Abstract
During heart development, chamber myocardium forms locally from the embryonic myocardium of the tubular heart. The atrial natriuretic factor (ANF) gene is specifically expressed in this developing chamber myocardium and is one of the first hallmarks of chamber formation. We investigated the regulatory mechanism underlying this selective expression. Transgenic analysis shows that a small fragment of the ANF gene is responsible for the developmental pattern of endogenous ANF gene expression. Furthermore, this fragment is able to repress cardiac troponin I (cTnI) promoter activity selectively in the embryonic myocardium of the atrioventricular canal (AVC). In vivo inactivation of a T-box factor (TBE)- or NK2-homeobox factor binding element (NKE) within the ANF fragment removed the repression in the AVC without affecting its chamber activity. The T-box family member Tbx2, encoding a transcriptional repressor, is expressed in the embryonic myocardium in a pattern mutually exclusive to ANF, thus suggesting a role in the suppression of ANF. Tbx2 formed a complex with Nkx2.5 on the ANF TBE–NKE, and was able to repress ANF promoter activity. Our data provide a potential mechanism for chamber-restricted gene activity in which the cooperative action of Tbx2 and Nkx2.5 inhibits expression in the AVC.
Keywords: Heart development, chamber formation, transgenic mice, ANF, Tbx2, Nkx2.5
The vertebrate heart is first formed as a linear tube, which subsequently loops and transforms into the definitive four-chambered heart. The events that lead to the formation of the mature heart have been described (Fishman and Chien 1997; Srivastava and Olson 2000), but the mechanisms that underlie the formation of the chambers are still largely undefined. The linear heart tube is patterned along three body axes and has an embryonic phenotype (i.e., ability to spontaneously dipolarise [automaticity], slow contraction, poor intercellular coupling, and poorly developed sarcoplasmic reticulum and sarcomeres). Positional information guides the localized development of different components of the heart. At specific sites of the looping tubular heart, trabeculated ventricular and atrial chamber myocardium is formed from this embryonic myocardium. In contrast to the embryonic myocardium, the chamber myocardium has lost its automaticity, has a fast contraction pattern reminiscent of the working myocardium of the mature heart, and is well coupled intercellularly (Moorman et al. 1998). The chamber myocardium specifically initiates the expression of gap-junction genes connexin (Cx) 40 and Cx43 required for intercellular coupling (Delorme et al. 1997), and other genes including ANF and Chisel (Christoffels et al. 2000; Palmer et al. 2001). Thus, chamber formation requires the localized initiation of a transcriptional differentiation program. The smooth-walled myocardium of the inflow tract (IFT), atrioventricular canal (AVC), and inner curvature and outflow tract (OFT) retains the embryonic myocardial phenotype longer, and concomitantly does not express Cx40, Cx43, ANF, and Chisel. These regions are crucial for septation and they also contribute to the formation of the nodal components of the conduction system (i.e., sino-atrial node, atrioventricular node, and atrioventricular junction myocardium), which share phenotypic characteristics with the embryonic myocardium (Moorman et al. 1998; Davis et al. 2001). As many cardiac malformations find their origin in the incorrect development of these embryonic regions, knowledge regarding the mechanisms behind the regulation of the site-specific differentiation program is essential.
The ANF gene is ideal to analyze the molecular mechanisms that may underlie the localized formation of atrial and ventricular chamber myocardium within the linear heart tube. First, although in the mature heart ANF gene expression is restricted to the atrial auricles, during development its expression is specific for the forming ventricular and atrial chambers. It therefore serves as a marker gene for the chamber myocardium (Christoffels et al. 2000). Second, the regulation of the ANF gene has been well characterized and serves as a paradigm for the regulatory mechanisms that control cardiac gene expression. Previously, a 0.7-kb upstream fragment of the ANF gene was shown to be sufficient for cardiac-specific gene expression in cultured cardiomyocytes and transgenic mice (Field 1988; Argentin et al. 1994; Knowlton et al. 1995), although the developmental pattern of the transgene was not reported. A number of general and cardiac-enriched transcription factors were shown to interact with this fragment. Of these, the NK2 homeobox factor Nkx2.5 and T-box factor Tbx5 were shown to be required for ANF gene expression in vivo (Lyons et al. 1995; Tanaka et al. 1999; Bruneau et al. 2001). Inactivation of either factor in Xenopus and mouse results in severely affected heart development. Moreover, mutations in the genes encoding these factors in human and mouse result in congenital cardiac malformations including septum defects and conduction disease (Basson et al. 1997; Li et al. 1997; Schott et al. 1998; Bruneau et al. 2001). In vitro studies showed that Tbx5 and Nkx2.5 associate and synergistically activate the ANF regulatory fragment (Bruneau et al. 2001; Hiroi et al. 2001). Although these studies have greatly advanced our understanding of the regulation of heart-specific gene expression, the mechanism for the chamber specificity remained unclear.
In this study we show that the 0.7-kb ANF fragment is responsible for the developmental pattern of the ANF gene. A part of this fragment was able to repress the activity of a cardiac troponin I (cTnI) promoter fragment specifically in the AVC. In vivo inactivation of an NK-2 homeobox factor binding element (NKE) or T-box factor binding element (TBE) within the ANF fragment did remove the repression in the embryonic myocardium of the AVC, whereas the activity in the chamber myocardium was not affected. Additional analysis showed that Tbx2 gene expression is restricted to the embryonic areas of the developing heart in a pattern complementary to ANF. Tbx2 and Nkx2.5 formed a complex on the TBE–NKE site within the ANF fragment, and Tbx2 was able to repress the activity of the ANF fragment. Our data suggest a novel mechanism for the site-specific formation of chamber myocardium by localized repression of the differentiation program within the embryonic heart.
Results
The ANF regulatory region is active in atrial and ventricular chamber myocardium
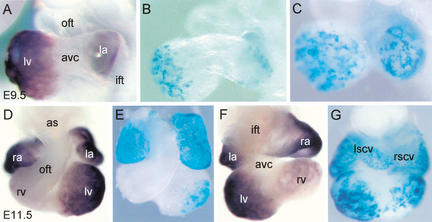
We first assessed whether the ANF regulatory region is capable of driving reporter gene expression specifically in the atrial and ventricular chamber myocardium of the developing heart. Therefore, we generated transgenic mice harboring this ANF regulatory region (−638/+70) coupled to the nlacZ reporter gene. Heart-specific reporter gene expression was analyzed by whole-mount X-gal staining of mouse (E9.5 and E11.5) embryos (Fig. 1). At E9.5, expression of the reporter gene was observed in the atrial and ventricular chamber myocardium, whereas expression was absent from the embryonic myocardium of AVC, inner curvature, and OFT (Table 1; Fig. 1B,C). At stage E11.5, nlacZ expression is still present in both atria and both ventricles, whereas the expression is higher in the LV as compared with the RV (Fig. 1E,G). At both stages, the transgene expression pattern is comparable with that of the endogenous ANF gene (Fig. 1A,D,F). The only exceptions were the right and left superior caval veins that express the transgene, but not the endogenous gene (Fig. 1F,G). Therefore, the 0.7-kb ANF regulatory region mimics the endogenous developmental expression pattern in the mouse heart and selectively demarcates the atrial and ventricular chamber myocardium.
Figure 1.
The 0.7-kb ANF regulatory region is responsible for the developmental pattern of the endogenous ANF gene. (A) A lateral view of the endogenous ANF gene expression in the heart of E9.5 mouse embryo. (B,C) A lateral view of the nlacZ reporter gene expression in the heart of E9.5 transgenic embryos. (D,F) A ventral (D) and dorsal (F) view of the endogenous ANF expression at E11.5. (E,G) A ventral (E) and dorsal (G) view of ANF transgene expression at E11.5. (ift) Inflow tract; (la) left atrium; (ra) right atrium; (avc) atrioventricular canal; (lv) left ventricle; (rv) right ventricle; (oft) outflow tract; (lscv) left superior caval vein; (rscv) right superior caval vein; (as) aortic sac.
Table 1.
Reporter gene expression data of ANFcTnI, ANF-cTnI, MLC2V-cTnI, ANFmutNKE-cTnI, ANFmutTBE-cTnI, and ANFmutTBE/NKE transgenic mice at E10.5
| Construct
|
IFT
|
LA
|
RA
|
AVC
|
LV
|
RV
|
OFT
|
Ectopic expression
|
|---|---|---|---|---|---|---|---|---|
| ANF 1a | ++ | +++ | +++ | − | ++ | − | − | Yes |
| ANF 2a | + | +++ | +++ | − | +++ | ++ | − | Yes |
| ANF 3a | − | − | − | − | − | − | − | Yes |
| cTnIa | − | − | +/− | + | − | − | − | No |
| cTnIa | − | − | +/− | ++ | − | − | − | No |
| cTnIa | − | + | ++ | ++ | ++ | + | − | Yes |
| cTnI | − | − | − | + | − | − | − | Yes |
| cTnI | − | − | + | + | +/− | − | − | No |
| cTnI | − | − | + | ++ | − | − | − | Yes |
| cTnI | − | +/− | ++ | ++ | + | − | − | Yes |
| cTnI | − | − | + | ++ | + | − | − | No |
| cTnI | − | − | ++ | +++ | ++ | − | − | No |
| cTnI | − | − | ++ | +++ | ++ | − | − | No |
| ANF-cTnIa | − | + | + | − | + | − | − | No |
| ANF-cTnIa | − | ++ | ++ | − | + | − | − | No |
| ANF-cTnIa | − | ++ | ++ | − | ++ | +/− | − | No |
| ANF-cTnIa | − | +++ | +++ | − | +++ | +/− | − | No |
| ANF-cTnI | − | ++ | ++ | − | ++ | − | − | Yes |
| ANF-cTnI | − | +++ | +++ | − | +++ | ++ | − | No |
| MLC2V-cTnIa | − | − | ++ | +++ | ++ | − | − | No |
| MLC2V-cTnIa | − | − | ++ | ++ | + | − | − | Yes |
| MLC2V-cTnI | − | +/− | + | + | + | − | − | Yes |
| MLC2V-cTnI | − | ++ | +++ | +++ | +++ | − | − | Yes |
| ANFmutNKE-cTnI | − | + | + | + | + | − | − | No |
| ANFmutNKE-cTnI | + | ++ | ++ | ++ | ++ | − | − | No |
| ANFmutNKE-cTnI | +/− | +++ | +++ | +++ | +++ | − | − | No |
| ANFmutNKE-cTnI | + | ++++ | ++++ | ++++ | ++++ | + | + | Yes |
| ANFmutNKE-cTnI | − | − | − | − | − | − | − | Yes |
| ANFmutTBE-cTnI | − | +/− | +/− | + | +/− | − | − | No |
| ANFmutTBE-cTnI | − | + | + | ++ | + | − | − | No |
| ANFmutTBE-cTnI | − | ++ | ++ | ++ | ++ | + | − | No |
| ANFmutTBE-cTnI | − | ++ | ++ | ++ | ++ | + | − | No |
| ANFmutTBE-cTnI | − | ++ | ++ | ++ | ++ | + | − | No |
| ANFmutTBE-cTnI | − | ++ | ++ | ++ | ++ | + | − | No |
| ANFmutTBE-cTnI | − | +++ | +++ | +++ | +++ | +++ | − | No |
| ANFmutTBE-cTnI | − | +++ | +++ | +++ | +++ | ++ | +/− | No |
| ANFmutTBE-cTnI | − | +++ | +++ | +++ | +++ | +++ | +/− | No |
| ANFmutTBE/NKE-cTnI | − | +++ | +++ | +++ | +++ | +++ | − | No |
| ANFmutTBE/NKE-cTnI | − | ++ | ++ | +++ | ++ | ++ | − | Yes |
| ANFmutTBE/NKE-cTnI | − | + | + | ++ | + | − | − | Yes |
An arbitrary scale of intensity was assigned. (++++) Very strong expression; (+/−) very weak expression; (−) no detectable staining.
The result from multiple embryos of a transgenic mouse line. Each embryo from a line showed an identical expression pattern. Others are single embryos derived from F0 screens. With the exception of the ANF construct, all constructs were flanked by insulators. (ift) Inflow tract; (la) left atrium; (ra) right atrium; (avc) atrioventricular canal; (lv) left ventricle; (rv) right ventricle; (oft) outflow tract.
The cTnI regulatory region is active in the embryonic myocardium of the atrioventricular canal
The observed absence of expression of the ANF transgenes in the embryonic myocardium of the AVC and OFT could result from lack of activation or from active repression in these regions. To discriminate between these two mechanisms, we searched for a minimal cardiac promoter region that is predominantly active in the embryonic myocardium. Coupled to the regulatory sequences of the ANF gene, this minimal cardiac promoter could be used as a read out for lack of activation or active repression in the embryonic myocardium. The cTnI gene is expressed in the entire myocardium (Vallins et al. 1990; Ausoni et al. 1991). The 356-bp promoter region (−230/+126), analyzed in transgenic mice, however, showed a variable pattern of expression, which always included the myocardium of the AVC (Di Lisi et al. 1998, 2000). Furthermore, only 6 of 16 transgenic mice showed expression (R. Di Lisi and S. Schiaffino, pers. comm.). To protect the small cTnI promoter region from position effects, it was flanked by insulator sequences from the chicken β-globin locus (Chung et al. 1993, 1997; Bell et al. 1999), which did not affect the activition of the cTnI promoter by various transcription factors in transient transfection assays (data not shown). As shown in Table 1, all insulated mouse lines and transgenic embryos expressed the transgene in the heart, and transgene expression was always present in the AVC (Fig. 2A). In addition, in 9 of 10 transgenic embryos, expression was extended to the RA and LV (Table 1; Fig. 2A). None of the transgenic embryos showed expression in the myocardium of IFT and OFT. Application of insulator sequences appeared to stabilize the transgene expression pattern and strongly increased the proportion of expressing transgenic mice (Z-test; P = 0.013, insulated vs. noninsulated). Therefore, insulators flanked all further constructs used in this study.
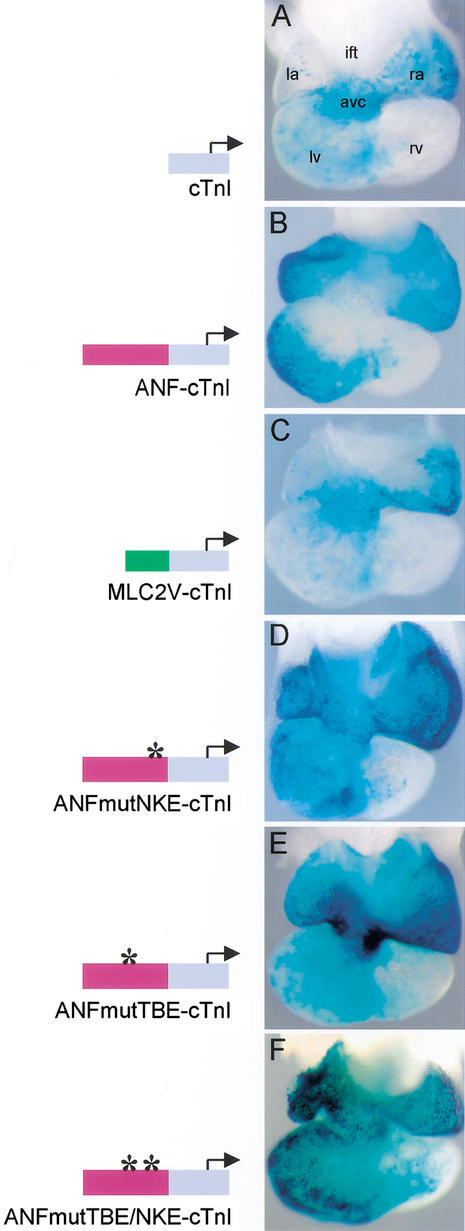
Figure 2.
. Localization of transgene expression in E10.5 mouse hearts. (A) The cTnI transgene is predominantly expressed in the primary myocardium of the AVC. (B) The ANF–cTnI transgene is solely expressed in the chamber myocardium. No transgene expression is present in the AVC myocardium. (C) MLC2V–cTnI transgenics show predominant expression in the primary myocardium of the AVC, similar to the pattern of the cTnI transgenes. (D) Mutation of the NKE at position −250 bp in the ANF regulatory region removes the repression in the AVC. (E) Mutation of the TBE at position −259 bp in the ANF regulatory region also removes the repression in the AVC. (F) Mutation of both the TBE and NKE removes the repression in the AVC as well. (ift) Inflow tract; (la) left atrium; (ra) right atrium; (avc) atrioventricular canal; (lv) left ventricle; (rv) right ventricle; (oft) outflow tract.
The 0.5-kb ANF regulatory region extinguishes cTnI promoter activity in the atrioventricular canal
The −638/−138-bp region of the ANF promoter (Durocher and Nemer 1998) was placed upstream of the otherwise identical cTnI construct and transgenic embryos were generated. All ANF–cTnI transgenic embryos showed a similar expression pattern in the heart (Table 1). At E10.5, transgene expression was observed in the chamber myocardium of the RA, LA, LV, and RV, with lower expression levels in the RV as compared with the LV. No transgene expression was observed in the myocardium of the IFT, AVC, inner curvature, and OFT (Fig. 2B). The expression pattern of these ANF–cTnI transgenes is very similar to the expression pattern of the transgenics that harbor the full 0.7-kb ANF regulatory region (cf. Figs. 1G and 2B). These observations suggest that the characteristic cTnI promoter activity in the AVC (Fig. 2A), is actively repressed by the presence of the 0.5-kb fragment of the ANF promoter. This, in turn, would require the presence of a repressor mechanism that is active in the AVC but not in the atrial and ventricular chamber myocardium.
To determine whether the repressive effects of the ANF regulatory sequences were specific, a third chimeric construct (MLC2V–cTnI) was made, in which we replaced the 0.5-kb ANF regulatory region by a 0.2-kb region (−250/−42) of the MLC2V promoter. This MLC2V promoter region confers right ventricular and OFT expression to a lacZ reporter gene in vivo (Ross et al. 1996), and also in our vector backbone (data not shown). Both MLC2V–cTnI transgenic lines gave similar expression patterns in the heart (Table 1). At E10.5, expression of the transgene was restricted to the RA, AVC, and LV, identical to the pattern of the cTnI transgenes (Fig. 2, cf. A and C). This indicates that the 0.2-kb MLC2V promoter region is not capable of imposing its activity onto the cTnI promoter or of extinguishing expression in the myocardium of the AVC. Therefore, the repression of AVC activity is specific for the ANF fragment.
Inactivation of a high-affinity NKE in the ANF regulatory region removes repression in the atrioventricular canal
Nkx2.5 is important in the control of ANF expression (Lyons et al. 1995; Durocher et al. 1996; Tanaka et al. 1999), and interacts with multiple binding elements (NKEs) within the ANF regulatory fragment, including a high-affinity NKE at position −250 bp (Durocher et al. 1997; Durocher and Nemer 1998; Lee et al. 1998; Shiojima et al. 1999; Hiroi et al. 2001). To analyze whether this NKE is involved in the repressive activity of the ANF fragment, we generated transgenic embryos with the ANF–cTnI construct, in which the NKE is inactivated by mutation. All transgenic embryos with the NKE mutation (ANFmutNKE–cTnI) did show nlacZ expression in the AVC (Table 1; Fig. 2D). Additionally, they showed expression in the RA, LA, LV, and RV similar to ANF (Fig. 1G) and ANF–cTnI transgenes (Fig. 2B). These results show that, in vivo, the NKE is not required for activation of expression in the chambers, but for repression in the AVC. It is not likely that the specific repression in the AVC is solely explained by the function of Nkx2.5, because this transcription factor is expressed in the entire heart (Komuro and Izumo 1993; Lints et al. 1993; Kasahara et al. 1998). Therefore, we assumed that the observed effect of the NKE mutation reflects an interaction between Nkx2.5 and other factors bound to neighboring elements.
Inactivation of a T-box binding element adjacent to the NKE removes repression in the atrioventricular canal
The ANF regulatory region contains a T-box binding element (TBE) in close vicinity (position −259 bp) to the NKE. This TBE is conserved between species, homologous to a T-half site (Kispert et al. 1995), and required for the activation by Tbx5 and Nkx2.5 in transfection assays (Bruneau et al. 2001; Hiroi et al. 2001). We generated transgenic embryos that have an inactivating mutation (Sinha et al. 2000) in the TBE within the ANF–cTnI transgene construct (ANFmutTBE–cTnI). Nine transgenic embryos were analyzed at E10.5 and revealed a similar transgene expression pattern in the heart (Table 1). Similar to the ANFmutNKE–cTnI transgenes, expression was present in the AVC as well as in the RA, LA, LV, and RV (Fig. 2E). These results show that the TBE is essential for the repression by ANF regulatory sequences in the embryonic myocardium of the AVC, but is not essential for activity in the chamber myocardium.
Both TBE and NKE were essential for the synergistic activation of the ANF fragment by Tbx5 and Nkx2.5 in transfection assays. However, inactivation of neither element visibly affected chamber activity in vivo. To investigate whether in vivo the TBE and NKE are redundant for ANF activity, transgenic embryos were generated in which both elements were inactivated (ANFmutTBE/NKE–cTnI). Three transgenic embryos were analyzed at E10.5 and revealed a similar transgene expression pattern in the heart (Table 1). Similar to the ANFmutNKE–cTnI and ANFmutTBE–cTnI transgenes, expression was present in the AVC as well as in the RA, LA, LV, and RV (Fig. 2F). These results show that both elements are dispensable for chamber activity.
The transcription factor Tbx2 is expressed in the embryonic myocardium
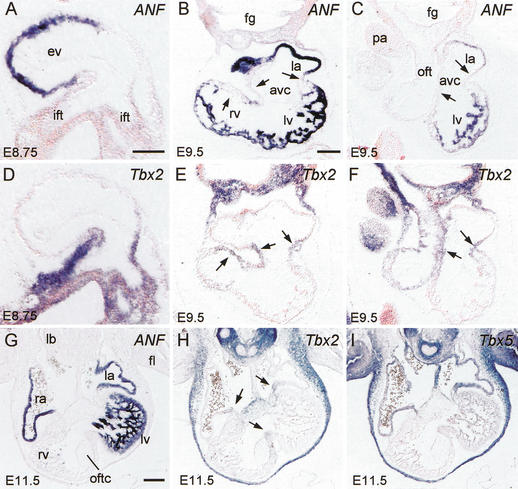
Tbx2 is a T-box factor family member that acts as a transcriptional repressor (Carreira et al. 1998; Jacobs et al. 2000; Sinha et al. 2000), and is expressed in the AVC of the chicken and mouse heart (Gibson-Brown et al. 1998; Yamada et al. 2000). To explore its possible involvement in ANF gene regulation, we analyzed the pattern of Tbx2 mRNA in the developing mouse heart by nonradioactive in situ hybridization on serial sections. At E8.75, the Tbx2 gene was present in the embryonic myocardium of the IFT and AVC (Fig. 3D). At this stage, ANF is selectively expressed in the ventricular myocardium and absent from the IFT (Fig. 3A). At E9.5, Tbx2 is expressed in the IFT, AVC, inner curvature, and in the OFT (Fig. 3E,F). No Tbx2 expression could be observed in the atrial and ventricular chamber myocardium (Fig. 3E,F). The pattern of ANF is strictly complementary to that of Tbx2, and is restricted to the chamber myocardium of both atria and ventricles and absent from the embryonic myocardium of the IFT, AVC, inner curvature, and OFT (Fig. 3B,C). At E11.5, Tbx2 is expressed in the AVC and OFT (Fig. 3H). The pattern is complementary to the pattern of ANF that is expressed in the atrial appendages and the LV (Fig. 3G).
Figure 3.
Nonradioactive in situ hybridization on serial sections shows complementary expression of endogenous ANF, Tbx2, and Tbx5 mRNA. (A–C,G) ANF expression at E8.75 (A), E9.5 (B,C), and E11.5 (G). (D–F,H) Tbx2 expression at E8.75 (D), E9.5 (E,F), and E11.5 (H). (I) Tbx5 expression at E11.5. Note the mutually exclusive pattern of expression of ANF and Tbx2. Arrows in B and E indicate the AVC and OFT region that is continuous at the inner curvature. Arrows in C, F, and H indicate the AVC region. (ift) Inflow tract; (la) left atrium; (ra) right atrium; (avc) atrioventricular canal; (lv) left ventricle; (rv) right ventricle; (oft) outflow tract; (ev) embryonic ventricle; (fg) foregut; (pa) pharyngeal arch; (oftc) outflow tract cushion; (lb) lung bud; (fl) forelimb. Bar, 100 μm.
At E11.5, the Tbx5 gene, encoding a transcriptional activator involved in ANF gene regulation (Bruneau et al. 2001; Ghosh et al. 2001; Hiroi et al. 2001), showed expression in the myocardium of RA, LA, AVC, and LV (Fig. 3I). Expression of both Tbx5 and ANF is virtually absent from the RV and OFT. The Tbx5 gene expression pattern overlaps that of ANF, Tbx5 being additionally expressed in the embryonic myocardium of the IFT, AVC, inner curvature, and in the atrial septum (Fig. 3G,I). Cx40 is, like ANF, also under control of Tbx5 (Bruneau et al. 2001). The expression of Cx40 (Delorme et al. 1997) is similar to the ANF expression pattern, being also complementary to the pattern of Tbx2 (data not shown).
Tbx2 and Nkx2.5 form a ternary complex with the ANF TBE–NKE
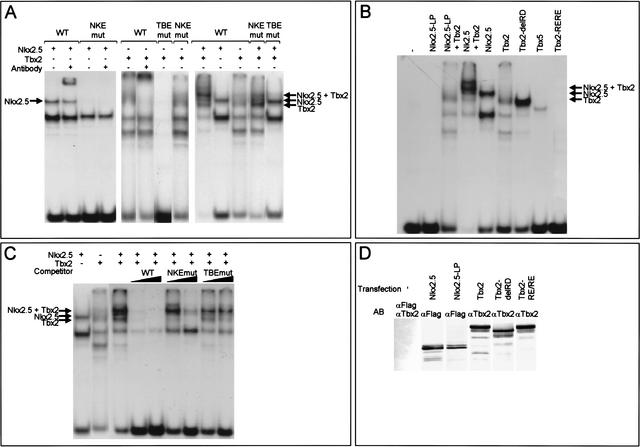
The requirement of the TBE and NKE for repression in the AVC and the complementarity in expression pattern between Tbx2 and ANF prompted us to study the interaction of Tbx2 and Nkx2.5 with the NKE–TBE site using electromobility shift assay (EMSA) experiments. Oligonucleotide probes were used that correspond to ANF promoter sequences −273 to −236 that harbor both the TBE and NKE (wild type), an intact TBE and a mutated NKE (NKEmut), or a mutated TBE and an intact NKE (TBEmut). Nkx2.5 as well as Tbx2 bound to the wild-type probe and could be supershifted using specific antibodies (Fig. 4A). Nkx2.5 binding was abolished by the NKE mutation (NKEmut probe), whereas Tbx2 binding was not affected (Fig. 4A). Tbx2 binding was abolished by the mutation in TBE (TBEmut probe; Fig. 4A), whereas Nkx2.5 binding was not affected (data not shown). Incubation of both Nkx2.5 and Tbx2 with the wild-type probe produced a larger ternary complex in addition to the Nkx2.5 and Tbx2–DNA complexes (Fig. 4A). Mutation of the TBE abolished ternary complex formation, whereas this ternary complex was still weakly present when the NKE was mutated (Fig. 4A). These results indicate that the TBE, and to a lesser extent, the NKE, are necessary for ternary complex formation. Nkx2.5–L176P, which contains a leucine to proline substitution within the Nkx2.5 DNA-binding domain that inactivates its DNA-binding ability (Grow and Krieg 1998), did not bind to the wild-type probe and did not form a ternary complex with Tbx2 (Fig. 4B). Tbx2–R122E/R123E, in which amino acids involved in DNA interaction were substituted, did not bind the wild-type probe (Fig. 4B), and did not form a complex with Nkx2.5 (data not shown). These results indicate that binding to the DNA of both Nkx2.5 and Tbx2 is necessary for ternary complex formation. Tbx2–delRD also shows binding to the TBE, indicating that the portion carboxy-terminal to the T-box that is involved in repression is not required for DNA binding (Fig. 4B). Compared with the full-length protein, binding of the truncated protein to the TBE is more efficient. A similar observation was made for C-terminal truncated versions of the Tbx5 protein, which were found to increase the affinity for the DNA (Ghosh et al. 2001). To test whether the ternary complex, once formed, was stable, competition assays were performed. A 100-fold excess of unlabeled wild-type probe successfully competed the ternary complex (Fig. 4C). In contrast, even a 1000-fold excess of NKEmut probe produced weak competion, and no competition was observed using the TBEmut probe as competitor (Fig. 4C). These results indicate that the ternary complex, once formed, is stable and is not disrupted by competion for binding with one of the two factors. Western blot analysis showed that nuclear extracts contained TBX2, TBX2-delRD, TBX2-R122E/R123E, FLAG-tagged Nkx2.5, and Nkx2.5-L176P protein (Fig. 4D).
Figure 4.
Tbx2 and Nkx2.5 form a ternary complex with the ANF TBE–NKE. (A) EMSAs were performed using nuclear extracts from HEK cells expressing Nkx2.5 or Tbx2. The wild-type probe contains the TBE–NKE, the NKEmut probe contains the TBE, and the mutated NKE as used in the transgene construct. The TBEmut probe contains the NKE and the mutated TBE as used in the transgene construct. Both Nkx2.5 and Tbx2 bind to the wild-type probe. Nkx2.5 does not bind the NKEmut probe and Tbx2 does not bind the TBEmut probe. When mixing together Nkx2.5 and Tbx2 extracts, an additional ternary complex is formed on the wild type, and to a lesser extent, on the NKEmut probe, whereas the complex is absent when using the TBEmut probe. (B) Replacing Nkx2.5 for Nkx2.5-L176P (NKx2.5-LP) or Tbx2 for Tbx2-R122E/R123E (Tbx2-RE/RE) shows that on the wild-type probe, the DNA-binding ability of both Nkx2.5 and Tbx2 is necessary for complex formation. The carboxy-terminal region of Tbx2 is not required for DNA binding (Tbx2-delRD). Tbx5 is also able to bind the wild-type probe. (C) Once formed, the Nkx2.5/Tbx2 complex is stable. The wild-type probe was incubated with nuclear extracts and cold competitor oligonucleotides (100- and 1000-fold excess) as indicated at top. Whereas a 100-fold excess of wild-type probe was sufficient to disrupt the complex, a 100-fold excess of NKEmut and a 1000-fold excess of TBEmut were not sufficient. (D) Western blots of nuclear extracts of HEK cells expressing FLAG–Nkx2.5, FLAG–Nkx2.5-LP, Tbx2, Tbx2-delRD and Tbx2-RE/RE.
The ANF promoter is a functional target of Tbx2
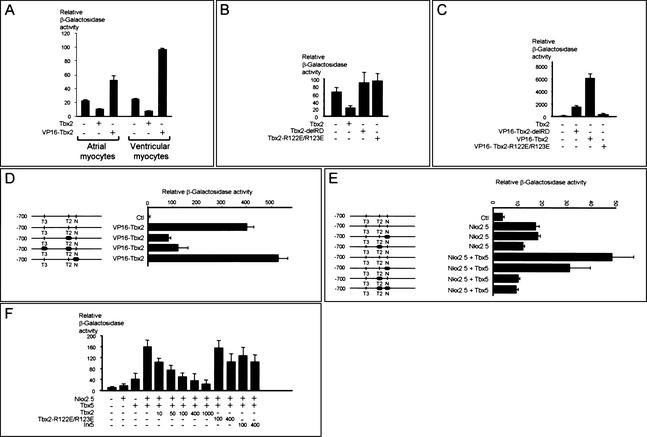
To study whether the ANF regulatory region is a functional target of Tbx2, cotransfections were performed with the 0.7-kb ANF promoter reporter construct. In atrial cultures, cotransfection of full-length Tbx2 resulted in a twofold decrease of ANF promoter activity, whereas cotransfection of Tbx2 without its repressor domain (RD) and fused to the transactivation domain of VP16 (VP16-Tbx2-delRD) gave a twofold increase in ANF promoter activity. Although the effect of cotransfecting these factors is similar in atrial and ventricular cardiomyocyte cultures, the differences are more pronounced in the ventricular cultures (Fig. 5A). In Cos-7 cells, ANF promoter activity decreased threefold upon cotransfection of Tbx2 (Fig. 5B). Tbx2-delRD was unable to repress the ANF promoter, indicating that the repressor domain is essential for the observed repression (Fig. 5B). Cotransfection of VP16-Tbx2-delRD resulted in a drastic increase in activity, which became even more pronouced when VP16-Tbx2 was used (Fig. 5C) that contains the full-length Tbx2 cDNA coupled to VP16. VP16-Tbx2-R122E/R123E was not able to activate the ANF promoter, showing that DNA binding of Tbx2 is essential for regulation of the ANF promoter (Fig. 5C). Together, these data show that the 0.7-kb ANF regulatory region is a target for Tbx2-mediated repression.
Figure 5.
The 0.7-kb ANF promoter is a functional target for Tbx2. (A) Transient transfections were carried out with the 0.7-kb ANF promoter in primary atrial and ventricular cardiomyocytes. Cotransfections show that Tbx2 repressed ANF promoter activity, whereas VP16-Tbx2-delRD activated the ANF promoter. The results are from one representative experiment (out of 3) done in duplicate. Error bars represent the difference between the duplicates. (B) Tbx2-mediated repression requires the Tbx2 repressor domain and interaction with the DNA. Cotransfection experiments were carried out with the 0.7-kb ANF promoter in Cos-7 cells. (C) Fusion of the VP16 transactivation domain to either the full-length Tbx2 (VP16-Tbx2) or to Tbx2, from which the carboxy-terminal end that includes the repression domain, was removed (VP16-Tbx2-delRD) resulted in strong activation of the ANF promoter region. VP16-Tbx2-R122E/R123E did not activate the ANF promoter region. (D) Cotransfection, using point mutations of the 0.7-kb ANF promoter in Cos-7 cells, shows that VP16-Tbx2 activates the ANF regulatory region via the TBE. The basal values of the mutated constructs were comparable with the control construct. (E) Nkx2.5 activation of the ANF promoter does not require the NKE. Synergistic activation of the ANF promoter by Tbx5 and Nkx2.5 requires an intact TBE and NKE. The basal values of the mutated constructs were comparable with the control construct. (F) Synergistic activity of Nkx2.5 and Tbx5 is reduced by Tbx2 in a dose-dependent manner, indicating that Tbx2 can efficiently compete with Tbx5 in the regulation of the ANF promoter. All results are from one representative experiment (out of 3) done in duplicate. Error bars represent the difference between the duplicates. (N) NKE located at position −250 bp; (T2) TBE located at position −259 bp; (T3) TBE located at position −485 bp.
To address whether Tbx2 and Nkx2.5 mediate ANF gene regulation via the TBE and NKE, respectively, an ANF reporter construct containing point mutations in the TBE at −259 bp was transfected in Cos-7 cells (Fig. 5D). Loss of the TBE site diminished the VP16-Tbx2-induced ANF promoter activity. Additional mutation of the TBE located at −485 bp did not further decrease promoter activity. The residual activation of the mutated ANF promoters possibly results from VP16–Tbx2 activation via a potential T-half site located at −90 bp (Bruneau et al. 2001). However, this site is not present in the ANF–cTnI transgene constructs.
Cotransfection of Nkx2.5 resulted in a threefold activation of the 0.7-kb ANF promoter (Fig. 5E). Mutation of the NKE located at −250 did not influence the inducibility of the ANF promoter by Nkx2.5. Possibly, this response is mediated by additional low-affinity NKEs located at −242 and −80 bp in the ANF promoter (Lee et al. 1998; Shiojima et al. 1999).
Previous studies have shown that the TBE is a functional binding site for the transcriptional activator Tbx5 and that the TBE–NKE is involved in synergistic activation of the ANF promoter by Tbx5 and Nkx2.5 (Bruneau et al. 2001; Hiroi et al. 2001). Because both Tbx5 and Tbx2 are expressed in the AVC, we tested whether Tbx2 can compete with Tbx5. The ANF promoter was transfected in Cos-7 cells and cotransfected with Tbx5, Nkx2.5, and increasing amounts of Tbx2 (Fig. 5F). As expected, Tbx5 and Nkx2.5 synergistically activated the ANF promoter (Fig. 5F). The activation depended on both the TBE and NKE (Fig. 5E). Adding as little as 10 to 100 ng of Tbx2 compared with 400 ng of Tbx5 and Nkx2.5 resulted in a loss of induction, indicating that Tbx2 can efficiently compete with Tbx5 in the regulation of ANF promoter activity (Fig. 5F). Adding larger amounts of Tbx2 resulted in an even higher reduction (Fig. 5F). The competition of Tbx2 is specific as both the Tbx2-R122E/R123E and the unrelated factor Irx5 were unable to interfere (Fig. 5F).
Discussion
Chimeric regulatory regions reveal active repression in the atrioventricular canal
In the developing heart, ANF displays a chamber-restricted pattern of expression that is recapitulated by the proximal 0.7-kb ANF regulatory region. Because the AVC activity of the cTnI promoter was extinguished in the ANF–cTnI transgenics, we conclude that the ANF regulatory region actively imposes repression on the cTnI promoter. By studying the 0.7-kb ANF regulatory region itself, and not in the context of chimeric constructs, this property would not have been revealed, and would not have prompted us to search for the repressor function within this region. Dysfunction of the cTnI promoter due to the composition of the chimeric construct is unlikely for a number of reasons. The combined promoter is active in the chambers, similar to the ANF promoter, showing that the construct is transcriptionally competent. When MLC2V sequences were placed upstream of the cTnI promoter, no interference with cTnI promoter activity was detected. When single-site mutations were made in the ANF–cTnI construct, the activity in the AVC was restored. Therefore, the AVC-specific extinction of transcription by ANF sequences can be attributed to an intrinsic repressor function.
The NKE and TBE are essential for repression in the atrioventricular canal
The removal of repression in the AVC by inactivation of the NKE or TBE site revealed that an NK2 factor, probably Nkx2.5, and a T-box factor are components of an inhibitory pathway. The pattern of Tbx2 gene expression, and the ability of Tbx2 to repress the ANF promoter and to bind to the ANF TBE–NKE site together with Nkx2.5 indicate that this TBE is a target for Tbx2. On the basis of these findings, we propose that Nkx2.5 and Tbx2 cooperatively repress the ANF promoter in the AVC. Preliminary data showed that the MLC2v promoter, active in the OFT and RV, is extinguished in the OFT by the 500-bp ANF regulatory region, suggesting that this pathway is also active in the OFT (P.E.M.H. Habets, A.F.M. Moorman, and V.M. Christoffels, unpubl.). In this repression mechanism, Nkx2.5 functions as a cardiac accessory factor for Tbx2, which in turn represses the ANF promoter in the AVC. The accessory function of Nkx2.5 is in line with the ability of this factor to cooperate with members of several classes of factors, including GATA factors, SRF, and Tbx5 in the regulation of cardiac genes (Grepin et al. 1994; Durocher et al. 1996, 1997; Morin et al. 2000, 2001; Bruneau et al. 2001; Hiroi et al. 2001). The hypothesis that cardiac compartment-specific gene expression/repression results from cooperativity between cardiac factors and compartment-restricted factors is strongly supported by our in vivo observations.
Nkx2.5 and Tbx5 were shown to be essential components of the activation pathway of the ANF gene in vivo (Lyons et al. 1995; Tanaka et al. 1999; Bruneau et al. 2001). Both factors activate transcription through multiple binding sites present within the ANF promoter (Lee et al. 1998; Shiojima et al. 1999; Bruneau et al. 2001; Hiroi et al. 2001). Furthermore, Nkx2.5 and Tbx5 were shown to activate the ANF promoter in synergy in transfection assays (Bruneau et al. 2001; Hiroi et al. 2001). Inactivation of the −259-bp TBE or −250-bp NKE, required for this synergy in transfections, did not visibly affect chamber activity, suggesting that neither site is essential for ANF promoter activity in vivo. Therefore, Nkx2.5 and Tbx5 achieve activation of the ANF promoter through the remaining elements, or through an indirect activation pathway. Our transient transfection results support a role for the remaining elements in activation, because the constructs in which either the NKE, TBE, or both were mutated could still be partially stimulated by VP16-Tbx2 or by Nkx2.5 and Tbx5 (Fig. 5D,E).
The repressive activity of Tbx2 on cardiac gene expression in the AVC might be relevant for the mechanism underlying the pathogenesis in Holt-Oram patients, Tbx5 mutant mice, and humans with a mutation in the NKX2.5 gene, which all have conduction disease including AV block (Basson et al. 1997; Li et al. 1997; Schott et al. 1998; Bruneau et al. 2001). The AV node and AV junctional myocardium are derived from the AVC (Moorman et al. 1998; Davis et al. 2001) that express Nkx2.5, Tbx5, and Tbx2. Beside affecting directly downstream gene expression in the AVC, reduction of Tbx5 or Nkx2.5 levels might cause an imbalance in the interaction with Tbx2 to regulate downstream genes. This, in turn, could affect the formation of the AV conduction system. The role of Tbx2 in formation of the conduction system merits further investigation. Furthermore, the wide variation in phenotype within Holt-Oram patients and patients with an NKX2.5 mutation suggests that polymorphic variations in the Tbx2 gene may contribute to this variable phenotype.
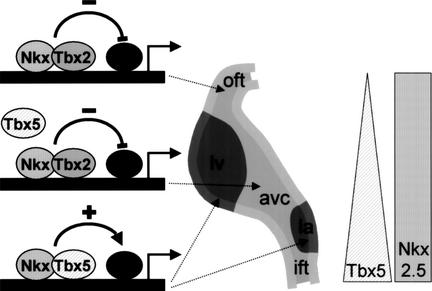
A potential mechanism for site-specific chamber formation: local repression of differentiation
To understand what role the inhibitory Tbx2/Nkx2.5 pathway might have in the formation of the four-chambered heart, it is important to appreciate that regional differences in differentiation within the tubular heart exist. The linear heart tube is patterned along the A-P, D-V, and L-R axis and has a nodal phenotype (high automaticity, slow contraction, slow conduction). Atrial and ventricular chamber myocardium forms at specific sites within the tubular heart during and after looping (de Jong et al. 1992; Christoffels et al. 2000). This chamber myocardium obtains a more mature phenotype (low automaticity, fast contraction, well-coupled cells, and a well-developed sarcoplasmic reticulum). The myocardium of the IFT, AVC, inner curvature, and OFT retains the nodal phenotype of the myocardium of the embryonic heart tube. These observations indicate that a transcriptional program responsible for differentiation is activated at specific sites in the tubular heart to form chamber myocardium. The IFT, AVC, inner curvature, and OFT escape the differentiation program until later in development and play an important role in the alignment of the chambers, in septation, and in the formation of the conduction system.
Genes for ANF, Chisel, and gap-junction proteins Cx40 and Cx43 are part of this differentiation program because they are specifically expressed in the forming chamber myocardium (Delorme et al. 1995, 1997; van Kempen et al. 1996; Christoffels et al. 2000; Palmer et al. 2001). ANF and Cx40 were shown to be targets of Tbx5 (Bruneau et al. 2001; Hiroi et al. 2001), and, also, Cx43 was shown to be a target for Tbx factors (Chen et al. 2001). In regions in which Tbx5 is (almost) absent, that is the OFT, and, later in development the RV, none of the downstream genes are expressed. The regions of the looped heart that express Tbx2, which functions as a repressor of transcription (Carreira et al. 1998; Jacobs et al. 2000; Sinha et al. 2000), remain embryonic irrespective of whether they express Tbx5. It is therefore tempting to speculate that Tbx2 expression in the IFT, AVC, inner curvature, and OFT is needed to escape the differentiation program (Fig. 6). The fact that Tbx2 and Tbx5 are coexpressed in the IFT, AVC, and inner curvature indicates that Tbx2 successfully competes with Tbx5 in the regulation of downstream genes. This implication is strengthened by our observation that Tbx2 forms a ternary complex with Nkx2.5 and the TBE–NKE site (Fig. 4A) and efficiently counteracts the synergistic activation by Tbx5 and Nkx2.5 (Fig. 5F). We propose that Tbx5 is involved in enforcing the chamber-specific transcription program, whereas Tbx2 counteracts the positive regulatory function of Tbx5 in specific regions of the heart. Both Tbx5 and Tbx2 cooperate with Nkx2.5, which functions as an accessory factor to restrict the T-box factor activities to cardiac genes.
Figure 6.
A potential mechanism for site-specific chamber formation by local repression of differentiation. Schematic representation of the transcriptional mechanisms involved in chamber formation. As part of an ongoing chamber formation program, Tbx5 and Nkx2.5 stimulate cardiac genes. Specific regions in the linear heart tube remain embryonic and do not develop into chamber myocardium due to the presence of Tbx2 in those regions. Nkx2.5 and Tbx2 form a repressor complex that suppresses genes that are part of the chamber differentiation program. The Tbx5 triangle and Nkx2.5 rectangle indicate Tbx5 and Nkx2.5 expression in the linear heart tube, respectively. Tbx2 is expressed in the primary myocardium of the inflow tract, atrioventricular canal, and outflow tract (light gray), whereas ANF is expressed in the chamber myocardium (dark gray). (ift) Inflow tract; (la) left atrium; (avc) atrioventricular canal; (lv) left ventricle; (oft) outflow tract.
Materials and methods
Transgene construction
All constructs used to generate transgenic mice (Table 1) contain a chimeric intron from the pCI vector (Promega), lacZ with a nuclear localization signal (nlacZ), and the polyadenylation signal from the bovine growth hormone gene. The ANF construct contains the −638/+70-bp ANF regulatory region, the cTnI construct contains the −230/+126-bp cTnI promoter region. ANF–cTnI is a chimeric construct in which the −638/−138 ANF sequence is fused to the −230/+126 cTnI promoter region. In the MLC2V–cTnI construct, the −250/−40 MLC2V sequence is fused to the −230/+126 cTnI promoter region. ANFmutNKE–cTnI is identical to the ANF–cTnI construct, with the exception of a 4-bp substitutional mutation of the NKE located at position −250 of the ANF promoter region (NKE, TTGAAGTGGG; NKEmut, TTGCCTCGGG) (Shiojima et al. 1999). The ANFmutTBE–cTnI construct is identical to the ANF–cTnI construct with the exception of a 4-bp substitutional mutation of the TBE located at position −259 of the ANF promoter region (TBE, TCTCACACCTT; TBEmut, TCTCTTTGCTT) (Sinha et al. 2000). The mutations were generated using the QuickChangeTM Site-Directed Mutagenesis kit (Stratagene). With the exception of the ANF–nlacZ construct, all constructs were flanked by a 1.2-kb SalI–BamHI tandem repeat of a chromosomal insulator sequence from the 5′ region of the chicken β-globin gene kindly provided by G. Felsenfeld (Chung et al. 1993).
Generation, identification, and analysis of transgenic mice
After removal of the vector sequences, the transgene constructs were injected into the pronuclei of zygotes of FVB mice and the injected zygotes were reimplanted into pseudopregnant foster mothers by use of standard techniques (Hogan et al. 1994). Animal care was according to guidelines as described (Christoffels et al. 1995). Constructs were analyzed in lines (ANF), F0 embryos (ANFmutNKE–cTnI and ANFmutTBE–cTnI ), or both F0 and lines (cTnI, ANF–cTnI, and MLC2V–cTnI). Positive embryos were scored by Southern blot and PCR on DNA prepared from the yolk sac. For Southern blot analysis, we used the nlacZ reporter gene (2-kb NcoI/SacI fragment) as a probe (Sambrook et al. 1989). For PCR analysis, primers specific to the nlacZ sequences were used (lacZ+, GCATCGAGCTGGGTAATAAGC GTTGGCAAT and lacZ−, ACTGCAACAACGCTGCTTCG GCCTGGTAAT) according to standard procedures (Sambrook et al. 1989). Embryos were stained for β-galactosidase activity as described (Franco et al. 2001).
Plasmid constructs and transfections
Cultures of primary atrial and ventricular cardiomyocytes were prepared from E17.5 Wistar rats as described (van Wamel et al. 2000). Cos-7 cells were grown under standard culture conditions in DMEM/F12 (GIBCO BRL) supplemented with 10% fetal calf serum. Cells were transfected with 4.4 μg of reporter construct, 10–1000 ng of expression plasmid or empty vector for compensation, and 200 ng of luciferase expression vector (CMV-Luc) as an internal control per 3-cm dish, using the calciumphosphate method (Sambrook et al. 1989). Cell extracts and luciferase assays were performed as described (Christoffels et al. 1995). β-Galactosidase activity was measured using the Galacto-Light kit (Tropix) according to the manufacturer's instructions. Light emission was measured in a Turner TD-20/20 luminometer. All results are from one representative experiment (out of 3) done in duplicate. Full-length mouse FLAG–Nkx2.5, kindly provided by Dr. R. Harvey (The Victor Chang Cardiac Research Institute, Darlinghurst, Australia), was cloned into pCI (Promega). FLAG–Nkx2.5–L176P was generated by PCR and subcloned into pCI (Promega). Full-length human TBX5 (Basson et al. 1999), kindly provided by Dr. C. Basson, was cloned into pcDNA3.1 (Clontech). Full-length human TBX2 and TBX2-delRD were cloned in pcDNA3.1 as described (Jacobs et al. 2000). TBX2R122E/R123E was generated by PCR and subcloned into pcDNA3.1 (Clontech).
Nonradioactive in situ hybridization
Whole-mount in situ hybridization and nonradioactive in situ hybridization on sections were performed as described (Moorman et al. 2001). The cDNA probes used were ANF (Zeller et al. 1987), Tbx2, Tbx5 (Chapman et al. 1996), and Cx40 (Delorme et al. 1997).
Electromobility shift assays
Nuclear extracts were prepared from HEK cells transfected with expression vectors for TBX2, TBX2–delRD, TBX2–R122E/R123E, TBX5, FLAG–Nkx2.5, and FLAG–Nkx2.5-L176P. Double-stranded oligonucleotides were synthesized and labeled with [α-32P]dATP using Klenow polymerase. Labeled probes were incubated and used in binding reactions as described and resolved on a 6% polyacrylamide gel (Espinas et al. 1994). Oligonucleotides used (complementary strand not shown, mutations underlined): Wild-type, TCTGCTCTTCTCACACCTTT GAAGTGGGGGCCTCTTG, TBE mutated (TBEmut), TCT GCTCTTCTCTTTGCTTTGAAGTGGGGGCCTCTTG, and NKE mutated (NKEmut) TCTGCTCTTCTCACACCTTT GCCTCGGGGGCCTCTTG.
Western blot analysis
Western-blot analysis was performed according to standard methods (Sambrook et al. 1989). Primary antibodies were a rabbit polyclonal raised against the amino terminus of human TBX2 (Jacobs et al. 2000), and an anti-FLAG antibody from ABR.
Acknowledgments
We thank Drs. R. Harvey, C. Basson, V. Papaioannou, and G. Felsenfeld for kindly providing FLAG–Nkx2.5, human TBX5 cDNA, the probes for mouse Tbx2 and Tbx5, and the β-globin insulators, respectively; Drs. R. Di Lisi and S. Schiaffino for sharing data; and P.A.J. de Boer, C. de Gier-de Vries, and W. Hoogaars for their expert technical support. M.C. was supported by a Telephon Foundation grant (no. 452/bi). This work was supported by grants from the Dutch Heart Foundation (NHS) M96.002 and NWO 902.16.243
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL v.m.christoffels@amc.uva.nl; FAX 31-20-6976177.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.222902.
References
- Argentin S, Ardati A, Tremblay S, Lihrmann I, Robitaille L, Drouin J, Nemer M. Developmental stage-specific regulation of atrial natriuretic factor gene transcription in cardiac cells. Mol Cell Biol. 1994;14:777–790. doi: 10.1128/mcb.14.1.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausoni S, de Nardi C, Moretti P, Gorza L, Schiaffino S. Developmental expression of rat cardiac troponin I mRNA. Development. 1991;112:1041–1051. doi: 10.1242/dev.112.4.1041. [DOI] [PubMed] [Google Scholar]
- Basson CT, Bachinsky DR, Lin RC, Levi T, Elkins JA, Soults J, Grayzel D, Kroumpouzou E, Traill TA, Leblanc-Straceski J, et al. Mutations in human TBX5 (corrected) cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet. 1997;15:30–35. doi: 10.1038/ng0197-30. [DOI] [PubMed] [Google Scholar]
- Basson CT, Huang T, Lin RC, Bachinsky DR, Weremowicz S, Vaglio A, Bruzzone R, Quadrelli R, Lerone M, Romeo G, et al. Different TBX5 interactions in heart and limb defined by Holt-Oram syndrome mutations. Proc Natl Acad Sci. 1999;96:2919–2924. doi: 10.1073/pnas.96.6.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, Conner DA, Gessler M, Nemer M, Seidman CE, et al. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106:709–721. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- Carreira S, Dexter TJ, Yavuzer U, Easty DJ, Goding CR. Brachyury-related transcription factor Tbx2 and repression of the melanocyte-specific TRP-1 promoter. Mol Cell Biol. 1998;18:5099–5108. doi: 10.1128/mcb.18.9.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman DL, Garvey N, Hancock S, Alexiou M, Agulnik SI, Gibson-Brown JJ, Cebra-Thomas J, Bollag RJ, Silver LM, Papaioannou VE. Expression of the T-box family genes, Tbx1-Tbx5, during early mouse development. Dev Dyn. 1996;206:379–390. doi: 10.1002/(SICI)1097-0177(199608)206:4<379::AID-AJA4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Chen J, Zhong Q, Wang J, Cameron RS, Borke JL, Isales CM, Bollag RJ. Microarray analysis of Tbx2-directed gene expression: A possible role in osteogenesis. Mol Cell Endocrinol. 2001;177:43–54. doi: 10.1016/s0303-7207(01)00456-7. [DOI] [PubMed] [Google Scholar]
- Christoffels VM, van den Hoff MJB, Moorman AFM, Lamers WH. The far-upstream enhancer of the carbamoylphosphate synthetase I gene is responsible for the tissue specificity and hormone inducibility of its expression. J Biol Chem. 1995;270:24932–24940. doi: 10.1074/jbc.270.42.24932. [DOI] [PubMed] [Google Scholar]
- Christoffels VM, Habets PEMH, Franco D, Campione M, de Jong F, Lamers WH, Bao ZZ, Palmer S, Biben C, Harvey RP, et al. Chamber formation and morphogenesis in the developing mammalian heart. Dev Biol. 2000;223:266–278. doi: 10.1006/dbio.2000.9753. [DOI] [PubMed] [Google Scholar]
- Chung JH, Whiteley M, Felsenfeld G. A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- Chung JH, Bell AC, Felsenfeld G. Characterization of the chicken β-globin insulator. Proc Natl Acad Sci. 1997;94:575–580. doi: 10.1073/pnas.94.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DL, Edwards AV, Juraszek AL, Phelps A, Wessels A, Burch JBE. A GATA-6 gene heart-region-specific enhancer provides a novel means to mark and probe a discrete component of the mouse cardiac conduction system. Mech Dev. 2001;108:105–119. doi: 10.1016/s0925-4773(01)00500-7. [DOI] [PubMed] [Google Scholar]
- de Jong F, Opthof T, Wilde AAM, Janse MJ, Charles R, Lamers WH, Moorman AFM. Persisting zones of slow impulse conduction in developing chicken hearts. Circ Res. 1992;71:240–250. doi: 10.1161/01.res.71.2.240. [DOI] [PubMed] [Google Scholar]
- Delorme B, Dahl E, Jarry-Guichard T, Marics I, Briand JP, Willecke K, Gros D, Théveniau-Ruissy M. Developmental regulation of connexin40 gene expression in mouse heart correlates with the differentiation of the conduction system. Dev Dyn. 1995;204:358–371. doi: 10.1002/aja.1002040403. [DOI] [PubMed] [Google Scholar]
- Delorme B, Dahl E, Jarry-Guichard T, Briand JP, Willecke K, Gros D, Théveniau-Ruissy M. Expression pattern of connexin gene products at the early developmental stages of the mouse cardiovascular system. Circ Res. 1997;81:423–437. doi: 10.1161/01.res.81.3.423. [DOI] [PubMed] [Google Scholar]
- Di Lisi R, Millino C, Calabria E, Altruda F, Schiaffino S, Ausoni S. Combinatorial cis-acting elements control tissue-specific activation of the cardiac troponin I gene in vitro and in vivo. J Biol Chem. 1998;273:25371–25380. doi: 10.1074/jbc.273.39.25371. [DOI] [PubMed] [Google Scholar]
- Di Lisi R, Sandri C, Franco D, Ausoni S, Moorman AFM, Schiaffino S. An atrioventricular canal domain defined by cardiac troponin I transgene expression in the embryonic myocardium. Anat Embryol. 2000;202:95–101. doi: 10.1007/s004290000102. [DOI] [PubMed] [Google Scholar]
- Durocher D, Nemer M. Combinatorial interactions regulating cardiac transcription. Dev Genet. 1998;22:262. doi: 10.1002/(SICI)1520-6408(1998)22:3<250::AID-DVG7>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Durocher D, Chen CY, Ardati A, Schwartz RJ, Nemer M. The atrial natriuretic factor promoter is a downstream target for Nkx- 2.5 in the myocardium. Mol Cell Biol. 1996;16:4648–4655. doi: 10.1128/mcb.16.9.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher D, Charron F, Warren R, Schwartz RJ, Nemer M. The cardiac transcription factors Nkx2.5 and GATA-4 are mutual cofactors. EMBO J. 1997;18:5687–5696. doi: 10.1093/emboj/16.18.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinas ML, Roux J, Ghysdael J, Pictet R, Grange T. Participation of Ets transcription factors in the glucocorticoid response of the rat tyrosine aminotransferase gene. Mol Cell Biol. 1994;14:4116–4125. doi: 10.1128/mcb.14.6.4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field LJ. Atrial natriuretic factor-SV40 T antigen transgenes produce tumors and cardiac arrhythmias in mice. Science. 1988;239:1029–1033. doi: 10.1126/science.2964082. [DOI] [PubMed] [Google Scholar]
- Fishman MC, Chien KR. Fashioning the vertebrate heart: Earliest embryonic decisions. Development. 1997;124:2099–2117. doi: 10.1242/dev.124.11.2099. [DOI] [PubMed] [Google Scholar]
- Franco D, de Boer PAJ, de Gier-de Vries C, Lamers WH, Moorman AFM. Methods on in situ hybridization, immunohistochemistry and β-galactosidase reporter gene detection. Eur J Morph. 2001;39:3–25. doi: 10.1076/ejom.39.1.3.7982. [DOI] [PubMed] [Google Scholar]
- Ghosh TK, Packham EA, Bonser AJ, Robinson TE, Cross SJ, Brook JD. Characterization of the TBX5 binding site and analysis of mutations that cause Holt-Oram syndrome. Hum Mol Genet. 2001;10:1983–1994. doi: 10.1093/hmg/10.18.1983. [DOI] [PubMed] [Google Scholar]
- Gibson-Brown JJ, Agulnik SI, Silver LM, Papaioannou VE. Expression of T-box genes Tbx2-Tbx5 during chick organogenesis. Mech Dev. 1998;74:165–169. doi: 10.1016/s0925-4773(98)00056-2. [DOI] [PubMed] [Google Scholar]
- Grepin C, Dagnino L, Robitaille L, Haberstroh L, Antakly T, Nemer M. A hormone-encoding gene identifies a pathway for cardiac but not skeletal muscle gene transcription. Mol Cell Biol. 1994;14:3115–3129. doi: 10.1128/mcb.14.5.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grow MW, Krieg PA. Tinman function is essential for vertebrate heart development: Elimination of cardiac differentiation by dominant inhibitory mutants of the tinman-related genes, XNkx2–3 and XNkx2.5. Dev Biol. 1998;204:187–196. doi: 10.1006/dbio.1998.9080. [DOI] [PubMed] [Google Scholar]
- Hiroi Y, Kudoh S, Monzen K, Ikeda Y, Yazaki Y, Nagai R, Komuro I. Tbx5 associates with Nkx2.5 and synergistically promotes cardiomyocyte differentiation. Nat Genet. 2001;28:276–280. doi: 10.1038/90123. [DOI] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the mouse embryo: A laboratory manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Jacobs JJL, Keblusek P, Robanus Maandag E, Kristel P, Lingbeek M, Nederlof PM, van Welsem T, van de Vijver MJ, Koh EY, Daley GQ, et al. Senescence bypass screen identifies Tbx2, which represses Cdkn2a (p19ARF) and is amplified in a subset of human breast cancers. Nat Genet. 2000;26:291–299. doi: 10.1038/81583. [DOI] [PubMed] [Google Scholar]
- Kasahara H, Bartunkova S, Schinke M, Tanaka M, Izumo S. Cardiac and extracardiac expression of Csx/Nkx2.5 homeodomain protein. Circ Res. 1998;82:936–946. doi: 10.1161/01.res.82.9.936. [DOI] [PubMed] [Google Scholar]
- Kispert A, Koschorz B, Herrmann BG. The T protein encoded by Brachyury is a tissue-specific transcription factor. EMBO J. 1995;14:4763–4772. doi: 10.1002/j.1460-2075.1995.tb00158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlton KU, Rockman HA, Itani M, Vovan A, Seidman CE, Chien KR. Divergent pathways mediate the induction of ANF transgenes in neonatal and hypertrophic ventricular myocardium. J Clin Invest. 1995;96:1311–1318. doi: 10.1172/JCI118166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro I, Izumo S. Csx: A murine homeobox-containing gene specifically expressed in the developing heart. Proc Natl Acad Sci. 1993;90:8145–8149. doi: 10.1073/pnas.90.17.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Shioi T, Kasahara H, Jobe SM, Wiese RJ, Markham BE, Izumo S. The cardiac tissue-restricted homeobox protein Csx/Nkx2.5 physically associates with the zinc finger protein GATA4 and cooperatively activates atrial natriuretic factor gene expression. Mol Cell Biol. 1998;18:3120–3129. doi: 10.1128/mcb.18.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QY, Newbury-Ecob RA, Terret JA, Wilson DI, Curtis AR, Yi CH, Gebuhr T, Bullen PJ, Robson SC, Strachan T, et al. Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat Genet. 1997;15:21–29. doi: 10.1038/ng0197-21. [DOI] [PubMed] [Google Scholar]
- Lints TJ, Parsons LM, Hartley L, Lyons I, Harvey RP. Nkx-2.5: A novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development. 1993;119:419–431. doi: 10.1242/dev.119.2.419. [DOI] [PubMed] [Google Scholar]
- Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeobox gene Nkx2.5. Genes & Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- Moorman AFM, de Jong F, Denyn MMFJ, Lamers WH. Development of the cardiac conduction system. Circ Res. 1998;82:629–644. doi: 10.1161/01.res.82.6.629. [DOI] [PubMed] [Google Scholar]
- Moorman AFM, Houweling AC, de Boer PAJ, Christoffels VM. Sensitive nonradioactive detection of mRNA in tissue sections: Novel application of the whole-mount in situ hybridization protocol. J Histo Cytochem. 2001;49:1–8. doi: 10.1177/002215540104900101. [DOI] [PubMed] [Google Scholar]
- Morin S, Charron F, Robitaille L, Nemer M. GATA-dependent recruitment of MEF2 proteins to target promoters. EMBO J. 2000;19:2046–2055. doi: 10.1093/emboj/19.9.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin S, Paradis P, Aries A, Nemer M. Serum response factor-GATA ternary complex required for nuclear signaling by a G-protein-coupled receptor. Mol Cell Biol. 2001;21:1036–1044. doi: 10.1128/MCB.21.4.1036-1044.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer S, Groves N, Schindeler A, Yeoh T, Biben C, Wang CC, Sparrow D, Barnett L, Jenkins NA, Copeland NG, et al. The small muscle-specific protein Csl modifies cell shape and promotes myocyte fusion in an insulin-like growth factor 1-dependent manner. J Cell Biol. 2001;153:985–997. doi: 10.1083/jcb.153.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross RS, Navankasattusas S, Harvey RP, Chien KR. An HF-1a/HF-1b/MEF-2 combinatorial element confers cardiac ventricular specificity and established an anterior-posterior gradient of expression. Development. 1996;122:1799–1809. doi: 10.1242/dev.122.6.1799. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schott J-J, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, Maron BJ, Seidman CE, Seidman JG. Congenital heart disease caused by mutations in the transcription factor NKX2.5. Science. 1998;281:108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- Shiojima I, Komuro I, Oka T, Hiroi Y, Mizuno T, Takimoto E, Monzen K, Aikawa R, Akazawa H, Yamazaki T, et al. Context-dependent transcriptional cooperation mediated by cardiac transcription factors Csx/Nkx-2.5 and GATA-4. J Biol Chem. 1999;274:8231–8239. doi: 10.1074/jbc.274.12.8231. [DOI] [PubMed] [Google Scholar]
- Sinha S, Abraham S, Gronostajski RM, Campbell CE. Differential DNA binding and transcription modulation by three T-box proteins, T, TBX1 and TBX2. Gene. 2000;258:15–29. doi: 10.1016/s0378-1119(00)00417-0. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Olson EN. A genetic blueprint for cardiac development. Nature. 2000;407:221–226. doi: 10.1038/35025190. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126:1269–1280. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- Vallins WJ, Brand NJ, Dabhade N, Butler-Browne GS, Yacoub MH, Barton PJR. Molecular cloning of human cardiac troponin I using polymerase chain reaction. FEBS Lett. 1990;270:57–61. doi: 10.1016/0014-5793(90)81234-f. [DOI] [PubMed] [Google Scholar]
- van Kempen MJA, Vermeulen JLM, Moorman AFM, Gros DB, Paul DL, Lamers WH. Developmental changes of connexin40 and connexin43 mRNA-distribution patterns in the rat heart. Cardiovasc Res. 1996;32:886–900. doi: 10.1016/0008-6363(96)00131-9. [DOI] [PubMed] [Google Scholar]
- van Wamel JET, Ruwhof C, van der valk-Kokshoorn PI, Schrier PI, van der Laarse A. Rapid gene transcription induced by stretch in cardiac myocytes and fibroblasts and their paracrine influence on stationary myocytes and fibroblasts. Eur J Physiol. 2000;439:781–788. doi: 10.1007/s004240000253. [DOI] [PubMed] [Google Scholar]
- Yamada M, Revelli JP, Eichele G, Barron M, Schwartz RJ. Expression of chick Tbx-2, Tbx-3, and Tbx-5 genes during early heart development: evidence for BMP2 induction of Tbx2. Dev Biol. 2000;228:95–105. doi: 10.1006/dbio.2000.9927. [DOI] [PubMed] [Google Scholar]
- Zeller R, Bloch KD, Williams BS, Arceci RJ, Seidman CE. Localized expression of the atrial natriuretic factor gene during cardiac embryogenesis. Genes & Dev. 1987;1:693–698. doi: 10.1101/gad.1.7.693. [DOI] [PubMed] [Google Scholar]