is now expressed at high levels in all seam cells. (C) In pry-1(mu38), a mab-5∷lacZ reporter gene is expressed in all Vn.p cells. (D) Staining with the DNA dye DAPI to visualize nuclei.
Discussion
Members of the Wnt family of secreted glycoproteins play an essential role in patterning developing tissues in species ranging from the simple metazoan Hydra to vertebrates. In C. elegans, one of the processes controlled by Wnt signaling is the expression of the Hox gene mab-5 in the Q neuroblast lineage. Detailed analysis of mutations that disrupt the expression of mab-5 in the Q cell lineage has suggested that a canonical Wnt pathway transduces the EGL-20/Wnt signal. In this study, we have investigated how this pathway is regulated. We have characterized a mutation, pry-1, which induces widespread ectopic expression of Wnt target genes (Maloof et al. 1999). We show that pry-1 encodes a protein that is distantly related to Axin/Conductin. Our results show that despite this sequence divergence, PRY-1 is a functional Axin homolog and that a conserved β-catenin destruction complex regulates the EGL-20/Wnt pathway.
A PRY-1/SGG-1/APR-1 complex negatively regulates BAR-1
The key effector of the Wnt pathway, β-catenin, is regulated by a complex consisting of Axin, APC, and GSK3β. In the absence of Wnt signaling, this complex enables GSK3β to phosphorylate β-catenin, which results in its destruction by the proteasome (for review, see Cadigan and Nusse 1997; Bienz and Clevers 2000). We present evidence that a similar complex regulates BAR-1/β-catenin stability in C. elegans. First, we show that the Axin-related protein PRY-1 is a negative regulator of the EGL-20/Wnt pathway. Loss of pry-1 function results in a bar-1 (Maloof et al. 1999) and pop-1-dependent activation of mab-5 expression. Furthermore, we found that overexpression of PRY-1 strongly inhibits mab-5 expression.
Second, we show that PRY-1 physically interacts with BAR-1, SGG-1/ GSK3β, and the APC-like protein APR-1. The interaction of PRY-1 with both SGG-1 and BAR-1 may allow the phosphorylation and down-regulation of BAR-1. The amino terminus of BAR-1 contains several conserved GSK3β phosphorylation sites (Eisenmann et al. 1998; Korswagen et al. 2000). Furthermore, overexpression of an amino-terminally truncated BAR-1 protein that lacks these phosphorylation sites results in a Wnt activation phenotype that is similar to the pry-1 phenotype (Gleason et al. 2002; data not shown). This suggests that BAR-1 is a substrate of SGG-1-mediated phosphorylation. We found that overexpression of SGG-1, like overexpression of PRY-1, induces a QL.d migration defect and inhibits the Wnt-1-induced activation of a Tcf reporter gene in mammalian cells. Although we cannot exclude that overexpression of SGG-1 may titrate out an essential Wnt pathway component, it is likely that overexpression of SGG-1 induces a QL.d migration defect by enhancing the phosphorylation and destruction of BAR-1. Taken together, our results indicate that in addition to a positive requirement in the MOM-2/Wnt pathway, SGG-1 may also function as a negative regulator of the EGL-20 pathway. The accompanying work by Gleason and colleagues (Gleason et al. 2002) suggests that APR-1 may have an analogous negative regulatory role. In the vulva precursor cells, a pathway similar to the Wnt pathway in the Q neuroblasts is required for the expression of the Hox gene lin-39 (Eisenmann et al. 1998). In pry-1 mutants, the expression of lin-39 is localized far outside of its normal expression domain (Maloof et al. 1999), and too many vulva precursor cells adopt the induced vulval fate, resulting in the generation of multiple vulva-like structures. apr-1(RNAi) strongly enhances the multivulva phenotype of pry-1(mu38), indicating that it functions together with PRY-1 as a negative regulator of a canonical Wnt pathway.
Third, the interaction of PRY-1 with MIG-5/Dsh indicates that the EGL-20/Wnt pathway negatively regulates the destruction complex through a Dishevelled, allowing the stabilization of BAR-1 and activation of target gene expression. The specific interaction of PRY-1 with MIG-5, but not DSH-1 or DSH-2, is in agreement with the observation that only MIG-5 is required for mab-5 expression in the QL lineage (Guo 1995; data not shown).
Finally, overexpression of pry-1 fully rescues the zebrafish axin1 mutation masterblind, showing that despite major sequence differences, PRY-1 and Axin1 are functionally similar. In addition, this suggests that PRY-1 can interact with the vertebrate components of the β-catenin destruction complex. We propose that a complex consisting of PRY-1, SGG-1, and APR-1 negatively regulates signaling through the β-catenin BAR-1.
pry-1 is a general repressor of Wnt target gene expression
In pry-1 mutants, the Hox genes lin-39, mab-5, and egl-5 are expressed far outside of their normal expression domains, resulting in defects in QR daughter cell migration, vulval fate specification, and the generation of sensory structures by the seam cell lineage (Maloof et al. 1999). The pry-1 phenotype is fully suppressed by mutations in bar-1/β-catenin and pop-1/Tcf. This indicates that a canonical Wnt pathway controls the expression of these Hox genes, and that PRY-1 functions as a general repressor of this pathway. Our observation that pry-1 is widely expressed throughout development is in agreement with such a general repressor function. Thus, pry-1 is expressed in the vulva precursor cells, in which it may restrict lin-39 expression (Gleason et al. 2002). We found that pry-1 is also expressed in the different stages of the Q cell lineage. Here, PRY-1 functions as a repressor of mab-5 expression in the QR lineage. A possible mechanism for the asymmetric expression of mab-5 between the QL and QR lineages would be a difference in pry-1 expression levels, with high PRY-1 levels in QR inhibiting mab-5. We did not observe such a difference in expression levels between the QL and QR lineages. This suggests that either the difference in sensitivity to EGL-20/Wnt and the ability to activate mab-5 between the QL and QR lineages is not mediated by differences in pry-1 expression levels or that overexpression of the PRY-1∷GFP fusion protein may have masked these differences. We favor the latter possibility, as the pry-1∷gfp transgene used in this study also inhibited mab-5 expression in the QL daughter cells.
We did observe a striking difference in pry-1 expression levels in the seam cell lineage. During the L1 stage, the seam cells divide in a stem cell-like fashion, generating an anterior daughter cell that fuses with the hypodermal syncytium hyp7, and a posterior daughter cell (Vn.p) that assumes the seam cell fate again. Contacts made between the Vn.p cells after cell division were found to be essential for correct fate specification within the seam cell lineage (Waring and Kenyon 1990; Austin and Kenyon 1994). Thus, the Hox gene mab-5 is only expressed in the posterior Vn.p cell V6.p. In more anterior Vn.p cells, the EGL-20/Wnt dependent expression of mab-5 is repressed, presumably via a mechanism that involves cell–cell contact between the Vn.p cells (Hunter et al. 1999). We noted that during the early L1 stage, pry-1 is mainly expressed in V5 and V6 (Fig. 6A). At the end of L1 (>8 h after hatching) however, we observed a striking difference, pry-1 is now expressed at high levels in all Vn.p cells (Fig. 6B). This up-regulation of pry-1 expression correlates with the establishment of cell–cell contacts between the Vn.p cells and suggests that pry-1 is required for the contact-induced repression of mab-5. Like disruption of the seam cell contacts, mutation of pry-1 results in ectopic mab-5 expression in the anterior Vn.p cells (Fig. 6C,D; Maloof et al. 1999). We speculate that intercellular signaling between the Vn.p cells may up-regulate pry-1 expression levels. High levels of PRY-1 may, in turn, actively inhibit the EGL-20/Wnt-dependent expression of mab-5 in the seam cells. The isolation and characterization of additional genes that, like pry-1, are required for the repression of mab-5, and other Wnt target genes will further elucidate how the canonical Wnt pathway is regulated in different cellular contexts.
Materials and methods
Nematode strains and culturing
General methods for culture, manipulation, and genetics of C. elegans were as described (Lewis and Fleming 1995). Unless indicated, strains were cultured at 20°C. Strains used in this study were Bristol N2 and LGI, pry-1(mu38) and pry-1(nc1) (Shioi et al. 2001); LGII, muIs32 [mec-7∷gfp]; LGIII, mab-5(e1239); LGIV, dpy-20(e1282), egl-20(n585); LGX, bar-1(ga80); and unassigned, muIs2[mab-∷:lacZ], huIs24[hs∷sgg-1; gpa-15∷gfp; dpy-20(+)] and huIs23[pry-1∷gfp; dpy-20(+)].
Yeast two-hybrid studies
C37A5.9 (pry-1) was isolated as a BAR-1 interacting protein in a yeast-two-hybrid screen. One of the isolated cDNAs was full length and contained a poly(A)-tract, as well as part of the trans-splice leader SL1. Full-length pry-1 cDNA (GenBank accession no. AF468834) or a fragment corresponding to amino acids 135–587 (lacking the RGS domain), was cloned into the Gal4 activation domain containing vector pPC86. Full-length cDNAs of sgg-1 (AF159950), C44H4.6 (Z79598), mig-5 (Z66500), dsh-1 (C34F11.9a, U46753), and dsh-2 (C27A6.2, U58760) were cloned by PCR on total C. elegans cDNA, and were checked by sequencing. Fragments corresponding to the regions indicated in Figure 2 were cloned into the Gal4 DNA-binding domain containing vector pPC97. Yeast two-hybrid assays were as described (Walhout et al. 2000).
RNA-mediated interference
A fragment of the pry-1 cDNA (position 407–1894) and full-length mig-5 cDNA were cloned into the RNAi feeding vector L4440. The constructs were transformed into E. coli HT115 and RNAi phenotypes were assayed as described (Kamath et al. 2001).
Transgenic animals
A genomic DNA fragment containing the complete C37A5.9/pry-1-coding sequence and 3.6 kb of upstream sequence was cloned from cosmids ZK849 and C37A5. This fragment was inserted in-frame into the gfp reporter gene containing vector pPD95.81 (A. Fire, pers. comm.). The pry-1∷gfp construct was injected at 100 ng/μL, with 100 ng/μL of the wild-type dpy-20 containing plasmid pMH86 into dpy-20(e1282). The extrachromosomal transgene was integrated using 40 Gy of γ-radiation from a 137Cs source. To generate a heat-shock inducible sgg-1 transgene, the full-length sgg-1 cDNA was cloned into the hsp16.2 vector pPD49.78. The hs∷sgg-1 construct was injected at 50 ng/μL together with 50 ng/μL gpa-15∷gfp and 50 ng/μL pMH86 into a dpy-20(e1282); muIs32[mec-7::gfp] strain. Transgenic L1 larvae were heat shocked for 2 h at 33°C.
Transfection experiments
Full-length cDNA of pry-1 and sgg-1 was cloned into pCDNA3.0 (Invitrogen). The 293-T cells were transfected using FuGENE6 (Roche) with 50 ng of a Wnt-1 expression plasmid, 100 ng pTATATOP or the negative control pTATAFOP, 5–300 ng of pCDNA3-pry-1 or pCDNA3-sgg-1 and 2 ng of the internal transfection control pTKRenilla. DNA concentrations were normalized to 1 μg using pCS2-lacZ (van de Wetering et al. 1997). Luciferase assays were as recommended by the manufacturer (Promega). Luciferase measurements were normalized for transfection efficiency using the renilla control.
Rescue of the zebrafish mutation masterblind
Full-length pry-1 cDNA was cloned into the T7TS vector and capped synthetic mRNA was prepared using a T7 mMessage Machine kit from Ambion. A total of 25 pg of pry-1 mRNA or 150 pg of axin1 mRNA was injected into one-cell stage embryos as described (van de Water et al. 2001). As a control, 275 pg of a frame-shifted pry-1 mRNA was injected. Embryos were either wild type, or derived from an incross between mbl carriers.
Acknowledgments
We thank David Eisenmann and Michael Herman for critically reading the manuscript, Shin Takagi for the pry-1 allele nc1, Andy Fire for heat-shock promoter and GFP vectors, and Marc van de Wetering for advice on transfection experiments. The Caenorhabditis elegans Genetics Center provided some of the strains used in this study. This work was supported by the Royal Netherlands Academy of Arts and Sciences (H.K.) and Pioneer and Program grants of NWO Medische Wetenschappen to H.C.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL rkors@niob.knaw.nl; FAX 31-30-251-6464.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.981802.
References
- Austin J, Kenyon C. Cell contact regulates neuroblast formation in the Caenorhabditis eleganslateral epidermis. Development. 1994;120:313–323. doi: 10.1242/dev.120.2.313. [DOI] [PubMed] [Google Scholar]
- Behrens J, Jerchow BA, Wurtele M, Grimm J, Asbrand C, Wirtz R, Kuhl M, Wedlich D, Birchmeier W. Functional interaction of an Axin homolog, conductin, with β-catenin, APC, and GSK3β. Science. 1998;280:596–599. doi: 10.1126/science.280.5363.596. [DOI] [PubMed] [Google Scholar]
- Bienz M, Clevers H. Linking colorectal cancer to Wnt signaling. Cell. 2000;103:311–320. doi: 10.1016/s0092-8674(00)00122-7. [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Nusse R. Wnt signaling: A common theme in animal development. Genes & Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- Calvo D, Victor M, Gay F, Sui G, Luke MP, Dufourcq P, Wen G, Maduro M, Rothman J, Shi Y. A POP-1 repressor complex restricts inappropriate cell type-specific gene transcription during Caenorhabditis elegansembryogenesis. EMBO J. 2001;20:7197–7208. doi: 10.1093/emboj/20.24.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Raich W, Agbunag C, Leung B, Hardin J, Priess JR. A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegansembryo. J Cell Biol. 1998;141:297–308. doi: 10.1083/jcb.141.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann DM, Maloof JN, Simske JS, Kenyon C, Kim SK. The β-catenin homolog BAR-1 and LET-60 Ras coordinately regulate the Hox gene lin-39 during Caenorhabditis elegansvulval development. Development. 1998;125:3667–3680. doi: 10.1242/dev.125.18.3667. [DOI] [PubMed] [Google Scholar]
- Gleason JE, Korswagen HC, Eisenmann DE. Activation of Wnt signaling bypasses the requirement for RTK/Ras signaling during C. elegansvulval induction. Genes & Dev. 2002;16:1281–1290. doi: 10.1101/gad.981602. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C. “mig-5, a gene that controls cell fate determination and cell migration in C. elegans, is a member of the Dsh family.” PhD thesis. Baltimore, MD: Johns Hopkins University; 1995. [Google Scholar]
- Hamada F, Tomoyasu Y, Takatsu Y, Nakamura M, Nagai S, Suzuki A, Fujita F, Shibuya H, Toyoshima K, Ueno N, et al. Negative regulation of Wingless signaling by D-Axin, a Drosophilahomolog of Axin. Science. 1999;283:1739–1742. doi: 10.1126/science.283.5408.1739. [DOI] [PubMed] [Google Scholar]
- Harris J, Honigberg L, Robinson N, Kenyon C. Neuronal cell migration in C. elegans: Regulation of Hox gene expression and cell position. Development. 1996;122:3117–3131. doi: 10.1242/dev.122.10.3117. [DOI] [PubMed] [Google Scholar]
- Hedgepeth CM, Deardorff MA, Rankin K, Klein PS. Regulation of glycogen synthase kinase 3β and downstream Wnt signaling by Axin. Mol Cell Biol. 1999;19:7147–7157. doi: 10.1128/mcb.19.10.7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisenberg CP, Houart C, Take-Uchi M, Rauch GJ, Young N, Coutinho P, Masai I, Caneparo L, Concha ML, Geisler R, et al. A mutation in the Gsk3-binding domain of zebrafish Masterblind/Axin1 leads to a fate transformation of telencephalon and eyes to diencephalon. Genes & Dev. 2001;15:1427–1434. doi: 10.1101/gad.194301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman M. C. elegansPOP-1/TCF functions in a canonical Wnt pathway that controls cell migration and in a non-canonical Wnt pathway that controls cell polarity. Development. 2001;128:581–590. doi: 10.1242/dev.128.4.581. [DOI] [PubMed] [Google Scholar]
- Hsu W, Zeng L, Costantini F. Identification of a domain of Axin that binds to the serine/threonine protein phosphatase 2A and a self-binding domain. J Biol Chem. 1999;274:3439–3445. doi: 10.1074/jbc.274.6.3439. [DOI] [PubMed] [Google Scholar]
- Hunter CP, Harris JM, Maloof JN, Kenyon C. Hox gene expression in a single Caenorhabditis eleganscell is regulated by a caudal homolog and intercellular signals that inhibit Wnt signaling. Development. 1999;126:805–814. doi: 10.1242/dev.126.4.805. [DOI] [PubMed] [Google Scholar]
- Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3β and β-catenin and promotes GSK-3β-dependent phosphorylation of β-catenin. EMBO J. 1998;17:1371–1384. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani T, Ninomiya-Tsuji J, Nagai S, Nishita M, Meneghini M, Barker N, Waterman M, Bowerman B, Clevers H, Shibuya H, et al. The TAK1-NLK-MAPK-related pathway antagonizes signalling between β-catenin and transcription factor TCF. Nature. 1999;399:798–802. doi: 10.1038/21674. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2:R2.1–R2.10. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. A gene involved in the development of the posterior body region of C. elegans. Cell. 1986;46:477–487. doi: 10.1016/0092-8674(86)90668-9. [DOI] [PubMed] [Google Scholar]
- Kishida S, Yamamoto H, Ikeda S, Kishida M, Sakamoto I, Koyama S, Kikuchi A. Axin, a negative regulator of the Wnt signaling pathway, directly interacts with adenomatous polyposis coli and regulates the stabilization of β-catenin. J Biol Chem. 1998;273:10823–10826. doi: 10.1074/jbc.273.18.10823. [DOI] [PubMed] [Google Scholar]
- Kishida S, Yamamoto H, Hino S, Ikeda S, Kishida M, Kikuchi A. DIX domains of Dvl and Axin are necessary for protein interactions and their ability to regulate β-catenin stability. Mol Cell Biol. 1999;19:4414–4422. doi: 10.1128/mcb.19.6.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelle MR, Horvitz HR. EGL-10 regulates G protein signaling in the C. elegansnervous system and shares a conserved domain with many mammalian proteins. Cell. 1996;84:115–125. doi: 10.1016/s0092-8674(00)80998-8. [DOI] [PubMed] [Google Scholar]
- Korinek V, Barker N, Willert K, Molenaar M, Roose J, Wagenaar G, Markman M, Lamers W, Destree O, Clevers H. Two members of the Tcf family implicated in Wnt/β-catenin signaling during embryogenesis in the mouse. Mol Cell Biol. 1998;18:1248–1256. doi: 10.1128/mcb.18.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korswagen HC, Herman MA, Clevers HC. Distinct β-catenins mediate adhesion and signalling functions in C. elegans. Nature. 2000;406:527–532. doi: 10.1038/35020099. [DOI] [PubMed] [Google Scholar]
- Lewis JA, Fleming JT. Basic culture methods. Methods Cell Biol. 1995;48:3–29. [PubMed] [Google Scholar]
- Li L, Yuan H, Weaver CD, Mao J, Farr GH, III, Sussman DJ, Jonkers J, Kimelman D, Wu D. Axin and Frat1 interact with Dvl and GSK, bridging Dvl to GSK in Wnt-mediated regulation of LEF-1. EMBO J. 1999;18:4233–4240. doi: 10.1093/emboj/18.15.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Thompson S, Priess JR. pop-1 encodes an HMG box protein required for the specification of a mesoderm precursor in early C. elegansembryos. Cell. 1995;83:599–609. doi: 10.1016/0092-8674(95)90100-0. [DOI] [PubMed] [Google Scholar]
- Lin R, Hill RJ, Priess JR. POP-1 and anterior-posterior fate decisions in C. elegansembryos. Cell. 1998;92:229–239. doi: 10.1016/s0092-8674(00)80917-4. [DOI] [PubMed] [Google Scholar]
- Maloof JN, Whangbo J, Harris JM, Jongeward GD, Kenyon C. A Wnt signaling pathway controls Hox gene expression and neuroblast migration in C. elegans. Development. 1999;126:37–49. doi: 10.1242/dev.126.1.37. [DOI] [PubMed] [Google Scholar]
- Mao J, Wang J, Liu B, Pan W, Farr GH, III, Flynn C, Yuan H, Takada S, Kimelman D, Li L, et al. Low-density lipoprotein receptor-related protein-5 binds to Axin and regulates the canonical Wnt signaling pathway. Mol Cell. 2001;7:801–809. doi: 10.1016/s1097-2765(01)00224-6. [DOI] [PubMed] [Google Scholar]
- Meneghini MD, Ishitani T, Carter JC, Hisamoto N, Ninomiya-Tsuji J, Thorpe CJ, Hamill DR, Matsumoto K, Bowerman B. MAP kinase and Wnt pathways converge to downregulate an HMG-domain repressor in Caenorhabditis elegans. Nature. 1999;399:793–797. doi: 10.1038/21666. [DOI] [PubMed] [Google Scholar]
- Natarajan L, Witwer NE, Eisenmann DM. The divergent Caenorhabditis elegansβ-catenin proteins BAR-1, WRM-1 and HMP-2 make distinct protein interactions but retain functional redundancy in vivo. Genetics. 2001;159:159–172. doi: 10.1093/genetics/159.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocheleau CE, Downs WD, Lin R, Wittmann C, Bei Y, Cha YH, Ali M, Priess JR, Mello CC. Wnt signaling and an APC-related gene specify endoderm in early C. elegansembryos. Cell. 1997;90:707–716. doi: 10.1016/s0092-8674(00)80531-0. [DOI] [PubMed] [Google Scholar]
- Rocheleau CE, Yasuda J, Shin TH, Lin R, Sawa H, Okano H, Priess JR, Davis RJ, Mello CC. WRM-1 activates the LIT-1 protein kinase to transduce anterior/posterior polarity signals in C. elegans. Cell. 1999;97:717–726. doi: 10.1016/s0092-8674(00)80784-9. [DOI] [PubMed] [Google Scholar]
- Ruvkun G, Hobert O. The taxonomy of developmental control in Caenorhabditis elegans. Science. 1998;282:2033–2041. doi: 10.1126/science.282.5396.2033. [DOI] [PubMed] [Google Scholar]
- Sakanaka C, Weiss JB, Williams LT. Bridging of β-catenin and glycogen synthase kinase-3β by Axin and inhibition of β-catenin-mediated transcription. Proc Natl Acad Sci. 1998;95:3020–3023. doi: 10.1073/pnas.95.6.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salser SJ, Kenyon C. Activation of a C. elegansAntennapedia homologue in migrating cells controls their direction of migration. Nature. 1992;355:255–258. doi: 10.1038/355255a0. [DOI] [PubMed] [Google Scholar]
- Schlesinger A, Shelton CA, Maloof JN, Meneghini M, Bowerman B. Wnt pathway components orient a mitotic spindle in the early Caenorhabditis elegansembryo without requiring gene transcription in the responding cell. Genes & Dev. 1999;13:2028–2038. doi: 10.1101/gad.13.15.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin TH, Yasuda J, Rocheleau CE, Lin R, Soto M, Bei Y, Davis RJ, Mello CC. MOM-4, a MAP kinase kinase kinase-related protein, activates WRM-1/LIT- 1 kinase to transduce anterior/posterior polarity signals in C. elegans. Mol Cell. 1999;4:275–280. doi: 10.1016/s1097-2765(00)80375-5. [DOI] [PubMed] [Google Scholar]
- Shioi G, Shoji M, Nakamura M, Ishihara T, Katsura I, Fujisawa H, Takagi S. Mutations affecting nerve attachment of Caenorhabditis elegans. Genetics. 2001;157:1611–1622. doi: 10.1093/genetics/157.4.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley MJ, Sara E, Paterson H, Naylor S, Cook D, Jayatilake H, Fryer LG, Hutchinson L, Fry MJ, Dale TC. Interaction of Axin and Dvl-2 proteins regulates Dvl-2-stimulated TCF- dependent transcription. EMBO J. 1999;18:2823–2835. doi: 10.1093/emboj/18.10.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spink KE, Polakis P, Weis WI. Structural basis of the Axin-adenomatous polyposis coli interaction. EMBO J. 2000;19:2270–2279. doi: 10.1093/emboj/19.10.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- van de Water S, van de Wetering M, Joore J, Esseling J, Bink R, Clevers H, Zivkovic D. Ectopic Wnt signal determines the eyeless phenotype of zebrafish masterblindmutant. Development. 2001;128:3877–3888. doi: 10.1242/dev.128.20.3877. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, et al. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- Walhout AJ, Sordella R, Lu X, Hartley JL, Temple GF, Brasch MA, Thierry-Mieg N, Vidal M. Protein interaction mapping in C. elegansusing proteins involved in vulval development. Science. 2000;287:116–122. doi: 10.1126/science.287.5450.116. [DOI] [PubMed] [Google Scholar]
- Waring DA, Kenyon C. Selective silencing of cell communication influences anteroposterior pattern formation in C. elegans. Cell. 1990;60:123–131. doi: 10.1016/0092-8674(90)90722-q. [DOI] [PubMed] [Google Scholar]
- Whangbo J, Kenyon C. A Wnt signaling system that specifies two patterns of cell migration in C. elegans. Mol Cell. 1999;4:851–858. doi: 10.1016/s1097-2765(00)80394-9. [DOI] [PubMed] [Google Scholar]
- Whangbo J, Harris J, Kenyon C. Multiple levels of regulation specify the polarity of an asymmetric cell division in C. elegans. Development. 2000;127:4587–4598. doi: 10.1242/dev.127.21.4587. [DOI] [PubMed] [Google Scholar]
- Willert K, Logan CY, Arora A, Fish M, Nusse R. A Drosophila Axin homolog, Daxin, inhibits Wnt signaling. Development. 1999;126:4165–4173. doi: 10.1242/dev.126.18.4165. [DOI] [PubMed] [Google Scholar]
- Zeng L, Fagotto F, Zhang T, Hsu W, Vasicek TJ, Perry WL, III, Lee JJ, Tilghman SM, Gumbiner BM, Costantini F. The mouse Fused locus encodes Axin, an inhibitor of the Wnt signaling pathway that regulates embryonic axis formation. Cell. 1997;90:181–192. doi: 10.1016/s0092-8674(00)80324-4. [DOI] [PubMed] [Google Scholar]