Abstract
Many regulons controlled by alternative σ factors, including σS and σ32, are poorly induced in cells lacking the alarmone ppGpp. We show that ppGpp is not absolutely required for the activity of σS-dependent promoters because underproduction of σ70, specific mutations in rpoD (rpoD40 and rpoD35), or overproduction of Rsd (anti-σ70) restored expression from σS-dependent promoters in vivo in the absence of ppGpp accumulation. An in vitro transcription/competition assay with reconstituted RNA polymerase showed that addition of ppGpp reduces the ability of wild-type σ70 to compete with σ32 for core binding and the mutant σ70 proteins, encoded by rpoD40 and rpoD35, compete less efficiently than wild-type σ70. Similarly, an in vivo competition assay showed that the ability of both σ32 and σS to compete with σ70 is diminished in cells lacking ppGpp. Consistently, the fraction of σS and σ32 bound to core was drastically reduced in ppGpp-deficient cells. Thus, the stringent response encompasses a mechanism that alters the relative competitiveness of σ factors in accordance with cellular demands during physiological stress.
Keywords: Stringent response, σ factor competition, RpoD, RpoS, RpoH, Rsd
Cells of Escherichia coli elicit stringent control of ribosome production during the transition from exponential growth to stationary phase (Sands and Roberts 1952; Stent and Brenner 1961). The effector molecule of the stringent control modulon is the alarmone guanosine tetraphosphate, ppGpp (Cashel and Gallant 1969; Lazzarini et al. 1971; Ryals et al. 1982a,b,c; Baracchini and Bremer 1988). The production of this nucleotide is dependent on the (p)ppGpp synthetases PSI and PSII encoded by the relA and spoT genes, respectively (Xiao et al. 1991). The alarmone ppGpp binds to the β and β‘ subunits of core RNA polymerase (E) (Chatterji et al. 1998; Toulokhonov et al. 2001) and thereby inhibits superfluous rRNA biosynthesis during growth inhibition (e.g., Travers 1976; Gourse et al. 1986; Ohlsen and Gralla 1992; Heinemann and Wagner 1997; Zhou and Jin 1998). Mechanisms suggested to explain this regulation of rRNA synthesis include ppGpp-dependent alterations in the initiation pathway that traps RNA polymerase (Heinemann and Wagner 1997), a reduced ability of the RNA polymerase to form an open complex (Ohlsen and Gralla 1992), and a reduction in the stability of the promoter-Eσ70–ppGpp open complex (Gourse et al. 1998). Because the rrnP1 promoters form intrinsically unstable open complexes with Eσ70, such promoters may be argued to be especially sensitive to the destabilizing effects of ppGpp (Gourse et al. 1998). Consistent with this idea, it has been shown that RNA polymerase (RNAP) mutants that suppress the requirement for ppGpp in vivo form unstable complexes with stable RNA promoters in vitro (Zhou and Jin 1998).
The alarmone ppGpp can also act as a positive effector of gene expression, and some σ70-dependent genes require this nucleotide for their induction during growth arrest (Xiao et al. 1991; Nyström 1994; Kvint et al. 2000b). In addition, many operons encoding amino acid biosynthetic pathways require ppGpp for their transcription, and E. coli cells lacking ppGpp are polyauxotrophs (Xiao et al. 1991). It has been suggested that the effect of ppGpp on such promoters is linked to ppGpp-dependent changes in core availability. According to a model by Zhou and Jin (1998), the rate-limiting step of promoters that are positively regulated by ppGpp is Eσ70 recruitment, and it is argued that these promoters would therefore be very sensitive to the concentration of free RNA polymerase. Thus, the accumulation of ppGpp is suggested to result in the dissociation of core from stringent rrnP1 promoters and the consequent increased availability of core leads to elevated initiation of transcription at promoters that have a relatively poor ability to recruit Eσ70 (Zhou and Jin 1998). Some aspects of this model have been supported recently by in vivo and in vitro transcription assays (Barker et al. 2001a,b).
To add to the role of ppGpp in the cell, genes requiring alternative σ factors have been shown to depend on ppGpp for their induction. For example, the inducers of the σ54-dependent promoters Po and Pu are only effective when ppGpp levels are elevated (Sze and Shingler 1999; Carmona et al. 2000; Sze et al. 2002). Similarly, mutant cells with no or low levels of ppGpp exhibit an attenuated and sluggish expression of σ32-dependent heat-shock genes (Grossman et al. 1984; VanBogelen and Neidhardt 1990; Jones et al. 1992). In addition, mutants lacking functional relA spoT or σS of the general stress response have been found to exhibit similar phenotypes. The σS transcription factor accumulates and directs the RNA polymerase to >50 genes upon conditions of cellular starvation and stress (Hengge-Aroni 2000). Mutants lacking σS exhibit an accelerated die-off during conditions of growth arrest (Lange and Hengge-Aronis 1991), and markedly elevated levels of oxidized proteins (Dukan and Nystrom 1998, 1999). The fact that σS itself requires ppGpp for its production (Gentry et al. 1993; Lange et al. 1995; Zgurskaya et al. 1997) initially appeared to fully explain the similarity between ΔrelA ΔspoT and rpoS phenotypes. However, it was later shown that σS-dependent genes require ppGpp for their induction, even in the presence of wild-type levels of σS (Kvint et al. 2000a). Thus, ppGpp appears to exert a dual control on the RpoS regulon by affecting both the levels of the required σ factor and its activity.
The exact role of ppGpp in alternative σ-factor function is not clear and could, conceivably, include control of promoter recognition and transcription initiation or act at the level of σ binding to core. In this work, we present evidence using (1) in vivo and in vitro competition assays and (2) quantification of σS and σ32 association with core so that the ability of σS and σ32 to compete with σ70 for core binding is facilitated in the presence of ppGpp, and that ppGpp requirement can be suppressed by σ70 underproduction. The data show that a ppGpp-dependent alteration in σ-factor competition for core binding is an integral part of the typical stringent response allowing alternative σ factors to operate successfully in concert with σ70 during cellular stress.
Results
The failure of relaxed mutants to induce the σS regulon can be alleviated by underproduction of σ70 or overproduction of Rsd
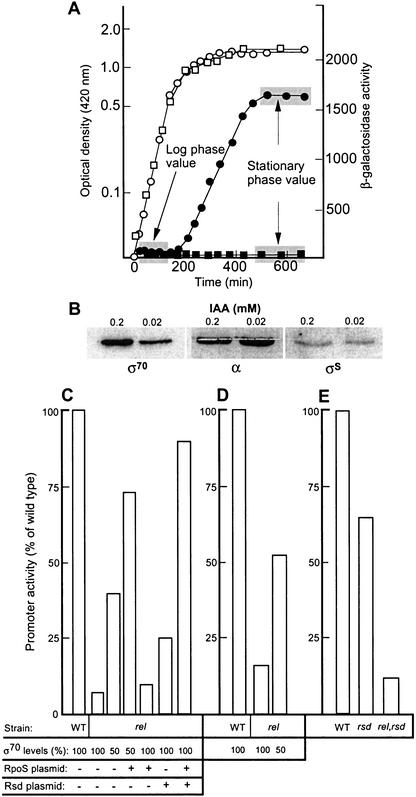
As seen in Figure 1A, expression of a σS-dependent model gene is induced during the transition phase and reaches a new steady state (stationary phase value) about 2 h after a brake point in the growth curve can be observed. The difference between the wild-type and ppGpp0 mutant is maximal at this time, whereas no significant difference can be observed during exponential growth (Fig. 1A). To facilitate easy and direct comparisons between expression levels in different strains, only the mean stationary phase values are shown in the subsequent experiments.
Figure 1.
Effects of σ70 underproduction and Rsd overproduction on σS-dependent gene expression. (A) PuspB–lacZ expression (closed symbols) and cell density (open symbols) in the wild-type (AF633, circles) and ppGpp0 (KK180, boxes) strains during growth and stationary phase. (B) Western blot immunoassay for detection of RpoD, RpoS, and α-subunit of RNAP in cells grown in different amounts of IAA as indicated (the levels of the β subunit were, like α, unaffected by 0.2 mM IAA; data not shown). (C) Expression of PuspB–lacZ in wild-type and relaxed strains (relA1 ΔspoT) underproducing RpoD, and/or overproducing RpoS and Rsd as indicated. The strains used are MJ429, MJ442, TN324, KK317, KK318, and KK316 (see Table 1 for details). (D) Expression of PkatE–lacZ in wild-type and relaxed strains (relA1 ΔspoT) underproducing RpoD. The strains used are TN321 and MJ432. (E) Expression of PuspB–lacZ in wild type (AF633), rsd∷Km (MJ234), and rsd∷Km ΔspoT (TN325). The data show expression levels after 1 h of glucose starvation. The phenotype rel (relA1 ΔspoT∷Km) indicated in the figure implies that the strains fail to accumulate ppGpp during the glucose starvation condition used.
To elucidate whether there is an absolute requirement of ppGpp for σS function or whether ppGpp facilitates σS competition for core binding, we determined the effect of σ70 underproduction on the expression from the σS-dependent promoters, PuspB and PkatE, in relaxed cells that fail to elevate ppGpp levels during carbon starvation (e.g., Kvint et al. 2000a). To approach this question, we used a genetic system for underproduction of σ70 in which expression of this σ factor is regulated by the trp promoter and can be controlled by the levels of IAA (indole-3-acrylic acid; an antagonist of the Trp repressor; Lonetto et al. 1998). We first determined the concentration of IAA (0.2 mM) that generated σ70 levels corresponding to wild-type σ70 levels and resulted in the correct growth rate in the growth medium used (data not shown). We also confirmed that the kinetics and magnitude of induction of σS and σ70-dependent genes were indistinguishable from that of wild type during entry of cells in glucose starvation-induced stationary phase by use of this concentration of IAA (data not shown). Growing cultures were diluted and split into two, such that one received 0.2 mM IAA (wild-type levels of σ70), whereas the other received 0.02 mM IAA (underproduction of σ70; see Fig. 1B) and the effect of this underproduction of σ70 on the expression of σS-dependent genes was determined. Note that underproduction of σ70 did not change the levels of the α subunit (Fig. 1B) or the β subunit (data not shown) of core, nor did σS levels change appreciably (Fig. 1B). In the experiment shown, we used the relA1 and ΔspoT∷Km alleles rather than ΔrelA∷Km and ΔspoT∷Cm because the Ptrp–rpoD fusion is linked to the Cm marker. However, we have confirmed that the behavior of the double ΔrelA ΔspoT deletion mutant and the relA1 ΔspoT∷Km mutant is indistinguishable with respect to the regulation of the promoters studied during the glucose starvation conditions used. As seen in Figure 1, C and D, underproduction of σ70 partly suppressed the lack of induction of PuspB and PkatE upon entry of relaxed cells into glucose starvation-induced stationary phase. It should be noted that the expression levels reached are clearly below those of wild-type cells. This result would be anticipated as σS levels are much lower in the relaxed mutant, and consequently σ70 underproduction alone could not be expected to accomplish a full suppression of σS-dependent expression. However, a stronger suppression was achieved when σ70 underproduction was performed with a strain carrying rpoS on a high copy number plasmid (Fig. 1C). Consistent with previous data (Kvint et al. 2000a), no effect was observed with the rpoS plasmid alone (Fig. 1C). As expected, σ70 underproduction did not up-regulate a σ70-dependent promoter (PuspA) requiring ppGpp for induction (data not shown).
The E. coli Rsd protein binds free σ70 and has been suggested to act as an anti-σ factor with a role in curtailing σ70-dependent transcription by blocking the access of σ70 to core RNA polymerase upon entry of cells into stationary phase (Jishage and Ishihama 1998). Because the rsd gene is induced in stationary phase in a ppGpp-dependent fashion (Jishage and Ishihama 1999), we entertained the idea that the reduced levels of Rsd observed in relaxed cells may thus reduce the ability of σS to compete for core binding. We tested this idea by elucidating the effect of overproducing Rsd on σS-dependent transcription in relaxed cells. As shown in Figure 1C, Rsd overproduction alone had a very small effect on uspB expression in the relaxed mutant. However, the effect was significantly enhanced when more RpoS was provided from the pMMKatF2 plasmid (Fig. 1C). Also, induction of the uspB gene was attenuated in the rsd mutant, but not to the same extent as in the ppGpp0 mutant (Fig. 1E). Thus, we conclude that the Rsd protein may be an important member of the stringent control modulon that allows σS to compete more successfully for core binding. However, the poor induction of the RpoS regulon in relaxed cells cannot be solely explained by the diminished concentration of Rsd in these cells.
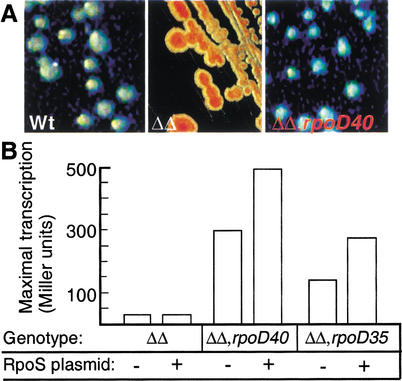
The rpoD40 and rpoD35 suppressor mutations partially restore σS-dependent promoter activity in a ppGpp0 mutant
Two new rpoD mutations, called rpoD35 and rpoD40 (see Table 1), have been identified recently, which suppress the ppGpp requirement of the σ54-dependent Po promoter. With respect to suppression of Po, the rpoD40 allele is a markedly better suppressor than the rpoD35 allele, and this phenotype has been shown to be attributable to reduced ability to bind and compete for core RNA polymerase (A. Laurie and V. Shingler, unpubl.). We transduced these rpoD suppressor alleles into KK358 (PuspB–lacZ ΔrelA ΔspoT). Using tetrazolium lactose plates, it became immediately obvious that the rpoD40 (Fig. 2A) and rpoD35 (data not shown) mutations also restored expression from the σS-dependent PuspB promoter in a ppGpp0 strain. Quantification of the promoter activity (Fig. 2B) showed that the rpoD40 allele is a better suppressor than rpoD35, and thus that the hierarchy observed with the σ54-dependent Po promoter is maintained at the σS-dependent PuspB promoter. We also introduced plasmid pMMKatF2 (overproducing σS) into the suppressor strains, and noted that the activity of PuspB was further elevated by σS overproduction (Fig. 2B).
Table 1.
Bacterial strains and plasmids
| Strain
|
Relevant genotype
|
Source/reference
|
|
|---|---|---|---|
| AF633 | MC4100 λφ (uspB–lacZ) | Farewell et al. 1998b | |
| AF634 | MC4100 λφ (uspA–lacZ) | Farewell et al. 1996 | |
| EC2922 | MG1655 Δlac relA∷Km spoT∷Cm aer-3075∷Tn10 rpoD-Y571H, (rpoD35) | V. Shingler | |
| EC2871 | MG1655 Δlac relA∷Km spoT∷Cm aer-3075∷Tn10 rpoD-▿DSA (536–538), (rpoD40) | V. Shingler | |
| KK153 | AF633/pMMKatF2 | This work | |
| KK180 | AF633 ΔrelA∷Km ΔspoT∷Cm | Kvint et al. 2000a | |
| KK315 | AF633 relA1 spoT∷Km | This work | |
| KK316 | KK315/pRsd/pMMKatF2 | This work | |
| KK317 | KK315/pRsd | This work | |
| KK318 | KK315/pMMKatF2 | This work | |
| KK357 | AF634 ΔrelA ΔspoT | This work | |
| KK358 | AF633 ΔrelA ΔspoT | This work | |
| KK373 | KK358 rpoD-▿DSA (536–538), (rpoD40) | This work | |
| KK374 | KK358/pMMKatF2 | This work | |
| KK375 | KK373/pMMKatF2 | This work | |
| KK384 | KK358 rpoD-Y571H, (rpoD35) | This work | |
| Kk385 | KK384/pMMKatF2 | This work | |
| KK390 | AF634/pKV1278 | This work | |
| KK391 | KK357/pKV1278 | This work | |
| MC4100 | F-araD139 Δ‘arqF–lac’ U169 rpsL 150 relA1 flbB5301 deoC1 ptsF25 rbsR | Lab. stock | |
| MJ234 | AF633 rsd∷Km | This work | |
| MJ265 | MC4100 λφ(mmpuspA4b–lacZ) | This work | |
| MJ271 | MC4100 λφ (katE–lacZ) | This work | |
| MJ285 | MJ265 ΔrelA∷Km ΔspoT∷Cm | This work | |
| MJ321 | MJ265/pMMKatF2 | This work | |
| MJ325 | MJ265/pKV1278 | This work | |
| MJ342 | AF633/pKV1278 | This work | |
| MJ352 | MJ265/pKVQ805 | This work | |
| MJ381 | MJ271/pKV1278 | This work | |
| MJ382 | SP887/pKV1278 | This work | |
| MJ429 | AF633 Ω(CamR) Ptrp–rpoD | This work | |
| MJ432 | MJ271 Ω(CamR) Ptrp–rpoD | This work | |
| MJ440 | MJ285/pKV1278 | This work | |
| MJ442 | MJ429 spoT∷Km | This work | |
| MJ500 | MJ285/pMMKatF2 | This work | |
| MO1000EL | λφ (katE–lacZ) | Ohnuma et al. 2000 | |
| TN321 | MJ432 spoT∷Km | This work | |
| TN322 | MC4100 λφ(fadD–lacZ)/pMMKatF2 | This work | |
| TN323 | TN322 ΔrelA∷Km ΔspoT∷Cm/pMMKatF2 | This work | |
| TN324 | MJ442/pMMKatF2 | This work | |
| TN325 | MJ234 ΔspoT∷Cm | This work | |
| TN326 | MC4100 λφ(fadD–lacZ)/pKV1278 | This work | |
| TN327 | TN326 ΔrelA∷Km ΔspoT∷Cm/pKV1278 | This work |
| Plasmid
|
Vector
|
Gene to be expressed
|
Source/reference
|
|---|---|---|---|
| pAT153 | L-O. Hedén | ||
| phis173 | Joo et al. 1997 | ||
| pJET40 | PdnaK, rna P1 | C. Gross | |
| pKV1278 | pTrc99A | rpoH | M. Kanamori, unpubl. |
| pKVQ805 | pTrc99A | rpoH173 (Q80R) | This work |
| pMJ261 | pTL61T | uspA promoter | This work |
| pMMKatF2 | pAT153 | rpoS | Mulvey et al. 1988 |
| pRsd | pACYC184 | rsd | Jishage and Ishihama 1999 |
| pTL61T | Linn and St. Pierre 1990 |
Figure 2.
Effects of RpoD mutations on PuspB–lacZ expression. (A) Colonies of wild type (AF633), ΔrelA ΔspoT (KK358), and ΔrelA ΔspoT rpoD40 (KK373) grown on tetrazolium lactose plates. (B) Maximal transcription of PuspB–lacZ in ppGpp0 strains carrying different rpoD alleles. The strains used are ΔrelA ΔspoT (KK358), ΔrelA ΔspoT/pMMKatF2 (KK374), ΔrelA ΔspoT rpoD40 (KK373), ΔrelA ΔspoT rpoD40/pMMKatF2 (KK375), ΔrelA ΔspoT rpoD35 (KK384), and ΔrelA ΔspoT rpoD35/pMMKatF2 (KK385). (ΔΔ) The ΔrelA ΔspoT genotype.
The mutations in the rpoD gene, such as rpoD40 and rpoD35, are not expected to directly affect promoters dependent on alternative σ factors. However, it is possible that these mutations, by reducing the ability of the σ70 to bind core, may thus also reduce their ability to compete for core. If this is the case, the underlying reason that these rpoD mutations restore expression from σS-dependent promoters in a ppGpp0 background may be similar to that of σ70 underproduction or Rsd overproduction, namely, a reduced potential to compete with alternative σ factors. To approach this notion, we set up a mixed in vitro transcription assay to directly analyze σ-factor competition.
ppGpp does not influence Eσ32 transcription from PdnaK in vitro
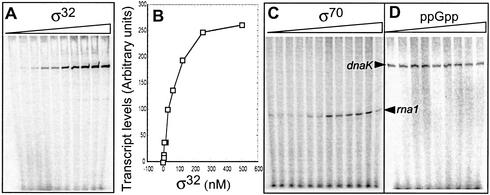
To set up a reliable in vitro transcription-competition (IVT) system, we used a σ32-dependent promoter (PdnaK), rather than a σS-dependent promoter, to monitor effects of σ-factor competition. This choice was based on the fact that a σ70-programmed holoenzyme has been shown to be able to initiate transcription from σS-dependent promoters in vitro (e.g., Kusano et al. 1996), an in vitro phenomenon we have also observed with PuspB and Pfic as templates (data not shown). In addition, by using a σ32-dependent promoter, we avoided possible promoter interference, as Eσ32 and Eσ70 recognizes markedly different promoters, and no cross activity has been observed between these systems in vitro (e.g., Gross et al. 1992).
To test the IVT system, we added a fixed amount of supercoiled DNA template (PdnaK and Prna1; 1.25 nM) and core (10 nM) with increasing amounts of σ32 in multiple round transcription reactions. As seen in Figure 3A, no transcription of the Eσ70-dependent promoter rna1 was observed using Eσ32. A saturation curve for this assay is shown in Figure 3B. To ensure that purified Eσ70 would not initiate transcription from the PdnaK promoter of the template, we repeated the experiment with reconstituted core RNAP and each of the individual σ70 (wild type, RpoD35, and RpoD40). No transcript derived from PdnaK could be detected with wild-type σ70 (Fig. 3C) or the two mutant σ70 (data not shown). In addition, we tested whether increasing amounts of ppGpp changed the transcriptional activity of PdnaK using Eσ32. We detected no direct effects of ppGpp (0 to 644 μM) on the transcriptional activity from the dnaK promoter (Fig. 3D).
Figure 3.
In vitro transcription (IVT) assay of PdnaK and rna P1 by σ32 and σ70 programmed RNAP. (A) Autoradiogram obtained from the transcription assay performed with increasing amounts of σ32 (0–500 nM) as indicated. (B) Quantification of the transcripts plotted as relative units. (C) Autoradiogram obtained from the transcription assay with increasing amounts (0–60 nM) of wild-type RpoD as indicated. (D) Autoradiogram obtained from transcription assay with fixed amounts of σ32 (250 nM) and increasing levels (0–644 μM) of ppGpp as indicated.
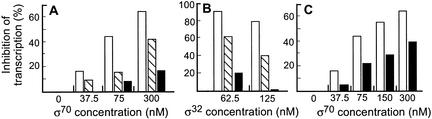
The σ70 mutant proteins compete poorly for RNAP core in vitro
To elucidate the relative competitiveness (inhibition of transcription from PdnaK) of the wild-type and mutant σ70s, we added increasing amounts of the different σ70s to our IVT reaction mix that contained a fixed concentration of σ32 (250 nM) that saturated PdnaK, and measured the relative level of the dnaK transcript. Increasing amounts of wild-type σ70 drastically inhibited transcription from dnaK (Fig. 4A). Inhibition by the RpoD35 protein was significantly lower than by wild-type σ70 and the RpoD40 protein was the least effective in inhibiting transcription from the dnaK promoter (Fig. 4A). In addition, we performed a similar experiment, but added increasing amounts of σ32 to a fixed amount of σ70 (wild-type RpoD at 10 nM, and RpoD35 and RpoD40 at 80 nM). The concentration of the different σ70s used in this experiment were chosen on the basis of the concentration of the σ that saturated the rna1 promoter (data not shown). Again, we found that the wild-type σ70 was much more competitive compared with the mutant σs and that RpoD35 was better than RpoD40 (Fig. 4B). Note also that the rpoD40 allele was more effective than rpoD35 in restoring in vivo σS-dependent expression in ppGpp0 strains (Fig. 2B).
Figure 4.
Inhibition of dnaK transcription by wild-type and mutant RpoD proteins in an IVT competition assay. (A) Transcription of dnaK in multiple round transcription reactions with 1.25 nM PdnaK-containing template (pJET40), 10 nM core RNAP, 250 nM σ32, and increasing amounts of wild-type or mutant σ70 proteins as indicated. The extent of inhibition (%) of dnaK transcription was related to transcription with no competing RpoD in the assay. (Open bars) Competition with wild-type RpoD; (hatched bars) competition with RpoD35; (solid bars) competition with RpoD40. (B) Same as A but increasing amounts of σ32 (as indicated) and fixed amounts of wt RpoD (20 nM) and mutant RpoD (80 nM) proteins. (C) Relative inhibition dnaK transcription by RpoD in the presence and absence of ppGpp (σ32 concentration fixed at 250 nM). (Open bars) indicate Competition in the absence of ppGpp; (solid bars) competition in the presence of 180 μM ppGpp.
Wild-type σ70 competitiveness is reduced in the presence of ppGpp
We next repeated the in vitro competition assay with wild-type σ70 and σ32 competing in the presence and absence of ppGpp. The experiment was performed as described above using a fixed amount of σ32 and increasing amounts of wild-type σ70, with and without ppGpp (180 μM). As seen in Figure 4C, the competitiveness of wild-type σ70 was markedly reduced by the addition of ppGpp. Thus, ppGpp has a positive effect on in vitro dnaK transcription under conditions of competition between σ32 and σ70, but have no observable effect when σ32 operates alone.
Sigma factors, σ32 and σS, compete less effectively in cells lacking ppGpp
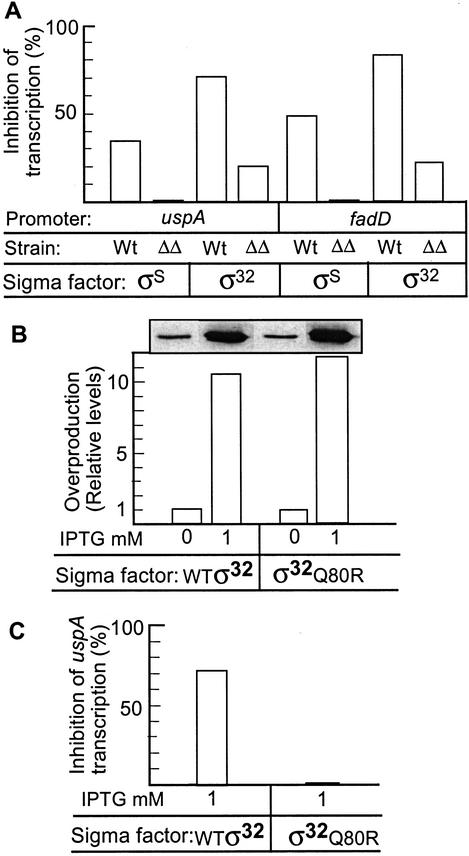
The data presented so far suggest that the poor ability of ppGpp0 mutants to induce regulons requiring alternative σ factors may be explained by a poor ability of these σ factors to compete with σ70 in the absence of ppGpp. Thus, the ability to down-regulate transcription from promoters requiring σ70 should be a measure of an alternative σ factor’s ability to successfully bind and compete for core binding in vivo. Therefore, we used lacZ fusions to elucidate the ability of σS and σ32 to reduce transcription from σ70-dependent genes in wild-type and ppGpp0 strains. First, we noted that overproduction of σS (fourfold overproduction measured by Western blotting; data not shown) caused such down-regulation of two model σ70-dependent promoters (uspA and fadD; Fig. 5A), consistent with previous results (Farewell et al. 1998a). However, this inhibition of transcription only occurred when cells entered stationary phase (high ppGpp levels), with no repression observable during exponential growth (data not shown). Next, we repeated the same experiment in a ppGpp0 mutant and noted that the same overproduction of σS failed to reduce expression from both uspA and fadD in stationary phase cells (Fig. 5A). We obtained the same results with relA1, ΔspoT∷Km mutants (data not shown). Similarly, we found that ectopic overproduction of σ32 was much more effective in inhibiting expression from the uspA and the fadD promoters in the wild-type background than the ppGpp0 mutant strain (Fig. 5A).
Figure 5.
Role of ppGpp on the in vivo competitiveness of σ32 and σS. (A) Relative inhibition (%) of Eσ70-dependent (PuspA–lacZ and PfadD–lacZ) expression by σS or σ32 overproduction (from pMMKatF2 or pKV1278) in a wild-type and a ΔrelA ΔspoT background. Overproduction of σ32 was achieved with 1 mM IPTG using the system described. The strains used for analysis of PuspA–lacZ expression were MJ321, MJ500, MJ325, and MJ440, and for PfadD–lacZ expression TN322, TN323, TN326, and TN327. (B) Extent of overproduction of wild-type σ32 and the mutant σ32Q80R by IPTG addition (1 mM). (C) Effects of overproducing the mutant σ32Q80R on the inhibition of uspA expression (MJ352). The effect of overproducing wild-type σ32 (strain MJ325) is shown for a comparison. (ΔΔ) The ΔrelA∷Km ΔspoT∷Cm genotype.
Overproduction of Q80R mutant σ32 (Fig. 5B), which exhibits a drastically reduced affinity for core (Joo et al. 1997) totally failed to repress the σ70-dependent promoter PuspA (Fig. 5C). This confirms that the observed inhibition of uspA and fadD transcription is an effect of σ32 out-competing σ70 for core binding.
Binding of σS and σ32 to core RNA polymerase is reduced in cells lacking ppGpp
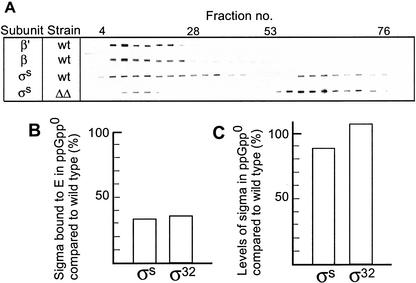
Next, we measured the concentration of σS and σ32 bound to core RNA polymerase in wild-type and ppGpp0 strains during carbon starvation. The cells carried plasmid-borne copies of the rpoS (pMMKatF2) and rpoH (pKV1278) genes to ensure similar expression levels in the wild-type and ppGpp0 strains. After harvesting, whole-cell lysates were fractionated by gel filtration and the concentration of σ factors and core subunits in each fraction was quantified by using monoclonal antibodies against σS, σ32, β, and β‘ subunits. σ factors were found in two separate portions of the fractions collected. In fractions 4–28, σ factors coeluted with the β and β‘ subunits of core RNA polymerase (interpreted as bound σs), whereas fractions 54–76 contained σ factors but no traces of core subunits (Fig. 6A). It became immediately evident that a significantly larger fraction of σS was bound to core in wild-type compared with ppGpp0 cells (Fig. 6A). Quantifications of Western blots showed that the concentration of σS and σ32 recovered in the core-bound fraction was reduced by ∼70% in the ppGpp0 strain compared with the wild type (Fig. 6B). These results cannot be attributed to reduced σS and σ32 content, as the presence of appropriate plasmids in the parental strains ensures that these do not differ significantly in this respect (Fig. 6C).
Figure 6.
Analysis of σ-core RNA polymerase interactions in ΔrelA ΔspoT and wild-type strains. (A) Whole-cell lysates from wild-type and ppGpp0 cells were fractionated by gel filtration and the relative concentration of RNA polymerase subunits was subsequently analyzed in the fractions by using monoclonal antibodies against the σS, σ32, β, and β‘ subunits. As indicated, σS coeluted with β, β‘ in early fractions (bound σ), and was also recovered in later fractions with no traces of core subunits (free σ). (B) Fraction (%) of σS and σ32 that coeluted with core (β and β‘ subunits) in ppGpp0 cells compared with wild-type cells, which was assigned a value of 100. (C) Levels (%) of σS and σ32 in ppGpp0 compared with wild type measured by Western blot analysis. Samples were taken 2 h into stationary phase and equal amounts of crude cell extracts were subjected to SDS-PAGE followed by Western blotting. Strains used were KK153, KK374, KK390, and KK391.
Discussion
We report here on a new function for ppGpp as a master regulator of transcription, that is, the regulation of σ-factor competition. This notion is based on data showing (1) that alternative σ factors compete significantly better against σ70 in the presence of ppGpp both in in vitro and in vivo transcription assays; (2) that the fraction of both σS and σ32 bound to core is reduced in stationary phase cells lacking ppGpp; (3) that the ppGpp requirement of genes transcribed by alternative σ factors is alleviated by specific rpoD alleles encoding σ70 proteins with reduced ability to compete for core; (4) that there is no absolute requirement of ppGpp for σS-dependent promoters, as underproduction of σ70 or overproduction of the anti-σ70 factor Rsd rescues the promoters in the absence of ppGpp accumulation. Thus, we suggest that elevated levels of ppGpp facilitate transcription of the general stress response and the heat-shock regulon by allowing the required σ factors, σS and σ32, respectively, to compete more successfully for available core.
Thus, ppGpp is priming the RNA polymerase in accordance with environmental signals such that the transcriptional apparatus will be primarily occupied with transcription of σ70-dependent housekeeping genes as long as the ppGpp levels are low, which signals that the nutritional status of the environment is favorable for growth. During growth arrest or growth under stress, elevated ppGpp levels allow the alternative σ factors to work in concert with σ70 by shifting the relative competitiveness of the σ factors. Such a role for ppGpp has, in fact, been hypothesized previously. Travers (1985) discussed the possibility that alarmones, such as ppGpp and AppppA, may act by loosening the protein–protein interactions between σ70 and core, thereby facilitating replacement of one σ by another. Alteration in the affinity of core for different σ factors was suggested also by VanBogelen and Neidhardt (1990) as a possible explanation for the sluggish and delayed induction of heat shock genes in ppGpp0 mutants. In addition, Hernandez and Cashel (1995) showed that ppGpp drastically reduces the fraction of σ70 bound to core, and put forward the idea that ppGpp may alter the competition between σ70 and alternative σ factors. Our data strongly supports such an idea.
Whereas the data above supports the notion that ppGpp facilitates the competition by alternative σ factors for available core, it is important to consider that ppGpp does not cause a full σ-factor replacement per se. In parallel to genes requiring alternative σ factors, many σ70-dependent genes are induced during a stringent response (e.g., Xiao et al. 1991; Cashel et al. 1996; Da Costa and Artz 1997; Diez et al. 2000; Kvint et al. 2000b). It has been proposed that the positive regulation of σ70-dependent promoters by ppGpp is linked to ppGpp-dependent effects on RNA polymerase availability. The rate-limiting step of such σ70-dependent promoters has been argued to be RNA polymerase recruitment and these promoters, therefore, would be very sensitive to the concentration of free RNA polymerase (Bartlett et al. 1998; Zhou and Jin 1998). The accumulation of ppGpp is suggested to result in the dissociation of RNA polymerase from stringent promoters (Bartlett et al. 1998; Zhou and Jin 1998) resulting in more RNA polymerase becoming available to initiate transcription at promoters that have a relatively poor ability to recruit RNA polymerase. Thus, despite the fact that ppGpp accumulation decreases the competitiveness of σ70 during stringency, many Eσ70-dependent promoters may well experience an increased Eσ70 availability, as a large fraction of the holoenzyme is no longer sequestered in transcribing stable RNA operons. Thus, the model for the positive control of ppGpp/σ70-dependent promoters (Zhou and Jin 1998) is fully compatible with the σ-factor competition model presented here. It cannot be excluded, however, that some σ70-dependent promoters are directly regulated by ppGpp and examples of such positive effect of ppGpp on gene expression has been achieved using a coupled in vitro transcription translation assay (Riggs et al. 1986; Choy 2000).
Likewise, our data does not exclude the possibility that ppGpp may have additional effects at the level of promoter recognition by EσS and Eσ32 or a subsequent step in transcription initiation. So far, however, we have not detected any direct effects of ppGpp on the transcription of EσS- or Eσ32-dependent promoters in in vitro transcription assays. In addition, we cannot exclude the possibility that the antagonism between σ factors σS and σ70 may also be linked also to promoter interference (Landini and Busby 1999). This concept is based on the argument that, in vivo, Eσ70 may be able to bind a σS-dependent promoter in a stable but nonproductive way and thereby inhibit binding of EσS. Such a mechanism may work in concert with σ factor competition between σ70 and σS but should not apply to the antagonism between σ70 and σ32, as the latter two σ factors recognize vastly different promoter structures. Moreover, the framework of the promoter interference model cannot explain the drastically reduced concentration of σS bound to core in ppGpp0 strains.
Finally, this work clearly supports the previous suggestion that RNA polymerase is limiting for transcription and that activation of the RpoS and RpoH regulons is critically dependent on the concentration of the housekeeping σ factor in vivo (e.g., Osawa and Yura 1981; Malik et al. 1987; Gross et al. 1992; Farewell et al. 1998a; Maeda et al. 2000). Similarly, recent work in the laboratory of V. Shingler has shown that the activity of σ54 is critically dependent on ppGpp and the concentration of σ70 in vivo. Thus, it appears that stringency is, in part, a general mechanism harnessed by the cell to alter the relative efficiency of σ factor binding to the core polymerase such that low ppGpp levels (conditions favorable for rapid growth) favor housekeeping functions, whereas high ppGpp levels allow alternative σ factors to operate in concert with σ70. In this respect, it is interesting that the E. coli anti-σ70 factor Rsd is itself under positive control by ppGpp (Jishage and Ishihama 1999). However, as shown here, the effects of ppGpp deficiency on σS activity (expression of σS-dependent genes) exceeds that of Rsd deficiency. Moreover, ppGpp alone clearly affected competition in vitro with reconstituted holoenzyme and competing σ factors. Future analysis, for example, using a plasmon resonance approach, may clarify the exact effects of ppGpp and Rsd on the kinetics of σ factor binding to core RNA polymerase and whether ppGpp acts by weakening σ70 core interaction and/or strengthening σS/σ32/σ54 core interaction.
Materials and methods
Bacterial strains and growth conditions
E. coli strains used in this work are listed in Table 1. Strain MO1005EL (Ohnuma et al. 2000) containing a transcriptional lacZ fusion to katE, was used to create the strain MJ271. The uspA promoter, containing the promoter and 4 bp upstream of the −35 region (mmuspA4b), fused to lacZ on pMJ261 was recombined onto phage λRS45, and recombinants were used to lysogenize MC4100 as described previously (Simons et al. 1987). For σ70 underproduction, the (CamR) Ptrp–rpoD (Lonetto et al. 1998) construct was introduced into different strains by phage P1-mediated transduction. EC2922 and EC2871 carrying the rpoD35 and rpoD40 mutations, respectively, were originally isolated as suppressors that restored Po-controlled transcription of a tetracycline resistance gene (and thus Tc resistance) in a ΔrelA ΔspoT ppGpp0 strain. The mutant rpoD alleles were subsequently linked to a Tn10 marker by P1-mediated transduction (A. Laurie and V. Shingler, unpubl.). The mutant isolation procedure will be published elsewhere. The strains KK357 and KK358 carrying deletions of the relA and spoT genes without resistance markers were essentially made as described earlier (Datsenko and Wanner 2000). The primer pairs, 3′-GTAGATA CAGTATATATCAATCTACATTGTAGATACGAGCAAATTT CGGCGTGTAGGCTGGAGCTGCTTC-5′, 3′TAGTTGCGATT TGCCGATTTCGGCSGGTCTGGTCCCTAAAGGAGAGGACGC ATATGAATATCCTCCTTAGT-5′ (for deletion of relA) and 3′-CCGTTACCGCTATTGCTGAAGGTCGTCGTTAATCACAAAG CGGGTCGCCCGTGTAGGCTGGAGCTGCTTC-5′, 3′-CTGG CGAGCATTTCGCAGATGCGTGCATAACGTGTTGGGTTCAT AAAACACATATGAATATCCTCCTTAGT-5′ (for deletion of spoT) were chosen so that the coding sequence was deleted for both genes. The elimination is leaving behind the (GTGTAG GCTGGAGCTGCTTCGAAGTTCCTATACTTTCTAGAGAAT AGGAACTTCGGAATAGGAACTAAGGAGGATATTCATATG) sequence from pKD3 in place of the disrupted gene(s) (Datsenko and Wanner 2000). Deletions were confirmed with sequencing. Cells were grown at 37°C aerobically in Lysogeny Broth (LB) or minimal M9 medium supplemented with glucose (0.08%), thiamine (10 mM) and all 20 amino acids in excess. When appropriate, the medium was supplemented with carbenicillin (50 μg/mL), chloramphenicol (30 μg/mL), tetracycline (20 μg/mL), or kanamycin (50 μg/mL).
Plasmids
Plasmids used in this work are listed in Table 1. To create the mutant σ32 (Q80R) expression plasmid pKVQ805, the NcoI–PstI fragment isolated from phis173 (Joo et al. 1997) was cloned into pKV1278 by fragment exchange. Primers uspA-top (5′-AATTC CGGATTGACGGATCATCCGGGTCGCTATAAGGTAAGG ATGGTCC-3′; EcoRI site underlined) and uspA-bottom (5′-TC GAGGACCATC C T TAC C T TATAGCGACCCGGATGATCC GTCAATCCGG-3′; XhoI site underlined) were annealed to generate the fragment containing the uspA promoter region sequence from −38 to +5 (called mmuspA4b) and the fragment was cloned into pTL61T (Linn and St. Pierre 1990) between EcoRI and XhoI to create pMJ261.
General methods
Gel electrophoresis was carried out using 11.5% SDS–polyacrylamide gels. Immunoblotting was performed according to standard procedures using mouse monoclonal antibodies specific for the relevant protein as primary antibody (Neoclone). For detection, we used the ECL-plus blotting kit (Amersham) using alkaline phosphatase-conjugated anti-mouse IgG as secondary antibody (Sigma). Blots were then quantified using the FUJI FILM LAS-1000 device and software.
In vitro synthesis and purification of ppGpp
Preparative-scale synthesis and purification of ppGpp was essentially as described previously (Carmona et al. 2000). In brief, synthesis of ppGpp was performed at 30°C using a His-tagged RelA protein (∼0.2 mg/mL) in a 5-mL reaction containing 2 mM ATP and GDP, and protease inhibitors (complete, Boehringer Mannheim) in buffer RB (50 mM Tris-acetate at pH 8.0, 15 mM magnesium acetate, 60 mM potassium acetate, 30 mM ammonium acetate, 0.2 mM EDTA, 15% methanol). The reaction was terminated after 12 to 16 h by addition of ice-cold formic acid to 1 M, and followed by centrifugation at 8000 rpm at 4°C for 15 min. The supernatant was diluted (1:6) with 50 mM triethylamine acetate (pH 7.7) and applied to a 25-ml DEAE-Bio-Gel column (Bio-Rad) equilibrated with the same buffer. The column was batch eluted with 50 mM (25 mL), 100 mM (25 mL), 150 mM (25 mL), 200 mM (200 mL), and 350 mM (200 mL) triethylamine acetate (pH 7.7) and collected as 12.5-mL fractions. Peak fractions of pure ppGpp were pooled, lyophilized, and stored at −80°C until used. Purity of the preparations was monitored by thin-layer chromatography on polyethyleneimine cellulose plates (Merck), using 1.5 M KH2PO4 (pH 3.4) as chromatographic buffer. Concentrations of ppGpp were determined spectrophotometrically at A260 using the molar extinction coefficient of 13,700.
β-galactosidase activity
Relative β-galactosidase levels were assayed according to the protocol of Miller (1972) with modifications (Albertson and Nyström 1994). The activity is expressed as Miller units; 1000 × A420nm/(A420nm × reaction time × volume). All experiments were repeated several times in to ensure reproducibility and the variation was <10%.
In vitro transcription assays
Multiple round transcriptions were performed at 37°C essentially as described previously (Claverie-Martin and Magasanik 1992). Reactions were performed (20 μL) in a transcriptional buffer containing 50 mM Tris-HCl (pH 7.5), 50 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol (DTT), 0.1 mM EDTA, and 0.275 mg of BSA/mL. The σ70 and the σ32 proteins were overproduced and purified as described by Fujita and Ishihama (1996). Different amounts of wild-type or mutant σ70 and/or σ32 were premixed for 5 min with RNAP core (10 nM; Epicentre Technologies) for holoenzyme formation. When ppGpp was used, RNAP core and ppGpp were premixed for 5 min prior to the addition of σ factor/s. Circular DNA template (pJET40, 0.125 μg) containing PdnaK was added, and the incubation was continued for 20 min to allow open complex formation. Multiple round transcription was started by adding a mixture of ATP, GTP, and CTP (final concentration, 0.4 mM [each]), as well as UTP (final concentration, 0.06 mM), and [α-32P]UTP (5 μCi at >3000 Ci/mmole). After an additional 5 min at 37°C, heparin (0.1 mg/mL) was added, and 5 min later, the reactions were terminated by adding 4 μL of 6X stop buffer (150 mM EDTA, 1.05 M NaCl, 14 M Urea, 3% glycerol, 0.075% xylene cyanol, and 0.075% bromophenol blue). Samples were then analyzed on a 7-M urea-5% acrylamide sequencing gel and quantified using a Bio-Rad PhosphorImager.
Determination of σS and σ32 associated with core RNAP
Strains were grown in minimal M9 medium supplemented with glucose (0.08%), thiamine (10 mM), and all 20 amino acids in excess. Two hours into stationary phase, 50 mL of the cells was spun, down washed, and resuspended in 3-mL reconstitution buffer (10 mM Tris-HCl [pH 7.6 at 4°C], 0.1 mM DTT, 0.1 mM EDTA, 200 mM NaCl, and 5% glycerol [Maeda et al. 2000]). Crude cell extract were obtained using a 20 K French Pressure Cell (Spectronic Instruments). The extracts were subsequently centrifuged for 3 min at 14,000 rpm. A total of 500 μL of the supernatant was subjected to gel filtration through a HiLoad Superdex 200 prep grade column (bed volume 120 mL) with a smart system (Pharmacia biotech). Elution with reconstitution buffer was performed at a flow rate of 1 mL/min at 4°C into fractions of 0.7 mL. Aliquots (50 μL) of elution fractions were dot-blotted onto PVDF membranes and RNAP subunits were detected with specific antibodies as described above.
Acknowledgments
We thank C. Gross, M. Kanamori, L-O. Hedén, A. Ishihama, B. Wanner, and A. Farewell for their contribution of plasmids and strains essential to this work. We thank G. Björk for valuable discussions. This work was supported by grants from the Swedish Natural Science Research Council and the European Commission DG XII Framework IV Programme on Cell factories, Project BIO4-CT98–0167 to T.N. and from Swedish Natural Science Research Council and the SSF to V.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Thomas.Nystrom@gmm.gu.se; FAX 46-31-7732599.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.227902.
References
- Albertson NH, Nyström T. Effects of starvation for exogenous carbon on functional mRNA stability and rate of peptide chain elongation in Escherichia coli. FEMS Microbiol Lett. 1994;117:181–187. doi: 10.1111/j.1574-6968.1994.tb06762.x. [DOI] [PubMed] [Google Scholar]
- Baracchini E, Bremer H. Stringent and growth control of rRNA synthesis in Escherichia coli are both mediated by ppGpp. J Biol Chem. 1988;263:2597–2602. [PubMed] [Google Scholar]
- Barker MM, Gaal T, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J Mol Biol. 2001a;305:689–702. doi: 10.1006/jmbi.2000.4328. [DOI] [PubMed] [Google Scholar]
- Barker MM, Gaal T, Josaitis CA, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J Mol Biol. 2001b;305:673–688. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- Bartlett MS, Gaal T, Ross W, Gourse RL. RNA polymerase mutants that destabilize RNA polymerase-promoter complexes alter NTP-sensing by rrn P1 promoters. J Mol Biol. 1998;279:331–345. doi: 10.1006/jmbi.1998.1779. [DOI] [PubMed] [Google Scholar]
- Carmona M, Rodriguez MJ, Martinez-Costa O, De Lorenzo V. In vivo and in vitro effects of (p)ppGpp on the sigma(54) promoter Pu of the TOL plasmid of Pseudomonas putida. J Bacteriol. 2000;182:4711–4718. doi: 10.1128/jb.182.17.4711-4718.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashel M, Gallant J. Two compounds implicated in the function of the RC gene of Escherichia coli. Nature. 1969;221:838–841. doi: 10.1038/221838a0. [DOI] [PubMed] [Google Scholar]
- Cashel M, Gentry DR, Hernandez VJ, Vinella D. The stringent response. In: Neidhardt FC, editor. Escherichia coli and Salmonella: Cellular and molecular biology. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- Chatterji D, Fujita N, Ishihama A. The mediator for stringent control, ppGpp, binds to the β-subunit of Escherichia coli RNA polymerase. Genes Cells. 1998;3:279–287. doi: 10.1046/j.1365-2443.1998.00190.x. [DOI] [PubMed] [Google Scholar]
- Choy HE. The study of guanosine 5′-diphosphate 3′-diphosphate-mediated transcription regulation in vitro using a coupled transcription-translation system. J Biol Chem. 2000;275:6783–6789. doi: 10.1074/jbc.275.10.6783. [DOI] [PubMed] [Google Scholar]
- Claverie-Martin F, Magasanik B. Positive and negative effects of DNA bending on activation of transcription from a distant site. J Mol Biol. 1992;227:996–1008. doi: 10.1016/0022-2836(92)90516-m. [DOI] [PubMed] [Google Scholar]
- Da Costa XJ, Artz SW. Mutations that render the promoter of the histidine operon of Salmonella typhimurium insensitive to nutrient-rich medium repression and amino acid downshift. J Bacteriol. 1997;179:5211–5217. doi: 10.1128/jb.179.16.5211-5217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez A, Gustavsson N, Nystrom T. The universal stress protein A of Escherichia coli is required for resistance to DNA damaging agents and is regulated by a RecA/FtsK- dependent regulatory pathway. Mol Microbiol. 2000;36:1494–1503. doi: 10.1046/j.1365-2958.2000.01979.x. [DOI] [PubMed] [Google Scholar]
- Dukan S, Nystrom T. Bacterial senescence: Stasis results in increased and differential oxidation of cytoplasmic proteins leading to developmental induction of the heat shock regulon. Genes & Dev. 1998;12:3431–3441. doi: 10.1101/gad.12.21.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Oxidative stress defense and deterioration of growth-arrested Escherichia coli cells. J Biol Chem. 1999;274:26027–26032. doi: 10.1074/jbc.274.37.26027. [DOI] [PubMed] [Google Scholar]
- Farewell A, Diez AA, DiRusso CC, Nyström T. Role of the Escherichia coli FadR regulator in stasis survival and growth phase-dependent expression of the uspA, fad, and fab genes. J Bacteriol. 1996;178:6443–6450. doi: 10.1128/jb.178.22.6443-6450.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farewell A, Kvint K, Nyström T. Negative regulation by RpoS: A case of σ factor competition. Mol Microbiol. 1998a;29:1039–1051. doi: 10.1046/j.1365-2958.1998.00990.x. [DOI] [PubMed] [Google Scholar]
- ————— uspB, a new σS-regulated gene in Escherichia coli which is required for stationary-phase resistance to ethanol. J Bacteriol. 1998b;180:6140–6147. doi: 10.1128/jb.180.23.6140-6147.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Ishihama A. Reconstitution of RNA polymerase. Methods Enzymol. 1996;273:121–130. doi: 10.1016/s0076-6879(96)73011-2. [DOI] [PubMed] [Google Scholar]
- Gentry DR, Hernandez VJ, Nguyen LH, Jensen DB, Cashel M. Synthesis of the stationary-phase σ factor σ s is positively regulated by ppGpp. J Bacteriol. 1993;175:7982–7989. doi: 10.1128/jb.175.24.7982-7989.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse RL, de Boer HA, Nomura M. DNA determinants of rRNA synthesis in E. coli: Growth rate dependent regulation, feedback inhibition, upstream activation, antitermination. Cell. 1986;44:197–205. doi: 10.1016/0092-8674(86)90498-8. [DOI] [PubMed] [Google Scholar]
- Gourse RL, Gaal T, Aiyar SE, Barker MM, Estrem ST, Hirvonen CA, Ross W. Strength and regulation without transcription factors: Lessons from bacterial rRNA promoters. Cold Spring Harb Symp Quant Biol. 1998;63:131–139. doi: 10.1101/sqb.1998.63.131. [DOI] [PubMed] [Google Scholar]
- Gross CA, Lonetto M, Losick R. Bacterial sigma factors. In: Night SL, Yamamoto KR, editors. Transcriptional regulation. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 129–176. [Google Scholar]
- Grossman AD, Erickson JW, Gross CA. The htpR gene product of E. coli is a σ factor for heat-shock promoters. Cell. 1984;38:383–390. doi: 10.1016/0092-8674(84)90493-8. [DOI] [PubMed] [Google Scholar]
- Heinemann M, Wagner R. Guanosine 3′,5′-bis(diphosphate) (ppGpp)-dependent inhibition of transcription from stringently controlled Escherichia coli promoters can be explained by an altered initiation pathway that traps RNA polymerase. Eur J Biochem. 1997;247:990–999. doi: 10.1111/j.1432-1033.1997.00990.x. [DOI] [PubMed] [Google Scholar]
- Hengge-Aronis R. The general stress response in Escherichia coli. In: Storz G, Hengge-Aronis R, editors. Bacterial stress response. Washington, D.C.: ASM Press; 2000. pp. 161–179. [Google Scholar]
- Hernandez VJ, Cashel M. Changes in conserved region 3 of Escherichia coli σ 70 mediate ppGpp-dependent functions in vivo. J Mol Biol. 1995;252:536–549. doi: 10.1006/jmbi.1995.0518. [DOI] [PubMed] [Google Scholar]
- Jishage M, Ishihama A. A stationary phase protein in Escherichia coli with binding activity to the major sigma subunit of RNA polymerase. Proc Natl Acad Sci. 1998;95:4953–4958. doi: 10.1073/pnas.95.9.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Transcriptional organization and in vivo role of the Escherichia coli rsd gene, encoding the regulator of RNA polymerase sigma D. J Bacteriol. 1999;181:3768–3776. doi: 10.1128/jb.181.12.3768-3776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PG, Cashel M, Glaser G, Neidhardt FC. Function of a relaxed-like state following temperature downshifts in Escherichia coli. J Bacteriol. 1992;174:3903–3914. doi: 10.1128/jb.174.12.3903-3914.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo DM, Ng N, Calendar R. A σ32 mutant with a single amino acid change in the highly conserved region 2.2 exhibits reduced core RNA polymerase affinity. Proc Natl Acad Sci. 1997;94:4907–4912. doi: 10.1073/pnas.94.10.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano S, Ding Q, Fujita N, Ishihama A. Promoter selectivity of Escherichia coli RNA polymerase E σ 70 and E σ 38 holoenzymes. Effect of DNA supercoiling. J Biol Chem. 1996;271:1998–2004. doi: 10.1074/jbc.271.4.1998. [DOI] [PubMed] [Google Scholar]
- Kvint K, Farewell A, Nyström T. RpoS-dependent promoters require guanosine tetraphosphate for induction even in the presence of high levels of σS. J Biol Chem. 2000a;275:14795–14798. doi: 10.1074/jbc.C000128200. [DOI] [PubMed] [Google Scholar]
- Kvint K, Hosbond C, Farewell A, Nybroe O, Nyström T. Emergency derepression: stringency allows RNA polymerase to override negative control by an active repressor. Mol Microbiol. 2000b;35:435–443. doi: 10.1046/j.1365-2958.2000.01714.x. [DOI] [PubMed] [Google Scholar]
- Landini P, Busby SJ. Expression of the Escherichia coli ada regulon in stationary phase: Evidence for rpoS-dependent negative regulation of alkA transcription. J Bacteriol. 1999;181:6836–6839. doi: 10.1128/jb.181.21.6836-6839.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange R, Hengge-Aronis R. Growth phase-regulated expression of bolA and morphology of stationary- phase Escherichia coli cells are controlled by the novel σ factor σ S. J Bacteriol. 1991;173:4474–4481. doi: 10.1128/jb.173.14.4474-4481.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange R, Fischer D, Hengge-Aronis R. Identification of transcriptional start sites and the role of ppGpp in the expression of rpoS, the structural gene for the σ S subunit of RNA polymerase in Escherichia coli. JBacteriol. 1995;177:4676–4680. doi: 10.1128/jb.177.16.4676-4680.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzarini RA, Cashel M, Gallant J. On the regulation of guanosine tetraphosphate levels in stringent and relaxed strains of Escherichia coli. J Biol Chem. 1971;246:4381–4385. [PubMed] [Google Scholar]
- Linn T, St. Pierre R. Improved vector system for constructing transcriptional fusions that ensures independent translation of lacZ. J Bacteriol. 1990;172:1077–1084. doi: 10.1128/jb.172.2.1077-1084.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonetto MA, Rhodius V, Lamberg K, Kiley P, Busby S, Gross C. Identification of a contact site for different transcription activators in region 4 of the Escherichia coli RNA polymerase σ70 subunit. J Mol Biol. 1998;284:1353–1365. doi: 10.1006/jmbi.1998.2268. [DOI] [PubMed] [Google Scholar]
- Maeda H, Fujita N, Ishihama A. Competition among seven Escherichia coli σ subunits: Relative binding affinities to the core RNA polymerase. Nucleic Acids Res. 2000;28:3497–3503. doi: 10.1093/nar/28.18.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Zalenskaya K, Goldfarb A. Competition between σ factors for core RNA polymerase. Nucleic Acids Res. 1987;15:8521–8530. doi: 10.1093/nar/15.20.8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Mulvey MR, Sorby PA, Triggs-Raine BL, Loewen PC. Cloning and physical characterization of katE and katF required for catalase HPII expression in Escherichia coli. Gene. 1988;73:337–345. doi: 10.1016/0378-1119(88)90498-2. [DOI] [PubMed] [Google Scholar]
- Nyström T. Role of guanosine tetraphosphate in gene expression and the survival of glucose or seryl-tRNA starved cells of Escherichia coli K12. Mol Gen Genet. 1994;245:355–362. doi: 10.1007/BF00290116. [DOI] [PubMed] [Google Scholar]
- Ohlsen KL, Gralla JD. Interrelated effects of DNA supercoiling, ppGpp, and low salt on melting within the Escherichia coli ribosomal RNA rrnB P1 promoter. Mol Microbiol. 1992;6:2243–2251. doi: 10.1111/j.1365-2958.1992.tb01400.x. [DOI] [PubMed] [Google Scholar]
- Ohnuma M, Fujita N, Ishihama A, Tanaka K, Takahashi H. A carboxy-terminal 16-amino-acid region of σ(38) of Escherichia coli is important for transcription under high-salt conditions and σ activities in vivo. J Bacteriol. 2000;182:4628–4631. doi: 10.1128/jb.182.16.4628-4631.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa T, Yura T. Effects of reduced amount of RNA polymerase σ factor on gene expression and growth of Escherichia coli: studies of the rpoD450 (amber) mutation. Mol Gen Genet. 1981;184:166–173. doi: 10.1007/BF00272900. [DOI] [PubMed] [Google Scholar]
- Riggs DL, Mueller RD, Kwan HS, Artz SW. Promoter domain mediates guanosine tetraphosphate activation of the histidine operon. Proc Natl Acad Sci. 1986;83:9333–9337. doi: 10.1073/pnas.83.24.9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals J, Little R, Bremer H. Control of RNA synthesis in Escherichia coli after a shift to higher temperature. J Bacteriol. 1982a;151:1425–1432. doi: 10.1128/jb.151.3.1425-1432.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Control of rRNA and tRNA syntheses in Escherichia coli by guanosine tetraphosphate. J Bacteriol. 1982b;151:1261–1268. doi: 10.1128/jb.151.3.1261-1268.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Temperature dependence of RNA synthesis parameters in Escherichia coli. J Bacteriol. 1982c;151:879–887. doi: 10.1128/jb.151.2.879-887.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sands MK, Roberts RB. The effects of a tryptophan-histidine deficiency in a mutant of Echerichia coli. J Bacteriol. 1952;63:505–511. doi: 10.1128/jb.63.4.505-511.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons RW, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Stent GS, Brenner S. A genetic locus for the regulation of ribonucleic acid synthesis. Proc Natl Acad Sci. 1961;47:2005–2014. doi: 10.1073/pnas.47.12.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze CC, Shingler V. The alarmone (p)ppGpp mediates physiological-responsive control at the σ 54-dependent Po promoter. Mol Microbiol. 1999;31:1217–1228. doi: 10.1046/j.1365-2958.1999.01264.x. [DOI] [PubMed] [Google Scholar]
- Sze CC, Bernardo LM, Shingler V. Integration of global regulation of two aromatic-responsive σ(54)-dependent systems: A common phenotype by different mechanisms. J Bacteriol. 2002;184:760–770. doi: 10.1128/JB.184.3.760-770.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulokhonov II, Shulgina I, Hernandez VJ. Binding of the transcription effector ppGpp to Escherichia coli RNA polymerase is allosteric, modular, and occurs near the N terminus of the β‘-subunit. J Biol Chem. 2001;276:1220–1225. doi: 10.1074/jbc.M007184200. [DOI] [PubMed] [Google Scholar]
- Travers A. Modulation of RNA polymerase specificity by ppGpp. Mol Gen Genet. 1976;147:225–232. doi: 10.1007/BF00267575. [DOI] [PubMed] [Google Scholar]
- ————— σ Factors in multitude. Nature. 1985;313:15–16. doi: 10.1038/313015b0. [DOI] [PubMed] [Google Scholar]
- VanBogelen RA, Neidhardt FC. Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc Natl Acad Sci. 1990;87:5589–5593. doi: 10.1073/pnas.87.15.5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]
- Zgurskaya HI, Keyhan M, Matin A. The σ S level in starving Escherichia coli cells increases solely as a result of its increased stability, despite decreased synthesis. Mol Microbiol. 1997;24:643–651. doi: 10.1046/j.1365-2958.1997.3961742.x. [DOI] [PubMed] [Google Scholar]
- Zhou YN, Jin DJ. The rpoB mutants destabilizing initiation complexes at stringently controlled promoters behave like “stringent” RNA polymerases in Escherichia coli. Proc Natl Acad Sci. 1998;95:2908–2913. doi: 10.1073/pnas.95.6.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]