Abstract
The availability of a sister chromatid, and thus the cell cycle phase in which DNA double-strand breaks (DSBs) occur, influences the choice between homologous recombination (HR) or nonhomologous end joining (NHEJ). The sequential activation and destruction of CDK–cyclin activities controls progression through the cell cycle. Here we provide evidence that the major Schizosaccharomyces pombe CDK, Cdc2–cyclin B, influences recombinational repair of radiation-induced DSBs during the G2 phase at two distinct stages. At an early stage in HR, a defect in Cdc2 kinase activity, which is caused by a single amino acid change in cyclin B, affects the formation of Rhp51 (Rad51sp) foci in response to ionizing radiation in a process that is redundant with the function of Rad50. At a late stage in HR, low Cdc2–cyclin B activity prevents the proper regulation of topoisomerase III (Top3) function, disrupting a recombination step that occurs after the assembly of Rhp51 foci. This effect of Cdc2–cyclin B kinase on Top3 function is mediated by the BRCT-domain-containing checkpoint protein Crb2, thus linking checkpoint proteins directly with recombinational repair in G2. Our data suggest a model in which CDK activity links processing of recombination intermediates to cell cycle progression via checkpoint proteins.
Keywords: Homologous recombination, cdc2, cyclin B, Rqh1, Top3, Crb2, checkpoint
DNA double-strand breaks (DSBs) pose a considerable threat to genomic integrity and cell survival. If not repaired, a single DSB can result in cell death (Rudin and Haber 1988). DSBs arise both spontaneously, for example, during DNA replication, and in response to exogenous agents such as ionizing radiation (IR). The two major DSB repair pathways in eukaryotic cells are nonhomologous end joining (NHEJ) and homologous recombination (HR). NHEJ results in the ligation of broken DNA ends irrespective of their sequence information. This process is potentially error-prone and can result in loss of genetic information. In contrast, HR uses the genetic information stored in the sister chromatid (or potentially the homologous chromosome) to repair DSBs with high fidelity.
Genetic studies in Saccharomyces cerevisiae have established that HR is performed by the products of the RAD52 epistasis group of genes (Paques and Haber 1999). Once a DSB is formed, the ends are resected by a 5′–3′ exonuclease in a manner partially dependent on the Rad50–Mre11–Xrs2 protein complex (Ivanov et al. 1994). This results in 3′-ended single-strand DNA (ssDNA) tails, which are channeled into different homology-dependent recombination pathways. In a minor pathway, 3′ tails can undergo intrachromosomal single-strand annealing (SSA) (Ivanov et al. 1996), which results in the deletion of the intervening sequence. The major pathway, however, is invasion of a homologous DNA region on a sister chromatid or on a homologous chromosome by the 3′ tails. Invasion by one of the two ssDNA tails generates a unidirectional replication fork, which can migrate down the template chromosome, copying the genetic information, a process termed DNA-break induced replication (BIR) (Kraus et al. 2001). Invasion by both ssDNA tails results in the formation of a double Holliday Junction (HJ), which, when resolved, results in gene conversion with or without a crossover event. SSA, BIR, and gene conversion are all dependent on Rad52, but only the latter pathway requires assembly of the Rad51 nucleoprotein filament, which promotes homologous pairing and strand exchange (Paques and Haber 1999).
The availability of the sister chromatid, and therefore the cell cycle phase in which the DSB occurs, determines the choice of the appropriate repair pathway. The molecular mechanisms underlying this channeling event are unknown. In G1, DSBs are rejoined by NHEJ, SSA, or BIR (in a diploid), whereas after DNA replication, the sister chromatid serves as an information donor for HR. The cell cycle is controlled by the sequential action of CDK–cyclin complexes, and it would thus be logical if these were also to influence the channeling of DSBs into different repair pathways. However, evidence for such a function has not been reported. There are several ways in which CDKs could channel DSBs into specific repair pathways. For example, CDKs could regulate the expression of key repair enzymes, thereby influencing the choice between HR and NHEJ. It has been proposed that Rad52 competes with proteins of the NHEJ pathway for binding to DSBs (Van Dyck et al. 1999). The increased expression of Rad52 in late S phase and in G2 (when HR is at its highest level) in human cells might thus affect the balance of repair events (Chen et al. 1997). Alternatively, CDKs could directly phosphorylate proteins involved in the response to DNA damage. CDK-dependent phosphorylation has been reported for several proteins involved in the DSB response, including single-strand binding protein (RPA) (Dutta and Stillman 1992; Niu et al. 1997), DNA ligase I (Aoufouchi et al. 1995), Srs2 DNA helicase (Liberi et al. 2000), the tumor suppressor protein BRCA1 (Ruffner et al. 1999), and the Schizosaccharomyces pombe checkpoint protein Crb2 (Esashi and Yanagida 1999).
Crb2 is the S. pombe analog of the canonical S. cerevisiae Rad9 checkpoint protein and contains two C-terminal BRCT motifs (Saka et al. 1997; Wilson et al. 1997). Cdc2–cyclin B kinase has been shown to phosphorylate Crb2 during metaphase, just before destruction of cyclin B (Esashi and Yanagida 1999). Later in the cell cycle, this modification is an essential prerequisite for the checkpoint-dependent hyperphosphorylation of Crb2 by the Rad3 protein kinase in response to DNA damage.
In this work, we provide evidence showing that a functional Cdc2–cyclin B kinase complex is required for successful HR. A point mutation in cdc13, encoding the major G2-cyclin in S. pombe, reduces CDK activity and renders cells sensitive to IR. The increase in sensitivity is not caused by a checkpoint defect, but relates primarily to an impaired function of topoisomerase III (Top3). Loss of Top3 activity, which acts downstream of, or in parallel with, the Rqh1 DNA helicase, permits the use of an alternative Rqh1-dependent mechanism that results in hyperrecombination and cell death following IR. Our data suggest that CDK targets Top3 indirectly, via the checkpoint protein Crb2. Although Crb2, Rqh1, and Top3 all act at a late stage in HR after formation of Rhp51 (Rad51 homolog) foci, we also find that CDK activity plays a second role at an early stage. CDK activity positively regulates a function that is required for the assembly of Rhp51 foci in a redundant manner with Rad50. Our findings show that a functional Cdc2–cyclin B kinase complex is necessary to protect cells from the genotoxic consequences of IR and suggest a mechanism by which Crb2 acts at the interface between cell cycle regulation, checkpoint function, and DSB repair.
Results
A point mutation in cdc13 renders S. pombe cells sensitive to IR
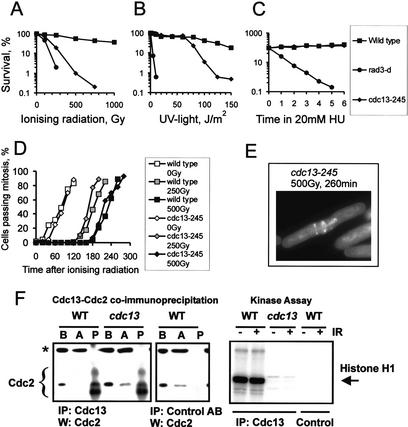
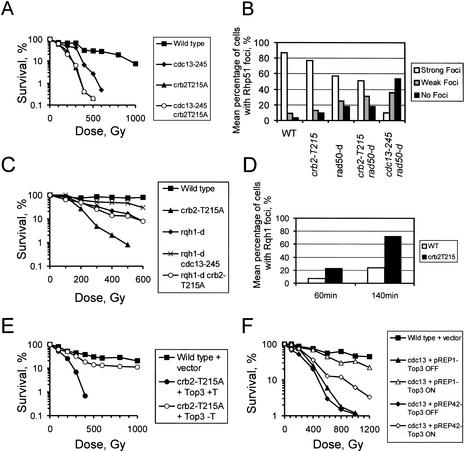
To identify S. pombe proteins involved in the response to DNA damage that are also essential for cell growth, we screened a collection of temperature-sensitive mutants for sensitivity to a variety of DNA-damaging agents. Under permissive conditions, one mutant (TC-245) was highly sensitive to ionizing radiation (IR), mildly sensitive to UV light, and insensitive to DNA replication blocks caused by nucleotide depletion (Fig. 1A–C). The mutation mapped to the cdc13 gene (cdc13-245), which encodes the major G2-cyclin in S. pombe (Booher and Beach 1988; Hagan et al. 1988). Sequence analysis shows that the phenotype results from a single amino acid replacement at position 255 (E255R) within the cyclin box (data not shown). Because loss-of-function mutants in cdc13 undergo DNA re-replication (Hayles et al. 1994) , we analyzed the DNA content of an asynchronous cdc13-245 culture under permissive conditions. FACS analysis revealed a 2C DNA content identical to wild-type controls, showing that cdc13-245 cells do not re-replicate under these conditions and that the IR sensitivity is not a result of an increase in G1 cells (data not shown).
Figure 1.
Low Cdc2–cyclin B kinase activity renders S. pombe cells sensitive to IR without affecting the DNA damage checkpoint. (A–C) The sensitivity of cdc13-245 mutant cells to IR, to UV-induced DNA damage, and to S-phase arrests caused by nucleotide depletion. The rad3-d strain is checkpoint-deficient and was used as a control. (D) G2-synchronized cells were exposed to 0 Gy, 250 Gy, or 500 Gy of IR, and progression through mitosis was analyzed under permissive conditions. A checkpoint-deficient mutant (rad3-d) was used as a negative control (data not shown). (E) An example of a cdc13-245 cell undergoing aberrant mitosis after IR. (F, left panels) Immunoprecipitation. Cdc2 kinase was precipitated from soluble extracts (prepared from untreated cells) using either an anti-Cdc13 antibody or an unrelated control antibody. (B) Before addition of the antibodies, 50 μg of soluble protein; (A) after precipitation, 50 μg of soluble protein; (P) pellet. The asterisk marks a nonspecific band. (Right panel) In vitro kinase assay. The anti-Cdc13 antibody was used to precipitate Cdc2–cyclin B kinase from untreated extracts and from extracts prepared after irradiation with a dose of 500 Gy. The precipitated material was incubated with histone H1 and ATP.
Cdc13 is known to regulate mitotic progression, and it is possible that the sensitivity of cdc13-245 cells is a result of checkpoint failure. To investigate this, we irradiated wild-type cells and cdc13-245 cells that had been synchronized in G2 by lactose gradient centrifugation and analyzed their subsequent cell cycle progression. Like wild-type cells, cdc13-245 cells transiently arrested entry into mitosis, showing that checkpoint failure is not responsible for the IR sensitivity (Fig. 1D). Interestingly, unlike wild-type cells, ∼40% of cdc13-245 cells entered a catastrophic mitosis upon recovery from the G2/M arrest, as indicated by nuclear abnormalities (Fig. 1E). This mitotic failure, despite normal G2 delay, is indicative of a defect in DNA repair.
The E255R substitution reduces Cdc2 kinase activity in vivo and in vitro
Under permissive conditions, cdc13-245 cells are slightly more elongated than wild-type cells (data not shown), which is indicative of a moderate delay over entry into mitosis. Because mitotic entry requires high Cdc2–Cdc13 kinase activity, this delay suggests that the E255R mutation reduces CDK activity in vivo. To characterize the effect of the E255R substitution under in vitro conditions, we coimmunoprecipitated Cdc2 from soluble wild-type and cdc13-245 extracts (prepared under permissive conditions from asynchronous cells) using a polyclonal anti-Cdc13 antibody. The efficient coprecipitation of Cdc2 from cdc13-245 extracts indicates that the E255R substitution did not affect the interaction between Cdc2 and Cdc13. However, the loss of histone H1 phosphorylation by Cdc2 in this in vitro assay (Fig. 1F) further suggests that the mutation in cyclin B impairs CDK activity.
The E255R substitution compromises homologous recombination
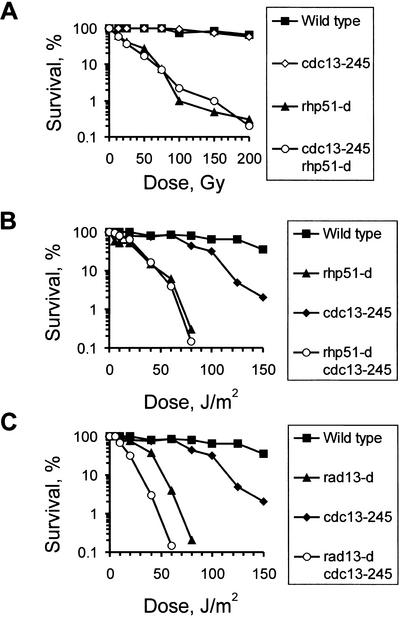
Given that Rad51 is essential for the repair of DSBs by HR (Muris et al. 1993; Paques and Haber 1999), we constructed a cdc13-245 rhp51-d double mutant. The double mutant displayed the same sensitivity as the rhp51-d single mutant (Fig. 2A), suggesting that low CDK activity affects the Rhp51-dependent DSB repair pathway. However, compared to rhp51-d mutants, which are completely deficient in HR, cdc13-245 cells are significantly less sensitive, especially to doses below 200 Gy (Fig. 2A). To verify the epistatic relationship between cdc13-245 and rhp51-d, we repeated our genetic analysis using UV light. S. pombe cells use two pathways to repair UV damage. Photoproducts are either removed by nucleotide excision repair (NER) or channeled into an Rhp51-dependent pathway after being removed by the UVDE enzyme (Murray et al. 1997; McCready et al. 2000). The cdc13-245 rhp51-d double mutant showed an identical UV sensitivity to that of the rhp51-d single mutant (Fig. 2B). In contrast, loss of NER (rad13 deletion) in a cdc13-245 background resulted in a double mutant that was significantly more UV sensitive than either of the two single mutants (Fig. 2C). These data are consistent with the involvement of HR in UV repair in fission yeast (McCready et al. 2000) and confirm the conclusion that low CDK activity compromises Rhp51-dependent HR.
Figure 2.
The cdc13-245 mutation affects homologous recombination. (A) cdc13-245 does not increase IR sensitivity of an rhp51-d mutant. (B) The cdc13-245 mutant is epistatic with rhp51 in recombinational repair of photoproducts. (C) Loss of NER renders cdc13-245 cells more sensitive to UV-induced DNA damage.
Low CDK activity impairs Rhp51 (Rad51 homolog) foci formation
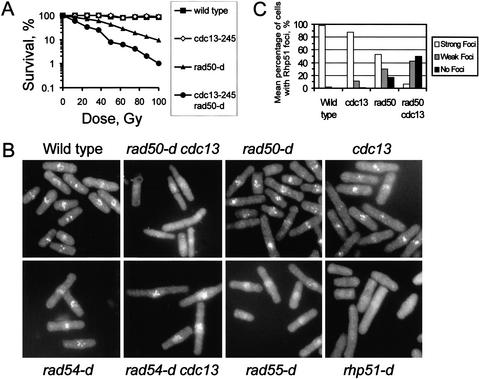
To map the function of Cdc2–cyclin B within the HR pathway, we combined the cdc13-245 allele with different mutants deficient in recombinational repair. The Rad50–Mre11–Xrs2 protein complex functions upstream of Rad51 and links processing of DNA ends with the availability of the sister chromatid (Hartsuiker et al. 2001). rad50-d cells are more radiation resistant than rhp51-d cells (Hartsuiker et al. 2001), indicating that the early functions in HR can be performed by redundant pathways. Interestingly, the rad50-d cdc13-245 double mutant is more sensitive to IR than the rad50-d single mutant (Fig. 3A). This increase implies that low CDK activity impairs HR at an early stage (such as end processing) in a manner redundant with the Rad50 function.
Figure 3.
The cdc13-245 mutation impairs an end-processing pathway that is redundant with Rad50. (A) The cdc13-245 mutation increases the IR sensitivity of rad50-d cells. (B) Indirect immunofluorescence microscopy of wild-type cells and the indicated mutant strains at 1 h postirradiation (500 Gy). A cross-reacting human anti-Rad51 antibody was used to detect Rhp51. Note that rad54-d cdc13-245 cells are proficient in the formation of Rhp51 foci, but a rad50-d cdc13-245 double mutant is impaired. (C) Mean percentage of various mutant cells showing Rhp51 foci at 1 h postirradiation.
To seek further evidence supporting this hypothesis at the cellular level, we used a cross-reacting anti-human Rad51 antibody to analyze the formation of Rhp51 foci in irradiated S. pombe cells. Human Rad51 accumulates at sites of DNA damage in postreplicative chromatin (Tashiro et al. 2000). Treatment of wild-type S. pombe cells with 500 Gy of IR resulted in the assembly of Rhp51 in bright nuclear foci within 1 h postirradiation (Fig. 3B). Irradiation of synchronized cells confirmed that foci formation occurred in G2 (data not shown; see below). Next, we visualized Rhp51 foci in different mutants to characterize the role of other recombination proteins. The formation of foci was dependent on the mediator protein Rhp55, which is thought to facilitate the formation of the Rad51 nucleoprotein filament, whereas it was independent of the synaptic factor Rad54, which functions later in HR to permit strand invasion. Cells deleted for rad50 showed only a moderate reduction in the focal assembly of Rhp51 (Fig. 3B). Although almost 100% of wild-type cells possessed Rhp51 foci 1 h postirradiation, only 53% ± 10% of rad50-d cells developed bright foci (Fig. 3C) and 17% ± 8% of rad50-d cells failed to produce any foci. The diminished ability of rad50-d cells to assemble Rhp51 foci is consistent with the higher radiation resistance of these cells when compared with an rhp51-d strain. Interestingly, in a rad50-d background, the ability to produce Rhp51 foci was largely dependent on an intact Cdc2–cyclin B kinase complex. At 1 h postirradiation, only 7% ± 4% of rad50-d cdc13-245 cells assembled bright wild-type-like foci and 53% ± 5% of cells were without foci (Fig. 3B,C). Western blot analysis confirmed that all mutants contained similar levels of Rhp51 protein (data not shown). These data provide evidence that CDK activity positively controls an early recombination function that is redundant with Rad50 and required for IR-induced focal assembly of Rhp51.
Loss of Rqh1 DNA helicase restores IR resistance in a cdc13-245 mutant
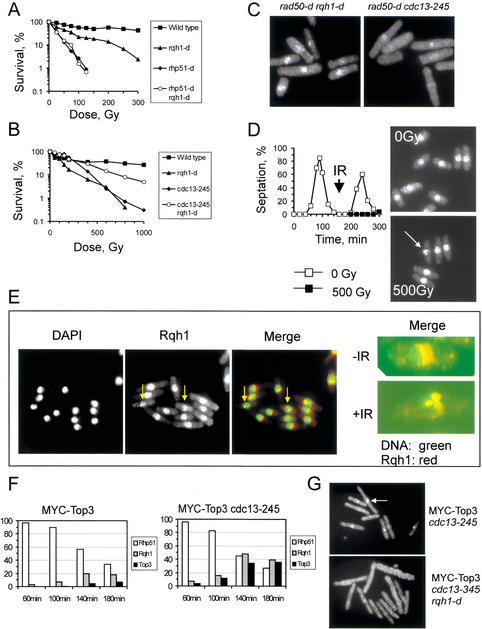
Rqh1 is the only RecQ-like DNA helicase in S. pombe (Murray et al. 1997; Stewart et al. 1997). During S phase, Rqh1 is thought to act at stalled replication forks during recombination-based repair (Murray et al. 1997). The link between Rqh1 and HR prompted us to test whether Rqh1 is also required for the response to IR in G2 cells. Survival analysis following IR revealed that rqh1-d cells are sensitive to IR and showed that Rqh1 acts in the same pathway as Rhp51 (Fig. 4A). Thus, both Rqh1 and Cdc13 contribute to successful recombinational repair of IR-induced DSBs via an Rhp51-dependent pathway. Surprisingly, deletion of rqh1 in a cdc13-245 background resulted in a significant restoration of IR resistance (Fig. 4B) compared with either the rqh1-d or cdc13-245 single mutants.
Figure 4.
Analysis of both Rqh1 foci and Top3 foci. (A) Rqh1 and Rhp51 act in the same recombination repair pathway. (B) Loss of Rqh1 DNA helicase restores IR resistance of a cdc13-245 mutant. Note the mutual rescue, which suggests that the cdc13-245 mutation can also prevent cell death in an rqh1-d background. This rescue could relate to the involvement of Cdc2–Cdc13 in an early recombination step such as end-processing. (C) Deletion of rqh1 in an rad50-d background does not affect assembly of Rhp51 in foci at 1 h postirradiation (500 Gy). (D, left) Kinetic analysis of cell cycle progression either untreated or with a dose of 500 Gy in the second G2 phase. (Right) Untreated (0 Gy) or irradiated (500 Gy) wild-type G2 cells from the same experiment. Rqh1 was visualized with an Rqh1-specific antibody. The arrow marks damage-induced Rqh1 foci at 1 h postirradiation. (E, left) DAPI, anti-Rqh1 antibody staining, and merge images showing representative cells irradiated in G2. Arrows indicate cells with foci. (Right) Single G2 cells either before (−IR) or 1 h after irradiation (+IR). In these merged images, chromatin is stained with DAPI (green) and Rqh1 is visualized by the anti-Rqh1 antibody (red). (F) Mean percentage of synchronized G2 cells showing Rhp51 foci, Rqh1 foci, or MYC-Top3 foci at the indicated periods of time after 500 Gy of IR. (G) Staining of the indicated asynchronous cells with the anti-MYC antibody at 140 min postirradiation (500 Gy). The arrow marks damage-induced MYC-Top3 foci.
This mutual rescue indicates that Cdc2–Cdc13 activity influences at least two distinct stages of HR. Because the cdc13-245 mutation suppresses loss of Rqh1, Cdc13 activity appears to control an early step that channels DNA structures into the Rqh1-dependent pathway. That Cdc13 activity is required for a second step later in HR becomes evident from the observation that inactivation of Rqh1 restores IR resistance in a cdc13-245 background. This rescue strongly suggests that Cdc13 activity is necessary to deal with recombination intermediates that are generated by Rqh1 DNA helicase. The fact that this late function is not blocked by the early defect indicates that the pathway is not linear and that Rqh1 receives DNA substrates from two parallel pathways, and only one of these is dependent on Cdc13.
This interpretation is consistent with our observation that Cdc13 activity is necessary to allow focal assembly of Rhp51 in a rad50-d background. To test whether Rqh1 shares this early function with Cdc13, we analyzed focal assembly of Rhp51 in a rad50-d rqh1-d double mutant. Deletion of rqh1 did not affect the appearance of Rhp51 foci in rad50-d cells 1 h postirradiation (Fig. 4C). This result implies that Rqh1 does not participate in the early step, but acts close to the late recombination function. Taken together, these data indicate that the pathways that provide DNA structures for Rqh1 are under control of Cdc13 and Rad50, respectively. This suggestion is supported by the observation that deletion of rad50 in a cdc13-245 rqh1-d background (the rqh1-d cdc13-245 rad50-d triple mutant) renders cells as IR sensitive as cdc13-245 rad50-d double-mutant cells (data not shown).
Impaired CDK activity stabilizes both Rqh1 foci and Top3 foci
The genetic interaction between cdc13-245 and rqh1-d indicates a function for Rqh1 not only during S phase but also in G2. To test this idea, we irradiated wild-type cells that were synchronized in G2 and analyzed the response of Rqh1. Neither protein levels nor electrophoretic mobility of Rqh1 changed upon IR (data not shown), but, in a proportion of cells, Rqh1 relocalized from a predominantly nucleolar distribution to discrete foci in the chromatin compartment 1 h postirradiation (Fig. 4D,E). This relocalization occurred in 5% ± 1% of wild-type cells after 60 min and increased to 20% ± 2% by 140 min postirradiation. To dissect the relationship between Rhp51 and Rqh1 foci, we monitored the kinetics of Rhp51, Rqh1, and Top3 assembly in nuclear foci upon irradiation of G2-synchronized cells (Fig. 4F). We included Top3 in our analysis, because the association between RecQ-like DNA helicases and Top3 is highly conserved (Harmon et al. 1999; Bennett and Wang 2001). The N-terminally MYC-tagged Top3 protein (L.V. Laursen, A.H. Andersen, and J.M. Murray, unpubl.), which we used in our study, had no obvious phenotype in either wild-type cells or in a cdc13-245 mutant background.
At 1 h postirradiation, almost 100% of wild-type cells possessed Rhp51 foci. The percentage of cell with Rhp51 foci declined steadily during the G2/M arrest period and dispersed as cells re-entered the cell cycle between 180 min and 200 min postirradiation (Fig. 4F; data not shown). In contrast, foci containing either Rqh1 or MCY-Top3 appeared toward the end of the arrest period, reaching a maximum at ∼140 min postirradiation (Fig. 4F). This late response to IR is consistent with the idea that both Rqh1 and Top3 function at a late stage in HR after formation of Rhp51 foci. Moreover, it suggests that both enzymes deal with recombination intermediates, which appear only in a limited number of cells shortly before the end of the arrest.
We repeated this analysis using G2-synchronized cdc13-245 cells and found that the number of cells assembling either Rqh1 or MYC-Top3 in nuclear foci upon IR increased significantly compared with wild-type cells. At 140 min postirradiation, the percentage of cells possessing Rqh1 foci rose from 20% ± 2% seen in wild-type cells to 48% ± 1%. Similarly, the number of cells showing MYC-Top3 foci increased from 5% ± 2% in wild-type cells to 35% ± 2% in cdc13-245 cells (Fig. 4F). The protein levels of both Rqh1 and MYC-Top3 did not change during the experiment (data not shown), and the kinetic of Rhp51 foci formation and subsequent dispersal was not affected by the cdc13-245 mutation.
To test whether the stabilization of these foci relates to our model that Cdc13 activity is required to process recombination intermediates that are generated by Rqh1, we analyzed formation of Top3 foci in an rqh1-d cdc13-245 background. Loss of Rqh1 prevented the focal assembly of Top3 in the double mutant (Fig. 4G), showing that assembly of Top3 foci is dependent on Rqh1 function and that low Cdc2–Cdc13 kinase activity stabilizes Top3 foci only when Rqh1 generates intermediates.
The checkpoint protein Crb2 acts in the same recombination pathway as cyclin B
The checkpoint protein Crb2 becomes phosphorylated at threonine 215 by Cdc2–cyclin B kinase during mid-mitosis. Phosphorylation of T215 is a prerequisite for subsequent Rad3-dependent hyperphosphorylation of Crb2 in response to DNA damage (Esashi and Yanagida 1999). The link between Crb2 and Cdc2–cyclin B prompted us to create the crb2-T215A allele at the endogenous crb2 locus in both a wild-type background and a cdc13-245 background. We observed that crb2-T215A cells are more IR sensitive than cdc13-245 cells and that the double crb2-T215A cdc13-245 mutant had the same IR sensitivity as the crb2-T215A single mutant, showing that both proteins act in the same pathway for IR survival (Fig. 5A).
Figure 5.
High levels of Top3 protein restore IR resistance in both the crb2-T215A mutant and the cdc13-245 mutant. (A) The inability to phosphorylate Crb2 at threonine 215 renders cells sensitive to IR. The crb2-T215A mutation is epistatic with the cdc13-245 mutation. (B) Mean percentage of the indicated asynchronous cells showing Rhp51 foci at 1 h postirradiation (500 Gy). (C) Deletion of rqh1 restores IR resistance in a crb2-T215A mutant. Note that, unlike the case seen for the rqh1-d cdc13-245 double mutant, the mutation in crb2 fails to improve survival of rqh1-d cells. This is consistent with the fact that Crb2 is not involved in an early step in recombination. (D) Mean percentage of asynchronous wild-type and crb2-T215A cells showing Rqh1 foci at the indicated periods of time upon 500 Gy of IR. (E) Overexpression of HIS6–top3 from a pREP1 plasmid restores IR resistance in a crb2-T215A mutant. (F) Overexpression of HIS6–top3 restores IR resistance of cdc13-245 cells.
Given that the mutation in cdc13 affects two steps in HR, we asked which of these steps might be compromised in the crb2-T215A mutant. For this purpose, we constructed a rad50-d crb2-T215A double mutant and an rqh1-d crb2-T215A double mutant. Upon IR, the rad50-d crb2-T215A double mutant assembled Rhp51 foci in a manner that was quite similar to that of the rad50-d single mutant (Fig. 5B), suggesting that the early Cdc13-dependent function is not affected by the crb2-T215A mutation. Interestingly, deletion of rqh1 suppressed the IR sensitivity of the crb2-T215A mutant, but, unlike in the case of the rqh1-d cdc13-245 double mutant, IR resistance was only restored to a level that is characteristic for the rqh1-d single mutant (Fig. 5C). This limited increase in IR resistance suggests that Crb2 function is restricted to the late step in HR that requires Rqh1 activity. Consistent with this suggestion, we found that the percentage of crb2-T215A cells containing Rqh1 foci rose from the wild-type level of 23% ± 3% to 72% ± 8% at 140 min postirradiation (Fig. 5D).
Top3 is an efficient multicopy suppressor of both cdc13-245 and crb2-T215A
During our strain construction, we noticed that MYC-top3 crb2-T215A double-mutant cells are inviable (data not shown). Although the MYC-top3 allele appears fully functional (cell morphology and damage resistance assays) in other genetic backgrounds, the N-terminal tag appears to impair a function that becomes essential in crb2-T215A cells. This genetic interaction prompted us to test whether overexpression of top3 could restore IR resistance in crb2-T215A cells. Indeed, high protein levels of Top3 proved very efficient in suppressing the sensitivity to IR (Fig. 5E). Because Crb2 and cyclin B appear to act in the same pathway, downstream of Rqh1, we repeated this experiment in cdc13-245 cells and obtained a similar result (Fig. 5F). This efficient rescue implies that both the cdc13-245 and crb2-T215A mutations impair Top3 activity. However, there is a potential alternative explanation for this rescue effect: as in the case of the deletion of rqh1, high Top3 protein levels could contribute to the increase in IR survival by impairing Rqh1 helicase function. To exclude this possibility, we overexpressed top3 in wild-type cells and found that high protein levels of Top3 did not increase the sensitivity of wild-type cells to IR (data not shown).
An obvious link between Cdc13 and Crb2 is the Cdc2-dependent phosphorylation of Crb2 at T215. Because the mutation in cdc13 impairs Cdc2 kinase activity, a reduced phosphorylation state of T215 could explain the observed genetic relationship. However, the use of an antibody (α-T125-P) that recognizes Crb2 only in its CDK-phosphorylated form revealed that the phosphorylation status of T215 was not significantly altered in a cdc13-245 background under permissive conditions (data not shown). Thus, the relationship between Crb2 and Cdc13 appears to be more indirect (see Discussion).
Mutations in cdc13 and crb2 cause Rqh1-dependent hyperrecombination
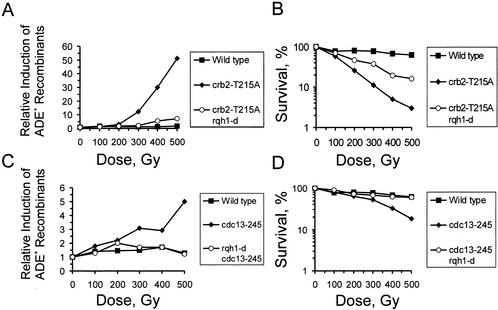
As a consequence of impaired Top3 activity in cdc13-245 and crb2-T215A mutants, Rqh1 DNA helicase could become hyperactive. Because such an increase in uncontrolled activity could result in more recombination events, we integrated a recombination substrate into wild-type, crb2-T215A, and cdc13-245 cells to measure spontaneous and IR-induced HR. The recombination substrate consists of two ade6 heteroalleles that are separated by 4.5 kb of DNA containing a ura4+ marker (Schuchert and Kohli 1988). Recombination between the heteroalleles restores a functional ade6 gene and allows growth on medium lacking adenine. Two types of event can be distinguished, gene conversion and crossover recombination, which results in loss of ura4+.
The spontaneous recombination frequency in a wild-type background was 2.8 ± 0.5 × 10−3 (0.7 × 10−3/2.1 × 10−3 conversion/crossover), and exposure to increasing doses of IR did not change this rate significantly (Fig. 6A,C). A similar rate of spontaneous recombination was observed in a crb2-T215A background, 2.2 ± 0.4 × 10−3 (0.6 × 10−3/1.6 × 10−3 conversion/crossover). However, in contrast to wild-type cells, the rate of recombination increased up to 50-fold when crb2-T215A cells were treated with increasing doses of IR (Fig. 6A). In agreement with the functional link between Cdc13 and Crb2, cells harboring the cdc13-245 allele showed a similar phenotypic trait, although IR-induced recombination increased only fivefold (Fig. 6C). Interestingly, the mutation in cdc13 deceased the spontaneous rate approximately fourfold, 0.8 ± 0.3 × 10−3 (0.5 × 10−3/0.3 × 10−3 conversion/crossover), when compared with the other two strains. This decrease in spontaneous recombination might be caused by the early defect in HR. In no case was there a significant difference in the ratio of conversion to crossover events in response to IR (data not shown).
Figure 6.
Both crb2-T215A cells and cdc13-245 cells show Rqh1-dependent hyperrecombination in response to IR. (A,C) Relative induction of the recombination rate between two ade6 heteroalleles in response to increasing doses of IR. (B,D) Cell survival during the recombination experiment. All data shown are the average of three independent experiments.
As predicted, the IR-induced hyperrecombination observed in crb2-T215A and cdc13-245 mutants was largely dependent on Rqh1 activity (Fig. 6A,C). Deletion of rqh1 significantly reduced the number of ade6+ recombinants in both mutant backgrounds at the same time as it suppressed the IR sensitivity (Fig. 6B,D). This increase in the survival rate indicates that Rqh1-dependent hyperrecombination may reflect the primary cause of IR sensitivity in both cdc13-245 cells and crb2-T215A cells.
Discussion
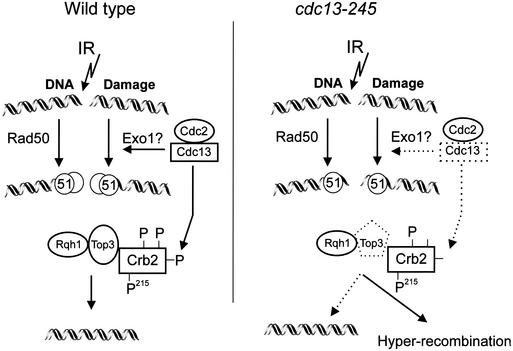
Our findings allow us to propose a model suggesting a functional link between Cdc2–cyclin B kinase activity and recombinational repair of DSB induced in G2 by IR (Fig. 7). In this model, Rqh1, in association with Top3, processes recombination intermediates that are channeled into this pathway by at least two different events. One of these early recombination steps is dependent on Cdc2–Cdc13 kinase activity, whereas the other step requires Rad50. In addition, Cdc2–Cdc13 kinase activity exerts a second function later in HR by facilitating the processing of recombination intermediates by Rqh1 and Top3. Interestingly, this late step also requires a functional Crb2 protein. Because Crb2 was so far only attributed to the checkpoint response and not to the repair response (Wilson et al. 1997; Esashi and Yanagida 1999), these data provide a new link between both pathways.
Figure 7.
A model indicating two functions for Cdc2–cyclin B kinase in HR. (Left panel) Cdc2–cyclin B kinase activity positively regulates an early step in HR that is redundant with Rad50. Both steps generate recombination intermediates that are processed by Rqh1 DNA helicase in association with topoisomerase III. Cdc2–cyclin B kinase activity exerts a second function together with Crb2 at a late step in HR. Both proteins are required to maintain Top3 activity until all recombination intermediates are processed. Cdc2–Cdc13 kinase activity may prevent premature dephosphorylation of Crb2, thereby supporting Top3 function. In the absence of Top3 activity, Rqh1 uses an alternative pathway resulting in hyperrecombination. (Right panel) Probable defects associated with the cdc13-245 mutation.
Is there an independent function for cyclin B?
In S. pombe, there is no evidence suggesting that cyclin B can function independently of Cdc2 kinase. Cdc13 appears to target Cdc2 to its nuclear destinations, and Cdc13 has to dissociate from Cdc2 to be degraded at the metaphase–anaphase transition (Decottignies et al. 2001). There are two plausible explanations for the IR sensitivity of cdc13-245 cells, which are not mutually exclusive. The mutation in cyclin B could either interfere with the localization of Cdc2 to structures within the nucleus, or it could reduce the kinase activity of the protein complex. Our data support the second explanation. Under permissive conditions, cdc13-245 cells slightly delay entry into mitosis (data not shown), which requires high Cdc2–cyclin B kinase activity. Such a decrease in Cdc2 activity would be consistent with the result of our in vitro kinase assay. The E255R substitution within the cyclin box of Cdc13 did not affect the interaction with Cdc2, but it dramatically decreased the phosphorylation of histone H1 (Fig. 1F). These in vitro data are in agreement with a previously published report showing that mutations of invariant residues in the cyclin box, which included an E255Q substitution, result in the accumulation of Cdc2–cyclin B kinase complexes with impaired activity (Zheng and Ruderman 1993).
A role for cyclins in the response to DNA damage in other systems
To our knowledge, there is only one report in the literature showing a role for cyclins in the response to DNA damage. Loss of the S-phase cyclins Clb5 and Clb6 renders Saccharomyces cerevisiae cells sensitive to MMS, UV-light, and IR (Meyn and Holloway 2000). In contrast to the normal checkpoint response of cdc13-245 cells (Fig. 1D), a clb5-d clb6-d double mutant bypasses the checkpoint arrest in metaphase and arrests instead with separated sister chromatids. Because sister chromatid cohesion is established when cells are passing through S phase (Uhlmann and Nasmyth 1998), loss of Clb5 and Clb6 could indirectly affect cell cycle arrest.
The repair defect caused by cdc13-245 is G2-specific
HR is not only required to repair DSBs in G2 but also during S phase to restart stalled replication forks (Michel et al. 2001). This dual function explains why mutants deficient in HR are sensitive to nucleotide depletion by HU treatment and lose viability when Okazaki fragment processing is defective (Muris et al. 1996). In this context, cdc13-245 is not a typical recombination mutant. cdc13-245 cells are not HU sensitive (Fig. 1C), and loss of Rad2 endonuclease, which is required for the processing of Okazaki fragments, is not synthetically lethal (data not shown). Hence, the repair defect in cdc13-245 mutant cells appears to be restricted to postreplicative stages in the cell cycle. A G2-specific function of Cdc13 is consistent with its expression profile, which rises during S phase and peaks in G2 (Moreno et al. 1989). A G2-specific role in recombination repair is also compatible with our observation (Fig. 1E) that ∼40% of cdc13-245 cells, when irradiated in G2, enter a catastrophic mitosis upon exit from the G2/M arrest.
Rqh1 DNA helicase is required to repair DSBs in G2
Rqh1 belongs to the highly conserved RecQ family of DNA helicases, which includes S. cerevisiae Sgs1 and the human BLM and WRN proteins, which are mutated in patients suffering from Bloom's and Werner's syndromes, respectively (Mohaghegh and Hickson 2001). A body of evidence suggests that these helicases function in S phase to prepare blocked replication forks for the recombinational repair machinery (Murray et al. 1997; Stewart et al. 1997; Doe et al. 2000). The data we present here show a further function for Rqh1 in the repair of DSBs in G2. Rqh1 relocalizes from a predominantly nucleolar distribution to foci in the chromatin compartment of the nucleus upon irradiation in G2 (Fig. 4E,F). In addition, loss of rqh1 improves the survival rate of cdc13-245 cells (Fig. 4B), strongly suggesting that Rqh1 acts upstream of, or parallel to, the recombination step dependent on Cdc2–cyclin B kinase activity. Several published observations support this suggestion: First, BLM is most highly expressed in late S phase and G2, when HR occurs. Second, BLM and Sgs1 both associate with Rad51. And, finally, BLM colocalizes with human Rad51 and binds to regions of ssDNA, when cells are irradiated in G2 (Bischof et al. 2001; Wu et al. 2001).
Our kinetic analysis of Rqh1 foci indicates that Rqh1 helicase acts late during the G2/M arrest. Such a late function is consistent with the finding that loss of Rqh1 did not affect focal assembly of Rhp51 in a rad50-d background (Fig. 4C). In vitro data obtained for Sgs1 and BLM helicases indicate that Rqh1 could process Holliday junctions. Both Sgs1 and BLM catalyze branch migration and unwind four-way junctions in vitro (Bennett et al. 1999; Karow et al. 2000).
What is the repair defect in a cdc13-245 mutant?
The correlation between DNA-damage-induced hyperrecombination and cell viability suggests that inappropriate recombination may be primarily responsible for the loss of IR resistance when Cdc2–cyclin B kinase activity is impaired. cdc13-245 cells show a fivefold induction in the recombination rate between two ade6 heteroalleles after a dose of 500 Gy (Fig. 6C). Based on the observations that loss of rqh1 both suppresses these recombination events (Fig. 6C) and restores IR resistance of cdc13-245 mutant cells (Fig. 6D), we can speculate that Rqh1 activity might be a target either directly or indirectly of Cdc2–Cdc13 kinase and is consequently deregulated in cdc13-245 cells. It has been suspected for some time that mutations in DNA topoisomerases stimulate recombination through a mechanism that involves RecQ-like DNA helicases (Wallis et al. 1989; Gangloff et al. 1994). In several systems, it has been proposed that helicases generate an intermediate that becomes recombinogenic if not processed by topoisomerases (Gangloff et al. 1999; Goodwin et al. 1999; Harmon et al. 1999).
Top3 is therefore a good candidate for such a function because of its interaction with RecQ-like DNA helicases and the fact that this is highly conserved (Bennett and Wang 2001). Our finding that overexpression of top3 efficiently restores radiation resistance in the cdc13-245 mutant (Fig. 5F) implies that Rqh1 DNA helicase becomes deregulated through a defect in Top3 function, and that excess Top3 can substitute for this loss of function. An alternative explanation would be that Top3 physically dissociates from Rqh1 in cdc13-245 cells, a possibility hard to test directly because the majority of Rqh1 and a proportion of Top3 proteins are insoluble in S. pombe G2 cells (data not shown). In S. cerevisiae, mutations in the N terminus of Sgs1, which promotes the interaction with Top3, are known to result in hyperrecombination (Mullen et al. 2001). Our unpublished experiments are not, however, consistent with the possibility that Cdc2–Cdc13 regulates Top3–Rqh1 interactions directly: we mutated the two highly conserved Cdc2 phosphorylation motifs in the N-terminal domain of Rqh1 without observing any obvious phenotypes. Furthermore, we were able to immunoprecipitate Rqh1 and Top3 from both soluble wild-type and cdc13-245 extracts before and after IR (data not shown). Such experiments, however, do not address the status of the insoluble Rqh1 and Top3, which may reflect the active component.
The role of Crb2 in the repair of DSBs
Support for the idea that checkpoint-dependent hyperphosphorylation of Crb2 is required for efficient Top3 activity comes from several experiments: Loss of rqh1 restores IR resistance in a crb2-T215A mutant (Fig. 5C), which cannot be hyperphosphorylated (data not shown; Esashi and Yanagida 1999), strongly indicating that Rqh1 DNA helicase generates harmful intermediates in these cells after IR treatment. At 140 min postirradiation, the number of crb2-T215A cells assembling Rqh1 foci increased from 23% ± 3% observed in wild-type cells to 72% ± 8% (Fig. 5D). Finally, overexpression of top3 very efficiently increased the survival rate of crb2-T215A cells in an rqh1-dependent manner (Fig. 5E; data not shown).
Although these results mirror the data obtained for cdc13-245 cells, our observation that the state of Cdc2-dependent phosphorylation of Crb2 at T215 was not significantly changed in a cdc13-245 background under permissive conditions (data not shown) suggests an indirect link between both proteins. We noticed repeatedly that, in response to IR, the Rad3-dependent modifications of Crb2 disappeared prematurely, although cells were still arrested in G2 (data not shown). This suggests that Cdc2–Cdc13 kinase activity exerts a regulatory role, thereby controlling either a phosphatase that dephosphorylates Crb2 or a kinase that modifies Crb2 upon IR. We speculate that premature loss of the IR-induced hyperphosphorylation of Crb2 terminates Top3 function before all recombination intermediates are processed.
Crb2 shares several features with the tumor suppressor protein BRCA1. Each protein contains two BRCT domains at the C terminus, both proteins are modified by CDKs, and, finally, both proteins become hyperphosphorylated in a checkpoint-dependent manner upon IR (Scully et al. 1997; Ruffner et al. 1999). BRCA1 seems to act as a scaffold protein to allow assembly of both checkpoint proteins and repair enzymes (Paull et al. 2000; Wang et al. 2000). Interestingly, among the proteins that bind to BRCA1 is the BLM DNA helicase (Wang et al. 2000). We suggest a similar scaffold role for Crb2. In response to IR, checkpoint-dependent hyperphosphorylation of Crb2 might promote an interaction with Top3 that is essential for Top3 activity. Consistent with this view, Crb2 interacts in the two-hybrid system with the BRCT domain of Cut5 (Saka et al. 1997) and the human homolog of Cut5, TopBP1, binds to topoisomerase II (Makiniemi et al. 2001).
The recent finding that BRCA1 binds directly to branched DNA structures and four-way junctions (Paull et al. 2001), the same DNA structures recognized by BLM and Sgs1 helicases, suggests an alternative, but not mutually exclusive, explanation for the function of Crb2: Crb2 could directly participate in the processing of recombination intermediates. Such a function would be consistent with the observation that the S. cerevisiae homolog of Crb2, Rad9, is involved in the formation of ssDNA following DNA damage near telomeres (Lydall and Weinert 1995; Booth et al. 2001).
Cdc2–cyclin B performs another function at an early stage in HR
The first step to be observed after DSB formation is a resection of the DNA ends resulting in 3′ ssDNA tails. Whether this resection is by a 5′–3′ exonuclease or by an endonuclease associated with a helicase is not known. However, loss of the Rad50–Mre11–Xrs2 protein complex retards the rate of 5′–3′ exonuclease activity in vivo (Ivanov et al. 1994). The fact that rad50-d cells still show end-processing suggests the existence of redundant pathways. This is consistent with our observation that deletion of rad50 only reduces, but does not abolish Rhp51 foci formation upon IR (Fig. 3B,C). Interestingly, in these cells assembly of Rhp51 foci is highly dependent on a functional Cdc2–cyclin B kinase (Fig. 3B,C), indicating that CDK activity controls an early step in HR that is redundant with Rad50. In S. cerevisiae, one endonuclease that appears to act as a backup system for Rad50 is Exo1 (Tsubouchi and Ogawa 2000). Given this information, we explored the role of Exo1 in S. pombe. Like cdc13-245 rad50-d cells, an exo1-d rad50-d double mutant was unable to assemble Rhp51 foci 1 h postirradiation (data not shown). However, we have been unable to find further evidence for a functional link between Exo1 and Cdc2–cyclin B kinase.
Conclusion
Our data provide a link between Cdc2–cyclin B activity, the BRCT-domain-containing checkpoint protein Crb2, and the control of DNA repair in G2. Intriguingly, the closest analog to Crb2 in mammalian cells is BRCA1, which is also known to be CDK-phosphorylated (Ruffner et al. 1999) and which has been implicated in channeling DSBs into different repair pathways (Moynahan et al. 1999). It is tempting to speculate that the control of recombinational repair by CDK complexes reflects a fundamental mechanism whereby the chromatin's replication status, and therefore the repair processes most appropriate for DSB repair, are ‘hard wired‘ into the chromatin. Mechanistically, we have implicated the BRCT-domain protein Crb2 in this process, where it appears to control a late stage of recombination by influencing the activity of Top3 and its associated RecQ-like DNA helicase, Rqh1.
Materials and methods
Genetics and cell biology techniques
Strains were constructed by standard genetic techniques. The protocols for checkpoint measurements, cell scoring, irradiation, lactose gradient centrifugation, centrifugal elutriation, and fluorescence-activated cell sorter (FACS) analysis are all described in Edwards and Carr (1997). Indirect immunofluorescence microscopy was performed according to the protocol previously published in Caspari et al. (2000), with the following changes: Both the anti-MYC antibody and the anti-hRad51 (Santa Cruz H-92) were diluted 1:100, whereas the anti-Rqh1 antibody was diluted 1:400. To determine the percentage of cells showing nuclear foci, we visually scored 1000 cells for each sample.
Isolation of the cdc13-245 mutant
Wild-type cells (leu1-32 ura4-D18 ade6-704 h−) were subjected to EMS (ethylmethane sulfonate) mutagenesis as described in Lawrence (1991). Mutants were grown on rich medium plates at 27°C and replica-plated onto two sets of plates, which were incubated at 27°C and at 36°C. Temperature-sensitive strains were isolated and tested for their sensitivity to various DNA-damaging agents. Sensitive strains were crossed three times with the wild-type strain to determine whether the sensitivity relates to a single mutation. The restrictive temperature for the cdc13-245 strain is 33°C. All experiments presented here were performed at the permissive temperature of 27°C.
Construction of the crb2-T215A mutant
Strain construction was based on the method described in Edwards et al. (1999). The crb2-T215A mutant was constructed as follows: The sequence between S43 and T215 (including both codons) was replaced by ura4+ in a wild-type background. The deletion mutant was transformed with an NdeI fragment containing the T215A single mutation. Strains that had replaced the ura4+ gene with the mutated crb2 fragment were counterselected on YEA plates containing 1 mg/mL 5-FOA (5-fluoroorotic acid).
Cell extracts, immunoprecipitation, and in vitro kinase assay
The protocols for total cell extracts, soluble extracts, and immunoprecipitation experiments are all described in Caspari et al. (2000). Insoluble extracts were prepared with the following changes: In buffer B, sodium phosphate was substituted by 100 mM HEPES (pH 7.0; called buffer D). Cells were broken with glass beads in a Ribolyser (Hybaid); 400 μL of buffer D was added after homogenization, and the cell homogenate was spun (Heraeus, 4K) into a new test tube. The protocol for the in vitro Cdc2 kinase assay is published in Moreno et al. (1989).
Recombination assay
Mitotic recombination was assayed by the recovery of Ade6+ recombinants from strains containing the recombination substrate described in Schuchert and Kohli (1988). The assay was performed according to the protocol published in Paulovich et al. (1998). To assay the recombination frequency in response to IR, cells were treated with the indicated doses of IR before being plated onto selective medium.
Acknowledgments
We thank Fumiko Esashi and Mitsuhiro Yanagida for providing the crb2 alleles used for integration and their help with the phospho-specific antibody. We are grateful to Stefania Francesconi for sending us the MYC-crb2 strain and to Edgar Hartsuiker for providing the rqh1-d rad50-d double mutant. Finally, we would like to thank Jacky Hales for the anti-Cdc13 antibodies. This work was funded by grants from the MRC to T.C. and A.M.C. (E160/121-4668), the Cancer Research Campaign to J.M.M.(SP2396/0201), and the HFSP to A.M.C. (RGO-178/2000-M).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL A.M.Carr@sussex.ac.uk; FAX 44-1273-678-121.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.221402.
References
- Aoufouchi S, Prigent C, Ford C, Thiebaud P, Philippe M, Theze N. Cyclin B/p34cdc2 triggers phosphorylation of DNA ligase I during Xenopus laevis oocyte maturation. Eur J Biochem. 1995;230:491–497. [PubMed] [Google Scholar]
- Bennett RJ, Wang JC. Association of yeast DNA topoisomerase III and Sgs1 DNA helicase: Studies of fusion proteins. Proc Natl Acad Sci. 2001;98:11108–11113. doi: 10.1073/pnas.201387098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RJ, Keck JL, Wang JC. Binding specificity determines polarity of DNA unwinding by the Sgs1 protein of S. cerevisiae. J Mol Biol. 1999;289:235–248. doi: 10.1006/jmbi.1999.2739. [DOI] [PubMed] [Google Scholar]
- Bischof O, Kim S-H, Irving J, Beresten S, Ellis NA, Campisi J. Regulation and localisation of the Bloom syndrome protein in response to DNA damage. J Cell Biol. 2001;153:367–380. doi: 10.1083/jcb.153.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booher R, Beach D. Involvement of cdc13+ in mitotic control in Schizosaccharomyces pombe: Possible interaction of the gene product with microtubules. EMBO J. 1988;7:2321–2327. doi: 10.1002/j.1460-2075.1988.tb03075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth C, Griffith E, Brady G, Lydall D. Quantitative amplification of single-stranded DNA (QAOS) demonstrates that cdc13-1 mutants generate ssDNA in a telomere to centromere direction. Nucleic Acids Res. 2001;29:4414–4422. doi: 10.1093/nar/29.21.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspari T, Dahlen M, Kanter-Smoler G, Lindsay HD, Hofmann K, Papadimitriou K, Sunnerhagen P, Carr AM. Characterization of Schizosaccharomyces pombe Hus1: A PCNA-related protein that associates with Rad1 and Rad9. Mol Cell Biol. 2000;20:1254–1262. doi: 10.1128/mcb.20.4.1254-1262.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Nastasi A, Shen Z, Brenneman M, Crissman H, Chen DJ. Cell cycle-dependent protein expression of mammalian homologues of yeast DNA double-strand break repair genes Rad51 and Rad52. Mutat Res. 1997;384:205–211. doi: 10.1016/s0921-8777(97)00020-7. [DOI] [PubMed] [Google Scholar]
- Decottignies A, Zarzov P, Nurse P. In vivo localisation of fission yeast cyclin-dependent kinase cdc2 and cyclin B cdc13 during mitosis and meiosis. J Cell Sci. 2001;114:2627–2640. doi: 10.1242/jcs.114.14.2627. [DOI] [PubMed] [Google Scholar]
- Doe CL, Dixon J, Osman F, Whitby MC. Partial suppression of the fission yeast rqh1− phenotype by expression of a bacterial Holliday junction resolvase. EMBO J. 2000;19:2751–2762. doi: 10.1093/emboj/19.11.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A, Stillman B. cdc2 family kinases phosphorylate a human cell DNA replication factor, RPA, and activate DNA replication. EMBO J. 1992;11:2189–2199. doi: 10.1002/j.1460-2075.1992.tb05278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards RJ, Carr AM. Analysis of radiation-sensitive mutants of fission yeast. Meth Enzymol. 1997;283:471–494. doi: 10.1016/s0076-6879(97)83038-8. [DOI] [PubMed] [Google Scholar]
- Edwards RJ, Bentley NJ, Carr AM. A Rad3–Rad26 complex responds to DNA damage independently of other checkpoint proteins. Nat Cell Biol. 1999;1:393–398. doi: 10.1038/15623. [DOI] [PubMed] [Google Scholar]
- Esashi F, Yanagida M. Cdc2 phosphorylation of Crb2 is required for reestablishing cell cycle progression after the damage checkpoint. Mol Cell. 1999;4:167–174. doi: 10.1016/s1097-2765(00)80364-0. [DOI] [PubMed] [Google Scholar]
- Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: A potential eukaryotic reverse gyrase. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S, de Massey B, Arthur L, Rothstein R, Fabre F. The essential role of yeast topoisomerase III in meiosis depends on recombination. EMBO J. 1999;18:1701–1711. doi: 10.1093/emboj/18.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin A, Wang S-W, Toda T, Norbury C, Hickson ID. Topoisomerase III is essential for accurate nuclear division in Schizosaccharomyces pombe. Nucleic Acids Res. 1999;27:4050–4058. doi: 10.1093/nar/27.20.4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I, Hayles J, Nurse P. Cloning and sequencing of the cylin-related cdc13+ gene and a cytological study of its role in fission yeast mitosis. J Cell Sci. 1988;91:587–595. doi: 10.1242/jcs.91.4.587. [DOI] [PubMed] [Google Scholar]
- Harmon FG, DiGate RJ, Kowalczykowski SC. RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: A conserved mechanism for control of DNA recombination. Mol Cell. 1999;3:611–620. doi: 10.1016/s1097-2765(00)80354-8. [DOI] [PubMed] [Google Scholar]
- Hartsuiker E, Vaessen B, Carr AM, Kohli J. Fission yeast Rad50 stimulates sister chromatid recombination and links cohesion with repair. EMBO J. 2001;20:6660–6671. doi: 10.1093/emboj/20.23.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayles J, Fisher D, Woollard A, Nurse P. Temporal order of S phase and mitosis in fission yeast is determined by the state of the p34(cdc2)–mitotic B cyclin complex. Cell. 1994;78:813–822. doi: 10.1016/s0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- Ivanov EL, Sugawara N, White CI, Fabre F, Haber JE. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3414–3425. doi: 10.1128/mcb.14.5.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov EL, Sugawara N, Fishman Lobell J, Haber JE. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow JK, Constantinou A, Li J-L, West SC, Hickson ID. The Bloom’s syndrome gene product promotes branch migration of Holliday junctions. Proc Natl Acad Sci. 2000;97:6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus E, Leung W-Y, Haber JE. Break-induced replication: A review and an example in budding yeast. Proc Natl Acad Sci. 2001;98:8255–8262. doi: 10.1073/pnas.151008198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CW. Classical mutagenesis techniques. Methods Enzymol. 1991;194:273–280. doi: 10.1016/0076-6879(91)94021-4. [DOI] [PubMed] [Google Scholar]
- Liberi G, Chiolo I, Pellicioli A, Lopes M, Plevani P, Muzi-Falconi M, Foiani M. Srs2 DNA helicase is involved in checkpoint response and its regulation requires a functional Mec1-dependent pathway and Cdk1. EMBO J. 2000;19:5027–5038. doi: 10.1093/emboj/19.18.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydall D, Weinert T. Yeast checkpoint genes in DNA damage processing: Implications for repair and arrest. Science. 1995;270:1488–1491. doi: 10.1126/science.270.5241.1488. [DOI] [PubMed] [Google Scholar]
- Makiniemi M, Hillukkala T, Tuusa J, Reini K, Vaara M, Huang D, Pospiech H, Majuri I, Westerling T, Makela TP, et al. BRCT domain-containing protein TopBP1 functions in DNA replication and damage response. J Biol Chem. 2001;276:30399–30406. doi: 10.1074/jbc.M102245200. [DOI] [PubMed] [Google Scholar]
- McCready SJ, Osman F, Yasui A. Repair of UV damage in the fission yeast Schizosaccharomyces pombe. Mutat Res. 2000;451:197–210. doi: 10.1016/s0027-5107(00)00050-6. [DOI] [PubMed] [Google Scholar]
- Meyn MA, Holloway SL. S-phase cyclins are required for a stable arrest at metaphase. Curr Biol. 2000;10:1599–1602. doi: 10.1016/s0960-9822(00)00863-0. [DOI] [PubMed] [Google Scholar]
- Michel B, Flores M-J, Viguera E, Grompone G, Seigneur M, Bidnenko V. Rescue of arrested replication forks by homologous recombination. Proc Natl Acad Sci. 2001;98:8181–8188. doi: 10.1073/pnas.111008798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohaghegh P, Hickson ID. DNA helicase deficiencies associated with cancer predisposition and premature aging disorders. Hum Mol Genet. 2001;10:741–746. doi: 10.1093/hmg/10.7.741. [DOI] [PubMed] [Google Scholar]
- Moreno S, Hayles J, Nurse P. Regulation of p34cdc2 protein kinase during mitosis. Cell. 1989;58:361–372. doi: 10.1016/0092-8674(89)90850-7. [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- Mullen JR, Kaliraman V, Brill SJ. Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001;154:1101–1114. doi: 10.1093/genetics/154.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muris DFR, Vreeden K, Carr AM, Broughton BC, Lehmann AR, Lohman PHM, Pastink A. Cloning the RAD51 homologue of Schizosaccharomyces pombe. Nucleic Acids Res. 1993;21:4586–4591. doi: 10.1093/nar/21.19.4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muris DFR, Vreeken K, Schmidt H, Ostermann K, Clever B, Lohman PHM, Pastink A. Homologous recombination in the fission yeast Schizosaccharomyces pombe: Different requirements for the rhp51+, rhp54+ and rad22+ genes. Curr Genet. 1996;31:248–254. doi: 10.1007/s002940050202. [DOI] [PubMed] [Google Scholar]
- Murray JM, Lindsay HD, Munday CA, Carr AM. Role of Schizosaccharomyces pombe RecQ homolog, recombination, and checkpoint genes in UV damage tolerance. Mol Cell Biol. 1997;17:6868–6875. doi: 10.1128/mcb.17.12.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Erdjument-Bromage H, Pan Z-Q, Lee S-H, Tempst P, Hurwitz J. Mapping of amino acid residues in the p34 subunit of human single-stranded DNA-binding protein phosphorylated by DNA-dependent kinase and Cdc2 kinase in vitro. J Biol Chem. 1997;272:12634–12641. doi: 10.1074/jbc.272.19.12634. [DOI] [PubMed] [Google Scholar]
- Paques F, Haber JE. Multiple pathways of double-strand-break-induced recombination in Saccharomyces cerevisiae. Mol Microbiol. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- Paull TT, Cortez D, Bowers B, Elledge SJ, Gellert M. Direct DNA binding by Brca1. Proc Natl Acad Sci. 2001;98:6086–6091. doi: 10.1073/pnas.111125998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulovich AG, Armour CD, Hartwell LH. The Saccharomyces cerevisiae RAD9, RAD17, RAD24 and MEC3 genes are required for tolerating irreparable, ultraviolet-induced DNA damage. Genetics. 1998;150:75–93. doi: 10.1093/genetics/150.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin N, Haber JE. Efficient repair of HO-induced chromosomal breaks in Saccharomyces cerevisiae by recombination between flanking homologous sequences. Mol Cell Biol. 1988;8:3918–3928. doi: 10.1128/mcb.8.9.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffner H, Jiang W, Craig AG, Hunter T, Verma IM. BRCA1 is phosphorylated at serine 1497 in vivo at a cyclin-dependent kinase 2 phosphorylation site. Mol Cell Biol. 1999;19:4843–4854. doi: 10.1128/mcb.19.7.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka Y, Esashi F, Matsusaka T, Mochida S, Yanagida M. Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes & Dev. 1997;11:3387–3400. doi: 10.1101/gad.11.24.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchert P, Kohli J. The ade6-M26 mutation of Schizosaccharomyces pombe increases the frequency of crossing over. Genetics. 1988;119:507–515. doi: 10.1093/genetics/119.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M, Feunteun J, Livingston DM. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- Stewart E, Chapman C, Al-khodairy F, Carr AM, Enoch T. rhq1+, a fission yeast gene related to the Bloom's and Werner's syndrome genes, is required for reversible S-phase arrest. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro S, Walter J, Shinohara A, Kamada N, Cremer T. Rad51 accumulation at sites of DNA damage and in postreplicative chromatin. J Cell Biol. 2000;150:283–291. doi: 10.1083/jcb.150.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H, Ogawa H. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:2221–2233. doi: 10.1091/mbc.11.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Nasmyth K. Cohesion between sister chromatids must be established during DNA replication. Curr Biol. 1998;8:1095–1101. doi: 10.1016/s0960-9822(98)70463-4. [DOI] [PubMed] [Google Scholar]
- Van Dyck E, Stasiak AZ, Stasiak A, West SC. Binding of double-strand breaks in DNA by human Rad52 protein. Nature. 1999;398:728–731. doi: 10.1038/19560. [DOI] [PubMed] [Google Scholar]
- Wallis JW, Chrebet G, Brodsky G, Rolfe M, Rothstein R. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989;58:409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes & Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- Wilson J, Wilson S, Warr N, Watts FZ. Isolation and characterization of the Schizosaccharomyces pombe rhp9 gene: A gene required for the DNA damage checkpoint but not the replication checkpoint. Nucleic Acids Res. 1997;25:2138–2146. doi: 10.1093/nar/25.11.2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Davies SI, Levitt NC, Hickson ID. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J Biol Chem. 2001;276:19375–19381. doi: 10.1074/jbc.M009471200. [DOI] [PubMed] [Google Scholar]
- Zheng X-F, Ruderman JV. Functional analysis of the P box, a domain in cyclin B required for the activation of Cdc25. Cell. 1993;75:155–164. [PubMed] [Google Scholar]